Abstract
Reproductive fitness in many animals relies upon a tight balance between the number of cells that proliferate in the germ line and the number of cells that enter meiosis and differentiate as gametes. In the Caenorhabditis elegans germ line, the GLP-1/Notch signaling pathway controls this balance between proliferation and meiotic entry. Here we describe the identification of the proteasome as an additional regulator of this balance. We show that a decrease in proteasome activity, through either genetic mutation or RNAi to core components of the proteasome, shifts this balance toward excess germ-line proliferation. We further demonstrate that there are likely two or more proteasome targets that contribute to excess germ-line proliferation when proteasome activity is reduced. One of these targets is likely a component or regulator of the Notch-signaling pathway, while the other functions on one of the two major redundant genetic pathways downstream of GLP-1/Notch signaling. We propose a model in which the proteasome degrades proteins that are necessary for proliferation as cells switch from proliferation to meiotic entry.
THE production of gametes is an essential process in any sexually reproducing species. In most of these species, proliferation of germ-line stem cells is needed first to generate the germ-line tissue and then to maintain a supply from which future gametes can be formed. If germ-line stem cell proliferation does not keep pace with gamete production, the population of stem cells will be depleted, resulting in sterility. Conversely, an increase in self-renewing proliferation of the germ-line stem cells at the expense of gamete production can also result in sterility, as well as tumor formation. With the large numbers of gametes that are formed over the life of some animals, it is imperative that the balance between proliferation and differentiation be fine-tuned; a slight shift in this balance will be amplified over time and greatly reduce the reproductive fitness of the animal. Much of our understanding of how the balance between germ-line proliferation and differentiation is controlled comes from study of classic genetic model organisms such as Drosophila and Caenorhabditis elegans. In both Drosophila males and females, the balance between proliferation and differentiation is a result of asymmetric division of the germ-line stem cells (Xie and Spradling 2000; Tulina and Matunis 2001). One daughter cell maintains contact with signaling somatic cells, resulting in retention of the stem cell property of the parent cell, while the other daughter cell is not in contact with the signaling somatic cells and begins the path to meiotic entry and differentiation (Xie and Spradling 2000; Tulina and Matunis 2001). In both male and hermaphrodite C. elegans, self-renewing proliferation of the germ-line stem cells is promoted by their proximity to a somatic signaling cell, the distal tip cell (DTC) (Kimble and White 1981). The major molecular signal promoting proliferation is transmitted through the conserved Notch-signaling pathway (Hansen and Schedl 2006; Kimble and Crittenden 2007). As part of this pathway, the membrane-bound LAG-2 ligand, which is expressed in the DTC, comes in contact with nearby proliferative cells that express the GLP-1/Notch receptor (Austin and Kimble 1987, 1989; Yochem and Greenwald 1989; Henderson et al. 1994; Tax et al. 1994). The interaction between ligand and receptor activates GLP-1/Notch signaling, which presumably culminates in the expression of genes necessary for proliferation and the inhibition of genes involved in meiotic entry. As germ cells move proximally, away from the influence of the DTC, the level of GLP-1/Notch signaling is thought to diminish, presumably allowing for the gene activity profile to switch, resulting in cells entering into meiotic prophase and eventually forming fully differentiated gametes. Numerous genetic screens for mutants that disrupt the balance between proliferation and differentiation in the C. elegans germ line, as well as phenotypic analysis of preexisting mutants, have identified many genes that control the balance between proliferation and differentiation (Hansen and Schedl 2006; Kimble and Crittenden 2007); most of these genes either modulate GLP-1/Notch signaling or are regulated by GLP-1/Notch signaling. Genetic epistasis analysis of these mutants has resulted in a genetic model (Figure 1A), which has been supported by substantial biochemical evidence. In this model, GLP-1/Notch signaling promotes the activities of two nearly identical proteins, FBF-1 and FBF-2, which are homologous to Drosophila Pumilio (Crittenden et al. 2002; Lamont et al. 2004). FBF-1 and FBF-2 then inhibit the activities of two major genetic pathways that function redundantly to inhibit proliferation and/or promote meiotic entry (Kadyk and Kimble 1998; Crittenden et al. 2002; Eckmann et al. 2004); we will refer to these two pathways as the gld-1 and gld-2 pathways on the basis of the founding genes of these two pathways (Francis et al. 1995b; Kadyk and Kimble 1998). When germ cells are close to the DTC, GLP-1/Notch signaling is active and the activities of the gld-1 and gld-2 pathways are inhibited. As germ cells move away from the DTC and GLP-1/Notch signaling is reduced, the activities of the genes in the gld-1 and gld-2 pathways increase, resulting in cells entering into meiotic prophase. The gld-1 and gld-2 pathways function redundantly: if a gene in only one of the pathways is mutated, the balance between proliferation and differentiation is very similar to wild type (Francis et al. 1995b; Kadyk and Kimble 1998). However, if the activities of both pathways are eliminated by mutating a gene in each of the two pathways, the balance is shifted toward proliferation and a germ-line tumor results (Kadyk and Kimble 1998; Eckmann et al. 2004; Hansen et al. 2004b). On the basis of the molecular identities of the genes in the gld-1 and gld-2 pathways and biochemical analysis, all known components of these pathways likely regulate mRNA metabolism. GLD-1 is a KH-domain-containing protein that binds to target mRNAs and inhibits translation (Jones and Schedl 1995; Jan et al. 1999; Kraemer et al. 1999; Lee and Schedl 2001; Xu et al. 2001; Marin and Evans 2003; Lee and Schedl 2004). The other gld-1 pathway gene, nos-3, encodes a protein with sequence similarity to Drosophila Nanos, which is a translational inhibitor (Tautz 1988; Wharton and Struhl 1991; Kraemer et al. 1999; Hansen et al. 2004b). In the gld-2 pathway, GLD-2 and GLD-3 bind to one another and function as a cytoplasmic polyadenylation complex, presumably to either stabilize or increase the translatability of target mRNAs (Wang et al. 2002; Eckmann et al. 2004). Although many mRNA targets of GLD-1 have been identified, these targets appear to relate to functions of GLD-1 other than the proliferation vs. meiotic entry decision (Jan et al. 1999; Lee and Schedl 2001, 2004; Xu et al. 2001; Marin and Evans 2003; Lakiza et al. 2005); no GLD-1 targets known to regulate the proliferation vs. meiotic entry decision have yet been identified, other than cross talk between the gld-1 and gld-2 pathways and negative feedback of glp-1 (Marin and Evans 2003; Hansen et al. 2004b; Suh et al. 2006).
Figure 1.—
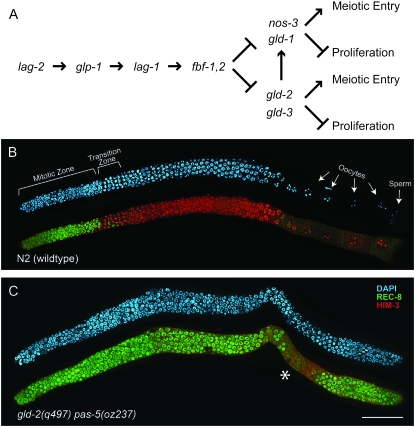
pas-5 is synthetic tumorous with gld-2. (A) The core genetic pathway regulating the proliferation vs. meiotic entry decision (Hansen and Schedl 2006). Cells in the distal end of the gonad, close to the DTC, have active GLP-1/Notch signaling. This is accomplished through interaction of the LAG-2 ligand with the GLP-1/Notch receptor, resulting in the activation of the LAG-1 transcription factor. Active GLP-1/Notch signaling promotes the activities of FBF-1 and FBF-2, which in turn inhibit the activities of the gld-1 and gld-2 redundant genetic pathways. Each of these pathways promote meiotic entry and/or inhibit proliferation. The gld-2 pathway also promotes the activity of the gld-1 pathway. A more complete description of the genetic pathway regulating the proliferation vs. meiotic entry decision, including a description of additional factors that interact with this core pathway, can be found elsewhere (Hansen and Schedl 2006; Kimble and Crittenden 2007). (B) Wild-type hermaphrodite gonad arm dissected 1 day after L4 and stained with DAPI (blue), anti-REC-8 antibodies that mark proliferative cells (green), and anti-HIM-3 antibodies that mark meiotic cells (red) (Zetka et al. 1999; Pasierbek et al. 2001; Hansen et al. 2004a). Cells are proliferative in the mitotic zone and enter meiotic prophase as they move into the transition zone. Cells continue to progress through meiotic prophase as they move more proximally and eventually differentiate as sperm or oocytes. (C) gld-2(q497) pas-5(oz237) hermaphrodite gonad arm dissected 1 day after L4 and stained as in B. Distal is to the left and proximal is to the right. Most cells in the gonad arm stain for anti-REC-8 antibodies and not for anti-HIM-3 antibodies, suggesting that they are mitotic. However, in some gonad arms some cells are negative for anti-REC-8 antibodies and positive for anti-HIM-3 antibodies (asterisk), suggesting that some cells do enter meiotic prophase. Photos in B and C were taken with the Apotome module to reduce background fluorescence. Bar, 50 μm.
The core genetic pathway described above is the basis of the regulation of the proliferation vs. meiotic entry decision in the C. elegans germ line. Other factors interact with this basic pathway to help regulate activity and contribute to the precise balance that must be maintained between proliferation and meiotic entry for reproductive fitness to be maximized (Hansen and Schedl 2006; Kimble and Crittenden 2007). Here we demonstrate that proteasome activity is involved in the regulation of the proliferation vs. meiotic entry decision in the C. elegans germ line. A decrease in proteasome activity enhances overproliferation in the germ line. Therefore, either one or more proteins are normally degraded by the proteasome as part of the switch from proliferation to meiotic entry. A mutation that reduces proteasome activity would allow these proteins to persist and promote ectopic proliferation. We demonstrate that the proteasome functions in two parts of the proliferation vs. meiotic entry genetic pathway, likely degrading different protein targets. One of these proteins is either a component or a regulator of GLP-1/Notch signaling, while the other functions in one of the two redundant pathways genetically downstream of GLP-1/Notch signaling.
MATERIALS AND METHODS
Nematode strains and culture:
Standard genetic manipulation and culture procedures of the C. elegans strains were followed with growth at 20° unless otherwise stated (Sulston and Hodgkin 1988). A list of all strains used in this study is provided in the supplemental material.
The following mutations were used: linkage group I (LGI)—smg-2(e2008), gld-2(q497null) (Kadyk and Kimble 1998), gld-1(q485null) (Francis et al. 1995a), unc-15(e73), unc-29(e193), pas-5(oz237) (this work), and unc-75(e950); LGII—gld-3(q730) (Eckmann et al. 2002) and nos-3(oz231null) (Hansen et al. 2004b); LGIII—unc-32(e189), lin-12(n379gf) (Greenwald et al. 1983), lin-12(n676gf) (Greenwald et al. 1983), glp-1(q35gf) (Mango et al. 1991), glp-1(q175null) (Austin and Kimble 1987), and glp-1(ar202gf) (Pepper et al. 2003); and LGV—sel-10(bc243null) (Jager et al. 2004).
Single nucleotide polymorphism mapping:
To narrow the physical region containing the oz237 mutation, single nucleotide polymorphism (SNP) mapping was used (Jakubowski and Kornfeld 1999; Wicks et al. 2001). Detailed information regarding individual SNPs can be found at http://www.wormbase.org and in the supplemental material. unc-29(e193) pas-5(oz237)/hT2[qIs48(myo-2∷gfp; pes-10∷gfp; ges-1∷gfp)] and pas-5(oz237) unc-75(e950)/hT2[qIs48(myo-2∷gfp; pes-10∷gfp; ges-1∷gfp)] hermaphrodites were mated to males from the CB4856 (Hawaiian, HA-8) strain (Wicks et al. 2001). Non-GFP L4 hermaphrodites were picked to individual plates and allowed to self-fertilize. The F2 generation was screened for unc-29(e193) or unc-75(e950) nonsterile/nonabnormally everted vulva recombinants. Homozygous recombinant progeny were tested for N2 or HA-8 DNA at SNPs in the region. In total, 125 unc-29 recombinants and 90 unc-75 recombinants were analyzed. From this analysis, the leftmost boundary of the critical region containing pas-5(oz237) was determined to be SNP F25H2[1] (10,539,544 bp) and the rightmost boundary was a SNP that we identified by sequencing HA-8 and N2 DNA at location 10,585,626 bp. This region on chromosome I contains 14 genes, including pas-5. All stated nucleotide positions correspond to WormBase freeze WS180.
To determine which of the 14 genes corresponds to the oz237 allele, we performed feeding RNA interference (RNAi) (Timmons and Fire 1998) for 13 of the 14 genes in both a glp-1(ar202gf) background and a gld-2(q497)/ccIs4251[myo-3∷gfp] unc-15(e73) background. We placed L4 animals on the feeding RNAi plate and scored the progeny [glp-1(ar202gf) and gld-2(q497) homozygotes] for germ-line overproliferation. None of the 13 genes showed overproliferation above background. Therefore, we sequenced the coding regions of seven genes (F25H2.4, F25H2.5, F25H2.7, F25H2.8, F25H2.9, F25H2.10, and F25H2.11) that showed germ-line phenotypes that may have masked an overproliferation phenotype (e.g., vacuoles in the germ line). We also sequenced the coding region of F29C6.1, which was the one gene not tested by RNAi. Only F25H2.9, pas-5, showed a difference in coding sequence from wild type.
Low-dose RNAi:
Template plasmids for synthesizing dsRNA contain the L4440 backbone, which has T7 promoters flanking the gene of interest (Timmons and Fire 1998; Fraser et al. 2000). The pas-5 and pbs-4 plasmids are commercially available (Geneservice; I-5G02 and I-1N17 for pas-5 and pbs-4, respectively). gfp dsRNA was generated from plasmid pPD128.110 (L4417) (Timmons and Fire 1998). dsRNA was injected into young adult hermaphrodites at 1 ng/μl concentration. Injected worms were allowed to recover for ∼4 hr and then individually plated. glp-1(ar202) animals injected with dsRNA were grown at 15° for 7–8 days and progeny were scored using DIC optics. gld-2(q497)/ccIs4251[myo-3∷gfp] unc-15(e73) animals injected with dsRNA were grown at 20° for 4–5 days. Homozygous gld-2(q497) (nongreen) progeny were scored for their germ-line proliferation phenotype.
Antibody staining:
Gonad dissection and antibody staining was performed as previously described (Jones et al. 1996). Dissected gonads were fixed in 3% formaldehyde, 0.1 m KPO4, pH 7.2, for 10–60 min followed by a postfix in 100% methanol at −20° for >10 min. Fluorescent images were captured on a Zeiss Imager Z.1 with an AxioCam MRm camera. Background fluorescence was minimized during image acquisition using the Apotome module (Zeiss). All 2° antibodies were preabsorbed with acetone worm powder in 30% goat serum in 1× PBS overnight at 4°. Anti-HIM-3 antibodies (Zetka et al. 1999) were used at a 1:500 dilution while anti-REC-8 antibodies (Pasierbek et al. 2001) were used at a dilution of 1:100.
Rescuing array:
The extrachromosomal arrays were generated as previously described (Mello et al. 1991). For the pas-5 rescuing arrays ugEx1 and ugEx2, four plasmids were co-injected: pRF4 (rol-6) (50 μg/ml) (Kramer et al. 1990; Mello et al. 1991), pJM67 (elt-2∷gfp) (20 μg/ml), pDH43 (pas-5) (20 μg/ml), and pBluescript KS+ (10 μg/ml). pDH43 was created by PCR amplifying wild-type pas-5 DNA. The primers used were F25H2.9BSNOTI (AAAGCGGCCGCGGTCTTCCTCACGATGTATATTTCC) and F25H2.9BSBAMHI (AAAGGATCCGAATACTGTAACTCCTGGTTGGG). The amplified product was cloned into the TOPO TA pCR-II vector and sequenced for mutations. The ugEx4 control array does not contain pDH43 [pas-5(+)]; rather, pBluescript KS+ was increased in concentration to keep the relative concentrations and ratios with the other plasmids similar to those in ugEx1 and ugEx2. All arrays were verified by PCR for presence (ugEx1 and ugEx2) or absence (ugEx4) of pas-5 pDH43 plasmid DNA. Transgenic animals were scored 1–3 days after L4 using DIC optics for their germ-line and vulval phenotypes. The phenotypes were scored blind in that the germ-line phenotype and the vulval phenotype were scored prior to determining if the animal carried the array by scoring for GFP in gut nuclei (ELT-2∷GFP).
Synchronization of pas-5; gld-3 tumorous animals:
pas-5(oz237); gld-3(q730); unc-32(e189) and pas-5(oz237); gld-3(q730); unc-32(e189) glp-1(q175) hermaphrodites were synchronized as previously described (Hansen et al. 2004a). In brief, pas-5(oz237)/hT2; gld-3(q730)/mIn1[mIs14 [myo-2∷gfp; pes-10∷gfp] dpy-10(128)]; unc-32(e189)/hT2 and pas-5(oz237)/hT2; gld-3(q730)/mIn1[mIs14 [myo-2∷gfp; pes-10∷gfp] dpy-10(128)]; unc-32(e189) glp-1(q175)/hT2 gravid hermaphrodites were bleached to isolate eggs, which were then resuspended in M9 buffer and shaken at 20° for ∼30 hr to arrest animals in the L1 stage. Animals were washed with M9 and placed on NGM plates (Sulston and Hodgkin 1988) and grown for specific lengths of time. Unc non-GFP animals were scored for the number of germ cells by DIC optics or dissected and stained as described above.
RESULTS
pas-5(oz237) identified in a tumorous screen with gld-2(q497):
A genetic screen was previously performed to identify factors that are involved in regulating the proliferation vs. meiotic entry decision in the C. elegans germ line (Hansen et al. 2004a). In this screen, mutations that cause overproliferation of the germ-line mitotic cells when gld-2 activity is also absent were isolated. gld-2 is a member of one of the two main redundant genetic pathways functioning downstream of GLP-1/Notch signaling (Kadyk and Kimble 1998) (Figure 1A). The balance between proliferation and differentiation in a gld-2 single mutant is similar to wild type. However, gld-2(0) homozygous animals do have germ-line overproliferation, resulting in a tumor, when they also lack the activity of a gld-1 pathway gene or have a mutation that increases GLP-1/Notch signaling (Kadyk and Kimble 1998; Hansen et al. 2004b). Therefore, mutations identified in the mutant screen as forming a synthetic tumor with gld-2(q497) are likely in genes that either function in the gld-1 pathway or negatively regulate GLP-1/Notch signaling. In this screen, we identified a single allele of a gene that we originally referred to as syt-2 (synthetic tumorous). We have since found this allele, oz237, to be an allele of pas-5 (proteasome alpha subunit; see below), which encodes a conserved component of the 20S proteasome (Groll et al. 2005; Kahn et al. 2008).
pas-5(oz237) disrupts the balance between proliferation and meiotic entry:
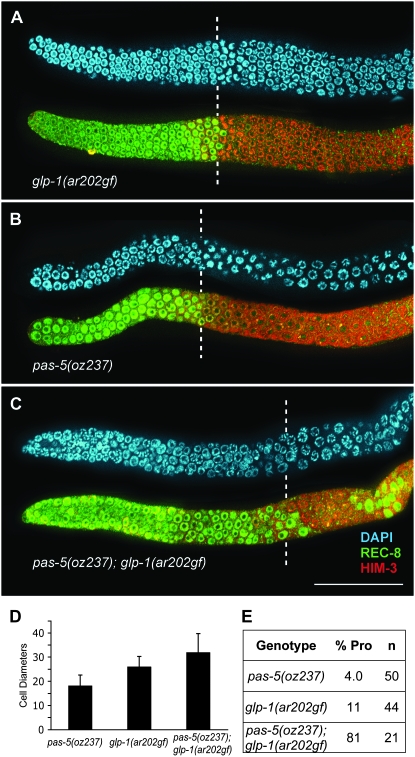
gld-2(q497) pas-5(oz237) animals have excess germ-line proliferation, forming a tumor (Figure 1, B and C). Proliferative cells are found throughout the gonad arm. Many of the gonad arms of gld-2(q497) pas-5(oz237) animals are completely tumorous (16/27), showing no evidence of meiotic entry as determined by anti-REC-8 and anti-HIM-3 antibody staining, which mark mitotic and meiotic cells, respectively (Zetka et al. 1999; Pasierbek et al. 2001; Hansen et al. 2004a). The remaining gonad arms (11/27) have proliferative cells throughout, but do have some scattered cells that stain positively for anti-HIM-3 antibodies, suggesting that they have entered meiotic prophase (Figure 1C, asterisk). While in wild-type animals the first cells to show evidence of meiotic entry are ∼20 cell diameters from the distal end (Crittenden et al. 1994; Hansen et al. 2004a), for those gld-2(q497) pas-5(oz237) gonads that show some anti-HIM-3-positive cells, on average the first anti-HIM-3-positive cell is 54 cell diameters from the distal end of the gonad (54 ± 18; n = 11). Therefore, the proliferative zone is much larger in these animals and very little meiotic entry is observed (Figure 1, B and C). Some germ-line tumors in C. elegans are not due to a disruption in the balance between proliferation and meiotic entry. For example, female germ cells in gld-1 single-mutant hermaphrodites enter meiotic prophase similar to wild type; however, the cells are unable to progress through meiotic prophase (Francis et al. 1995a). These cells then reenter the mitotic cell cycle and form a germ-line tumor. To determine whether pas-5(oz237) affects the balance between proliferation and meiotic entry, we determined its ability to enhance germ cell overproliferation caused by an increase in GLP-1/Notch signaling. We have previously demonstrated that null mutations in all genes regulating the proliferation vs. meiotic entry decision in the gld-1 and gld-2 pathways enhance overproliferation associated with the increase in Notch signaling caused by the glp-1(ar202gf) allele (Pepper et al. 2003; Hansen et al. 2004a). We found that pas-5(oz237) does enhance the overproliferation phenotype of glp-1(ar202gf) (Figure 2). pas-5(oz237); glp-1(ar202gf) animals have a larger mitotic zone than either single mutant (late-onset tumor), as well as an increased penetrance of proliferation in the proximal end of the gonad (Pro tumor) (Pepper et al. 2003) (Figure 2). Therefore, pas-5(oz237) does disrupt the balance between proliferation and meiotic entry.
Figure 2.—
pas-5(oz237) enhances the overproliferation phenotype of glp-1(ar202gf). (A–C) Gonad arms were dissected from hermaphrodites 1 day after the L4 stage and grown at 20°. The fixed gonad arms were stained with DAPI that shows nuclear morphology (blue), anti-REC-8 antibodies that mark mitotic cells (green), and anti-HIM-3 antibodies that mark meiotic cells (red). Only the distal portion of each gonad arm is shown with the distal-most end to the left. Dashed vertical lines show the position of the most distal meiotic (HIM-3 positive) cell. Photos were taken using the Apotome module (Zeiss) to reduce background fluorescence. Bar, 50 μm. (D) Summary chart of the size of the mitotic zone. The y-axis shows the size of the mitotic zone as measured in cell diameters from the distal end for each genotype of the animals shown on the x-axis. Error bars, 1 SD. The size of the mitotic zone in the pas-5(oz237); glp-1(ar202) double mutant is statistically larger than the sizes of the mitotic zones in pas-5(oz237) animals (P < 4.8 × 10−8, Student's two-tailed t-test, two-sample unequal variance) and glp-1(ar202gf) animals (P < 0.004). For each genotype, at least 20 gonad arms were counted. The size of the mitotic zone in wild-type animals is ∼20 cell diameters (Crittenden et al. 1994; Hansen et al. 2004a); therefore, the mitotic zone in pas-5(oz237) animals is slightly smaller than that of wild type. (E) Summary table showing the percentage of gonad arms that display ectopic proliferation in the proximal end of the gonad (Pro phenotype) (Pepper et al. 2003).
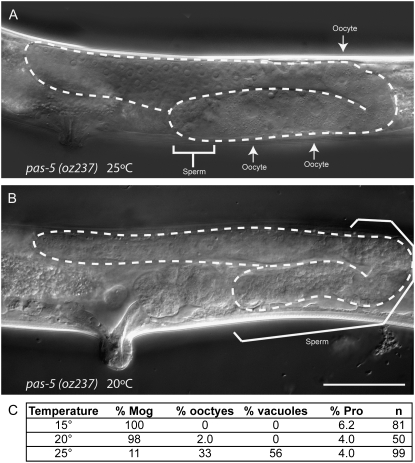
Very few pas-5(oz237) single-mutant animals display germ cell overproliferation (Figure 2). Therefore, pas-5 is not absolutely necessary for germ cells to switch from proliferation to meiotic entry and/or pas-5(oz237) does not fully reduce pas-5 function (see below). However, pas-5(oz237) single-mutant animals do display some other germ-line and somatic phenotypes. When grown at lower temperatures (15° or 20°), pas-5(oz237) hermaphrodites have a masculinized germ line (Mog) in which only sperm, not oocytes, are produced (average 432 ± 87.2 sperm/gonad arm in hermaphrodites 2–3 days after L4; n = 6) (Figure 3). Animals grown at higher temperatures (25°) do make the switch from spermatogenesis to oogenesis; however, the oocytes are often misshapen and viable embryos are not produced (Figure 3). pas-5(oz237) animals also have an abnormally everted vulva (Evl) and a low percentage of animals are multivulval (Muv) (see below). Therefore, pas-5 appears to have multiple germ-line and somatic functions.
Figure 3.—
pas-5(oz237) animals display other germ-line and somatic phenotypes. DIC images of pas-5(oz237) young adult hermaphrodites grown at 25° (A) or 20° (B). Animals grown at 25° make both sperm and oocytes (A and C), although the oocytes are often misshapen and no viable embryos are formed. Many animals also accumulate vacuoles in the germ line. Animals grown at 15° or 20° do not form oocytes, but rather make only sperm (B and C). pas-5(oz237) animals grown at all temperatures have an abnormally everted vulva. Independent of the other germ-line phenotypes displayed, a low percentage of animals also have ectopic proliferation in the proximal end of the gonad (Pro) (C). Bar, 50μm.
oz237 is an allele of pas-5:
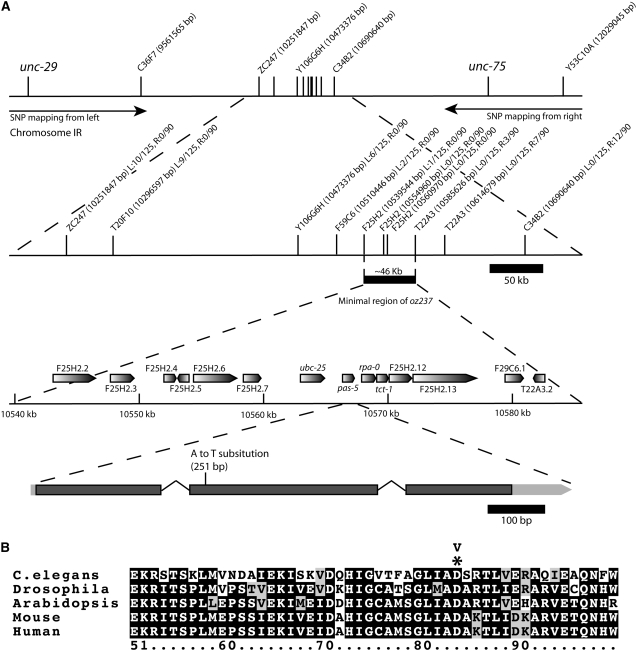
To identify the gene associated with the oz237 allele, we mapped the mutation using the abnormally everted vulva and sterile phenotypes. Using standard three-factor mapping, we placed oz237 between unc-29 and spe-9, close to dpy-24 and lin-11, on linkage group I (Figure 4). We then used SNP mapping, using unc-29 and unc-75 as visible markers, to further narrow the critical region containing oz237 to ∼46 kb containing 14 predicted genes (Figure 4). We sequenced candidate genes using DNA from oz237 mutant animals (see materials and methods) and found an A-to-T nucleotide substitution in the coding region of pas-5. This missense mutation is predicted to replace an aspartic acid, which is highly conserved across phyla, with a valine (Figure 4). pas-5 encodes a highly conserved α-subunit of the 20S proteasome (Groll et al. 2005; Kahn et al. 2008). To confirm that oz237 is an allele of pas-5, we rescued the oz237 mutant phenotype with two independent extrachromosomal arrays that each contain a genomic copy of pas-5 (Table 1). The arrays rescued the Mog phenotype of pas-5(oz237) single mutants and the overproliferation phenotype of pas-5(oz237); glp-1(ar202gf) double mutants (Table 1). A synthetic multivulval phenotype described below was also rescued by a pas-5(+) array (see supplemental material).
Figure 4.—
oz237 is an allele of pas-5, a highly conserved proteasome subunit. (A) Summary of SNP mapping data. Unc nonsterile recombinant animals were picked from unc-29(e193) pas-5(oz237)/HA-8 and pas-5(oz237) unc-75(e950)/HA-8 strains and the resulting animals were tested for N2 or HA-8 DNA at the SNP locations on the top line. The name of the cosmid containing the SNP is shown along with its nucleotide position (http://www.wormbase.org). From 125 unc-29 nonsterile recombinants and 90 unc-75 nonsterile recombinants, the minimal region containing the oz237 lesion was narrowed to ∼46 kb containing 14 genes. A summary of the SNP recombination data is shown next to each SNP on the second line from the top. “L” corresponds to the recombinants from the left (unc-29), while “R” corresponds to recombinants from the right (unc-75). At each SNP location the number of recombinants that retained N2 DNA at the SNP location are shown over the total number of recombinants. An A-to-T substitution was found in the pas-5 gene of oz237 animals. (B) A partial alignment of PAS-5 (top sequence) with homologs from other species. Protein sequences were aligned with the ClustalW program (Chenna et al. 2003). Identical amino acids are shown against a solid background, similar amino acids against a gray background, and nonsimilar amino acids against a white background. The asterisk shows the location of the missense mutation, which is predicted to replace an aspartic acid with a valine. A full alignment of these proteins is in the supplemental material. Accession numbers for proteins are NP_492765 (C. elegans), AAB93421 (Drosophila), NP_188046 (Arabidopsis), NP_036097 (mice), and AAV38522 (humans). PAS-5 shares 57, 57, 61, and 61% amino acid identity to Drosophila, Arabidopsis, mouse, and human proteins, respectively.
TABLE 1.
Transgenic rescue of pas-5(oz237)
| Transgene | Genotypea | Rescue (%)b | n |
|---|---|---|---|
| pas-5(+) ugEx1c | pas-5(oz237) | 12 | 122 |
| pas-5(+) ugEx2d | pas-5(oz237) | 42 | 52 |
| Control ugEx4e | pas-5(oz237) | 0 | 64 |
| Nonef | pas-5(oz237) | 0 | 109 |
| pas-5(+) ugEx1 | pas-5(oz237); glp-1(ar202gf) | 39 | 59 |
| pas-5(+) ugEx2 | pas-5(oz237); glp-1(ar202gf) | 6.7 | 134 |
| Control ugEx4 | pas-5(oz237); glp-1(ar202gf) | 0 | 54 |
| Nonef | pas-5(oz237); glp-1(ar202gf) | 0 | 76 |
Actual genotype for pas-5(oz237) animals is pas-5(oz237) unc-75(e950). Actual genotype for pas-5(oz237); glp-1(ar202gf) animals is pas-5(oz237) unc-75(e950); glp-1(ar202gf). Both strains were balanced with hT2 and the germ-line phenotypes of Unc progeny were analyzed.
pas-5(oz237) animals were categorized as rescued if they contained oocytes and/or embryos. All experiments were performed at 15°, at which temperature all pas-5(oz237) animals are Mog. pas-5(oz237); glp-1(ar202gf) animals were categorized as rescued if they did not have a full or proximal tumor.
This extrachromosomal array, ugEx1, contains four plasmids: pDH43[pas-5(+)], pRF4[rol-6(dm)], pJM67[elt-2∷gfp], and pBluescript KS+.
This extrachromosomal array, ugEx2, contains the same plasmids as ugEx1, but was obtained from an independent injection.
This extrachromosomal array, ugEx4, contains three of the plasmids contained in ugEx1 and ugEx2—pRF4[rol-6(dm)], pJM67[elt-2∷gfp], and pBluescript KS+—but lacks pDH43[pas-5(+)].
ugEx1 and ugEx2 segregate to only a portion of the progeny. As a control, we scored the germ-line phenotypes of animals containing the array and those that did not. The germ-line phenotype was scored blind; the presence of the array was determined by GFP expression (elt-2∷gfp) after the germ-line phenotype was scored.
To further support the hypothesis that oz237 is an allele of pas-5, we used RNAi against pas-5 to phenocopy the oz237 phenotype. RNAi against pas-5, with dsRNA delivered through either feeding or injection, resulted in an embryonic lethal phenotype. We reasoned that RNAi against pas-5, which encodes a core component of the proteasome, might reduce proteasome activity to a much greater degree than the likely reduction in proteasome activity due to the single amino acid change in pas-5(oz237) animals. Additionally, pas-5(oz237) mutants likely have maternal rescue, whereas RNAi can eliminate both maternal and zygotic mRNA. In other words, oz237 may have a much milder phenotype as compared to that produced by pas-5 RNAi. Therefore, we reduced the degree of knockdown caused by RNAi by performing low-dose RNAi; we reduced the injected dsRNA from a concentration of 100–1000 ng/μl to 1 ng/μl. As expected, due to the low concentration of dsRNA, a large proportion of the progeny showed no phenotype other than that provided by the genetic background; however, low-dose pas-5(RNAi) did enhance the overproliferation phenotype in both glp-1(ar202gf) and gld-2(q497) animals as compared to control RNAi (gfp) (Table 2). Therefore, RNAi directed against pas-5 does show similar phenotypes as the oz237 allele. The mapping data narrowing the critical region of oz237 to 14 genes, the identification of a missense mutation in a highly conserved amino acid of PAS-5, the rescue of the oz237 phenotype with a genomic pas-5 transgene, and the similar phenotypes of pas-5(RNAi) and the oz237 allele, all combined, confirm that oz237 is an allele of pas-5.
TABLE 2.
RNAi of proteasome subunits
| Phenotype (%)
|
||||||
|---|---|---|---|---|---|---|
| Genotypea | dsRNA | Wild typeb | Necroticc | Full tumord | Proe | Nf |
| Wild type | pas-5 | 71 | 29 | 0 | 0 | 300 |
| Wild type | gfp | 100 | 0 | 0 | 0 | 129 |
| glp-1(ar202gf) | pas-5 | 54 | 31 | 8.7 | 6.5 | 138 |
| glp-1(ar202gf) | pbs-4 | 76 | 8.6 | 9.5 | 6.0 | 116 |
| glp-1(ar202gf) | gfp | 93 | 0 | 0 | 6.8 | 133 |
| gld-2(q497) | pas-5 | 58 | 22 | 9.6 | 9.6 | 125 |
| gld-2(q497) | pbs-4 | 74 | 13 | 8.1 | 3.7 | 135 |
| gld-2(q497) | gfp | 91 | 0 | 0 | 9.1 | 110 |
Low-dose dsRNA was injected into young adult animals that were allowed to self-fertilize. glp-1(ar202gf) homozygous animals were injected and grown at 15°. gld-2(q497)/unc-15(e73) ccIs4251[myo-3∷Ngfp; myo-3∷Mtgfp] were injected and nongreen progeny were scored for their germ-line phenotype. gld-2(q497)/unc-15(e73) ccIs4251[myo-3∷Ngfp; myo-3∷Mtgfp] and N2 animals were injected and screened at 20°.
gld-2(q497) animals showed a phenotype no different from that of uninjected animals.
The gonads in these animals have very few cells, and cells that are present appear to be degrading.
Gonads in these animals completely lack sperm or oocytes, but rather have proliferative cells throughout the gonad arm.
Pro, proximal proliferation. Gonads in these animals contain sperm and/or oocytes, but also contain proliferative cells in the proximal end of the gonad.
Total number of progeny scored. Injected animals often yielded low numbers of progeny due to the RNAi.
oz237 partially reduces proteasome activity:
PAS-5 is a subunit of the 20S proteasome, which is a highly conserved multisubunit complex that is the primary component of the cell's protein degradation machinery (Groll et al. 2005). The basic structure of the 20S proteasome consists of four rings: the two inner (β) rings contain the proteolyticly active region of the proteasome. The two outer (α) rings are not proteolytic, but rather participate in substrate translocation into the central core of the 20S proteasome (Groll et al. 2005). PAS-5 is one of the seven highly conserved subunits of the α-ring (Groll et al. 2005; Kahn et al. 2008); therefore, PAS-5 is not directly involved in the degradation of proteins, but rather in the presenting of target proteins to the catalytic portion of the proteasome. We have already shown that low-dose RNAi against pas-5 has a similar phenotype as the oz237 allele, suggesting that the oz237 missense mutation reduces PAS-5 activity; however, this does not necessarily mean that the overall function of the proteasome is reduced. Rather, this mutation could simply affect the gating of proteins into the proteasome and could actually increase the degradation of certain proteins. Therefore, to determine if a general reduction in proteasome activity increases germ-line proliferation, we performed RNAi on a subunit of the β-ring (PBS-4), which is a component of the catalytic portion of the proteasome (Groll et al. 2005). As with pas-5 RNAi, when pbs-4 RNAi is performed at a typical concentration of 100–1000 ng/μl, a severe embryonic lethal phenotype is observed. However, low-dose RNAi against pbs-4, presumably only partially reducing PBS-4 levels, resulted in the same overproliferation synthetic interactions as pas-5 RNAi (Table 2). Low-dose RNAi to both pas-5 and pbs-4 enhances the overproliferation phenotypes of glp-1(ar202gf) and gld-2(q497) animals, similar to the enhancement observed with pas-5(oz237), and higher-dose RNAi to both results in a much stronger embryonic lethal phenotype; these data are consistent with the pas-5(oz237) allele partially reducing proteasome activity.
pas-5(oz237) influences another Notch-regulated cell fate decision:
Since proteasome activity is reduced in a pas-5(oz237) mutant, the enhancement of overproliferation is likely due to a target protein(s) not being degraded as efficiently as in wild-type animals and inappropriately promoting proliferation. A possible target of the proteasome whose decreased rate of degradation contributes to tumor formation is a Notch component or regulator. No direct readout of GLP-1/Notch signaling in the C. elegans germ line is currently available; therefore, we cannot directly test Notch signaling levels. However, we reasoned that if pas-5(oz237) is affecting Notch signaling levels, it may enhance increased Notch-signaling phenotypes in other Notch-regulated cell fate decisions. To test this, we assessed the ability of pas-5(oz237) to enhance the Muv phenotype of two weak gain-of-function alleles of lin-12. C. elegans has two Notch homologs, GLP-1 and LIN-12 (Yochem and Greenwald 1989). During the formation of the vulva, LIN-12 signaling provides the lateral signal between vulval precursor cells (VPCs) to specify VPCs P5.p and P7.p to adopt the secondary cell fate (Greenwald et al. 1983). Strong lin-12 gain-of-function alleles cause increased lateral signaling, resulting in all six VPCs adopting the secondary fate, whereas in wild-type animals only two VPCs adopt the secondary fate (Greenwald et al. 1983). Both lin-12(n379gf) and lin-12(n676gf) are weak gain-of-function alleles that cause only a low percentage of animals to be Muv (Greenwald et al. 1983). A reduction in the activities of negative regulators of Notch signaling enhances the multivulval phenotype of lin-12(n676gf) and lin-12(n379gf) (Sundaram and Greenwald 1993; Hubbard et al. 1997; Wen and Greenwald 1999; Yoo and Greenwald 2005). We found that pas-5(oz237) enhances the multivulval phenotype of both lin-12(n379gf) and lin-12(n676gf) (Table 3). A lin-11∷gfp secondary cell fate marker was used to confirm that the multivulval phenotype was due to an increase in secondary cells, consistent with an increase in Notch signaling (see supplemental materials) (Reddien et al. 2001; Gupta and Sternberg 2002). Therefore, a reduction in proteasome activity due to pas-5(oz237) enhances the multivulval phenotype of lin-12(gf) and the tumorous phenotype of glp-1(gf). This supports the model of a reduction in proteasome activity increasing the level of Notch signaling. Therefore, at least part of the overproliferation phenotype in pas-5(oz237) synthetic double mutants is likely due to an increase in Notch signaling. The protein(s) not properly degraded in pas-5(oz237) mutants, which could be in either the soma or the germ line, may be a component or a regulator of Notch signaling.
TABLE 3.
pas-5(oz237) enhances the Muv phenotype
| Genotype | Muv (pseudovulvae ≥2) (%)a | Muv (pseudovulvae ≥3) (%)b | n |
|---|---|---|---|
| pas-5(oz237)c | 2.3 | 0 | 89 |
| sel-10(bc243) | 0 | 0 | 43 |
| lin-12(n379gf) | 9.3 | 0 | 74 |
| lin-12(n676gf) | 4.2 | 2.1 | 95 |
| pas-5(oz237); sel-10(bc423)d | 42 | 12 | 67 |
| pas-5(oz237); lin-12(n379gf)e | 48 | 16 | 94 |
| pas-5(oz237); lin-12(n676gf)f | 68 | 38 | 95 |
“Pseudovulvae ≥2” is the percentage of animals that displayed two or more pseudovulvae.
“Pseudovulvae ≥3” is the percentage of animals that displayed three or more pseudovulvae.
pas-5(oz237) homozygotes were progeny from pas-5(oz237)/hT2 mothers.
pas-5(oz237); sel-10(bc423) homozygotes were progeny from pas-5(oz237)/hT2; sel-10(bc423) mothers.
pas-5(oz237); lin-12(n379gf) homozygotes were progeny from pas-5(oz237)/hT2; lin-12(n379gf)/hT2 mothers.
pas-5(oz237); lin-12(n676gf) homozygotes were progeny from pas-5(oz237)/hT2; lin-12(n676gf)/hT2 mothers.
pas-5(oz237) causes a synthetic multivulval phenotype with sel-10(null):
We have provided support for the model that decreased proteasome activity due to the pas-5(oz237) missense mutation increases Notch signaling in C. elegans. Proteasome-mediated protein degradation has previously been implicated in inhibiting Notch signaling. For example, SEL-10 is involved in targeting the intracellular domain of the Notch receptor for proteasome-mediated degradation (Hubbard et al. 1997; Gupta-Rossi et al. 2001; Oberg et al. 2001; Wu et al. 2001). SEL-10 is a component of an E3-ubiquitin ligase that is thought to ubiquitinate the cleaved intracellular portion of the Notch receptor (INTRA), targeting it for degradation by the proteasome (Hubbard et al. 1997; Gupta-Rossi et al. 2001; Oberg et al. 2001; Wu et al. 2001). C. elegans lacking sel-10 activity have increased Notch signaling, enhancing the multivulval phenotype of lin-12(gf) (Sundaram and Greenwald 1993). To determine if the presumed increase in Notch signaling in pas-5(oz237) animals is due to a decrease in sel-10-mediated degradation of INTRA, we analyzed pas-5(oz237); sel-10(bc243) double mutants in which sel-10(bc243) is a null allele (Jager et al. 2004). We found that pas-5(oz237); sel-10(bc243) animals have a synthetic multivulval phenotype, suggesting that the double mutant has a higher level of Notch signaling than that found in either single mutant (Table 3). Therefore, the increase in Notch signaling in pas-5(oz237) mutants cannot be due solely to a decrease in sel-10-mediated INTRA degradation. Either sel-10 may not be the only means by which INTRA is targeted to the proteasome (see discussion) or a component or positive regulator of Notch signaling, other than INTRA, is also degraded by the proteasome. The germ lines of pas-5(oz237); sel-10(bc243) animals have very few germ cells and the nuclei have abnormal morphology (see supplemental material). This does not appear to be a result of a disruption in the proliferation vs. meiotic entry decision. sel-10 likely is involved in other processes in the germ line that result in this phenotype. Indeed, SEL-10 is involved in targeting proteins other than Notch for degradation. For example, SEL-10 targets sex determination proteins in C. elegans (Jager et al. 2004) and contributes to the degradation of proteins such as c-Jun, cyclin E, c-Myc, and Aurora-A in other systems (Koepp et al. 2001; Moberg et al. 2001; Strohmaier et al. 2001; Mao et al. 2004; Nateri et al. 2004; Welcker et al. 2004; Yada et al. 2004). Therefore, pas-5(oz237); sel-10(bc243) animals have a synthetic germ-line phenotype, likely related to a target of SEL-10 other than Notch, which prevents the germ line from forming properly. Thus, this germ-line phenotype prevents us from analyzing the effect of pas-5(oz237); sel-10(bc243) on the proliferation vs. meiotic entry decision.
To determine if a decrease in INTRA degradation can explain the enhancement of overproliferation in pas-5(oz237) mutants, we determined whether glp-1(q35) forms a synthetic tumor with gld-2 and/or the other gld-2 pathway gene, gld-3. The mutation in glp-1(q35) is a premature stop codon, which results in a truncated GLP-1 protein that lacks the PEST domain involved in protein degradation (Rogers et al. 1986; Mango et al. 1991). The q35 mutation results in a gain-of-function multivulval phenotype, especially when nonsense-mediated decay, which degrades mRNAs with premature stop codons (Wagner and Lykke-Andersen 2002), is also eliminated through genetic mutation (Mango et al. 1991). The gain-of-function multivulval phenotype is thought to occur due to a decrease in INTRA degradation (Mango et al. 1991; Westlund et al. 1997). We reasoned that if pas-5(oz237) contributes to the synthetic tumorous phenotype with gld-2(q497) by decreasing INTRA degradation, then glp-1(q35) should also form a synthetic tumor with gld-2 or with the other gld-2 pathway gene, gld-3. We found that glp-1(q35) does not form a synthetic tumor with gld-2(q497) or gld-3(q730) (Table 4), suggesting that pas-5(oz237)'s contribution to overproliferation is not solely due to a decrease in INTRA degradation. However, since glp-1(q35) single-mutant animals do not display a germ-line gain-of-function phenotype, even when nonsense-mediated decay is eliminated (Mango et al. 1991), it is possible that glp-1(q35) does not cause the same increase in Notch signaling in the germ line as that observed in the soma. Therefore, the lack of overproliferation in the germ lines of gld-2(q497); glp-1(q35gf) and gld-3(q730); glp-1(q35gf) animals could also be due to glp-1(q35gf) not sufficiently increasing Notch signaling in the germ line.
TABLE 4.
Synthetic tumorous and epistasis analysis
| Genotype | Phenotypea |
|---|---|
| pas-5(oz237) | Not tumorous |
| gld-1(q485)b | Not tumorousc |
| gld-2(q497) | Not tumorousd |
| gld-3(q730) | Not tumorouse |
| nos-3(oz231) | Not tumorousf |
| glp-1(q175) | Glpg |
| glp-1(q35gf) | Not tumoroush |
| gld-1(q485) pas-5(oz237)i | Not tumorous |
| gld-2(q497) pas-5(oz237) | Tumorous |
| pas-5(oz237); gld-3(q730) | Tumorous |
| pas-5(oz237); nos-3(oz231) | Not tumorous |
| gld-2(q497) pas-5(oz237); glp-1(175)j | Glp |
| pas-5(oz237); gld-3(q730); glp-1(q175)k | Tumorous |
| gld-2(q497); glp-1(q35)l | Not tumorous |
| gld-3(q730); glp-1(q35)m | Not tumorous |
Phenotype refers only to the germ-line proliferation phenotype. Some genotypes yielded other germ-line phenotypes, but were still counted as “not tumorous” if the amount of proliferation was similar to wild type. Some single- and double-mutant animals labeled as “not tumorous” have slightly larger or smaller mitotic regions as compared to wild-type animals. Only those animals with proliferative cells throughout the germ line were labeled as “tumorous.” Glp means that animals show a phenotype similar to glp-1(0) in that all germ cells enter meiotic prophase prematurely and no proliferative cells are present in the late larval or adult germ line. In all cases, >20 animals were analyzed and in all cases 100% of the animals displayed the same general phenotype, unless otherwise noted.
gld-1(q485) hermaphrodites have a germ-line tumor that is not due to a defect in the proliferation vs. meiotic entry decision, but rather due to a defect in meiotic prophase progression of female germ cells (Francis et al. 1995b). Therefore, we have used gld-1(q485) males, which do not have tumorous germ lines, for this analysis.
A low percentage (∼2%) of gld-2(q497) homozygotes have proximal proliferation (Pro) (Kadyk and Kimble 1998).
Homozygous male progeny were analyzed.
Actual genotype gld-2(q497) pas-5(oz237); unc-32(e189) glp-1(175).
Actual genotype pas-5(oz237); gld-3(q730); unc-32(e189) glp-1(q175).
Actual genotype smg-2(e2008) gld-2(q497); unc-32(e189) glp-1(q35gf).
Actual genotype smg-2(e2008); gld-3(q730); unc-32(e189) glp-1(q35gf).
Notch-independent targets of the proteasome contribute to the overproliferation phenotype:
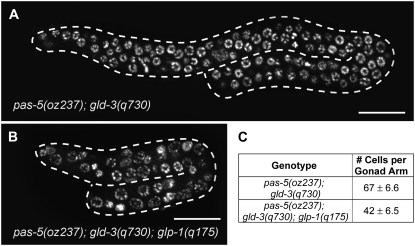
As mentioned above, pas-5(oz237) forms a synthetic tumor with gld-2(q497) and glp-1(ar202gf). We have shown that pas-5(oz237) enhances both glp-1(gf) and lin-12(gf) alleles, suggesting that the overproliferation phenotype may be due to an increase in GLP-1/Notch signaling. However, the lack of a tumor in the gld-2(q497); glp-1(q35gf) double mutant suggests that stabilization of INTRA is not sufficient to explain the synthetic tumorous phenotype with gld-2. To further test the model that proteasome activity inhibits GLP-1/Notch signaling in the regulation of the proliferation vs. meiotic entry decision, and to determine if the proteasome could also function downstream of GLP-1/Notch signaling in regulating this balance, we performed genetic epistasis analysis. We first found that pas-5(oz237) forms a synthetic tumor with both gld-2 pathway genes, gld-2 and gld-3, but not with either gld-1 pathway genes, gld-1 or nos-3 (Table 4). Therefore, if pas-5 functions downstream of GLP-1/Notch signaling, it likely functions in the gld-1 pathway. However, it is still possible that pas-5 could function upstream of GLP-1/Notch signaling, but that different relative “strengths” of the gld-1 and gld-2 pathways could cause one pathway, and not the other, to be synthetic tumorous with pas-5(oz237). Therefore, we tested for the dependence of the tumors in gld-2(q497) pas-5(oz237) and pas-5(oz237); gld-3(q730) animals on GLP-1/Notch signaling by analyzing triple mutants that lacked glp-1 activity. We found that gld-2(q497) pas-5(oz237); glp-1(q175) triple mutants have a Glp phenotype in which germ cells prematurely enter into meiotic prophase, depleting the population of proliferative cells (Table 4). Therefore, glp-1(q175) is epistatic to the gld-2(q497) pas-5(oz237) tumor, suggesting that the proteasome's contribution to tumor formation may occur, at least in part, genetically upstream of GLP-1/Notch signaling. This supports the model that the proteasome contributes to the regulation of the proliferation vs. meiotic entry decision by inhibiting GLP-1/Notch signaling. However, it should be emphasized that pas-5(oz237) is not a null allele; therefore, this result is not conclusive. It is still possible that pas-5 functions downstream of GLP-1/Notch signaling and that the Glp phenotype in gld-2(q497) pas-5(oz237); glp-1(q175) animals is due to a release of the GLP-1/Notch repression of proteasome activity that is still present in a pas-5(oz237) mutant. To further test if GLP-1/Notch signaling is necessary for pas-5(oz237) to enhance overproliferation, we determined if the tumor in pas-5(oz237); gld-3(q730) animals is dependent upon GLP-1/Notch signaling. Surprisingly, pas-5(oz237); gld-3(q730); glp-1(q175) triple-mutant animals have a tumorous germ line (Table 4; Figure 5). The tumor is smaller in pas-5(oz237); gld-3(q730); glp-1(q175) animals than in pas-5(oz237); gld-3(q730) animals; however, the germ cells continue to proliferate and the tumor increases in size over time (see supplemental material). Therefore, pas-5(oz237); gld-3(q730); glp-1(q175) animals form a germ-line tumor, unlike gld-2(q497) pas-5(oz237); glp-1(q175) animals, which are Glp (Table 4; Figure 5). The fact that pas-5(oz237); gld-3(q730); glp-1(q175) animals are tumorous argues against the inhibition of GLP-1/Notch signaling as the proteasome's sole function in regulating the proliferation vs. meiotic entry decision. It suggests that pas-5 may function downstream of GLP-1/Notch signaling in the gld-1 pathway. This result provides very strong evidence for the proteasome functioning downstream of GLP-1/Notch signaling because in the complete absence of GLP-1/Notch signaling a tumor is formed. However, the tumor in the pas-5(oz237); gld-3(q730) double mutant, with active Notch signaling, is much more robust than the tumor in the pas-5(oz237); gld-3(q730); glp-1(q175) triple mutant (Figure 5). The pas-5(oz237); gld-3(q730); glp-1(q175) tumor is very small, but it does continue to grow in size over time, making us confident in describing it as an overproliferative tumor (see supplemental material). However, the significantly larger size of the tumor when GLP-1/Notch signaling is active demonstrates that GLP-1/Notch signaling does contribute to tumor formation in pas-5(oz237) mutants. Therefore, our epistasis analysis supports a model in which the proteasome functions as a negative regulator of GLP-1/Notch signaling, as well as affects the activity of a component or regulator of the gld-1 pathway. Our analysis also demonstrates that the gld-2 and gld-3 genes are not equivalent in their roles in regulating the proliferation vs. meiotic entry decision since gld-2(q497) pas-5(oz237); glp-1(q175) and pas-5(oz237); gld-3(q730); glp-1(q175) have significantly different phenotypes (Table 4; see discussion).
Figure 5.—
GLP-1/Notch signaling contributes to the pas-5; gld-3 tumor. (A and B) Gonads from staged hermaphrodites were dissected, fixed, and stained with DAPI to visualize nuclear morphology. Animals of both genotypes were arrested as L1's and grown side by side for the same period of time until they reached the L4 stage and were dissected. More than 20 gonad arms for each genotype were dissected and stained. In all cases, the pas-5(oz237); gld-3(q730); glp-1(q175) gonad arms had far fewer germ cells than the pas-5(oz237); gld-3(q730) gonad arms. The distal end of each gonad is at the top left. Photos were taken using the Apotome module (Zeiss) to reduce background fluorescence. (A) Actual genotype pas-5(oz237); gld-3(q730); unc-32(e189). Bar, 20 μm. (B) Actual genotype pas-5(oz237); gld-3(q730); unc-32(e189) glp-1(q175). Bar, 20 μm. (C) Summary table of the number of germ cells in gonad arms of staged animals. Animals were staged as in A and B and were grown side by side until they reached the mid-L3 stage. Total germ cell numbers were determined at the same time point for both genotypes using DIC optics. Actual genotypes are pas-5(oz237); gld-3(q730); unc-32(e189) (n = 5) and pas-5(oz237); gld-3(q730); unc-32(e189) glp-1(q175) (n = 7). The range shown is 1 SD. The total cell numbers are statistically different (P < 1.2 × 10−4, Student's t-test, two-sample, two-tailed, unequal variance).
DISCUSSION
We have found that a decrease in proteasome activity enhances overproliferation in the C. elegans germ line. We found that a strong reduction in proteasome activity results in embryonic lethality, preventing a role in the proliferation vs. meiotic entry decision from being analyzed. However, by using an allele that only partially reduces proteasome activity, pleiotropic masking phenotypes could be avoided and a role for the proteasome in regulating the proliferation vs. meiotic entry decision was discovered. We found that reduced proteasome activity results in phenotypes associated with an increase in Notch signaling and that at least part of this increase is independent of SEL-10-mediated degradation of the intracellular portion of the Notch receptor. We further demonstrated that at least one target of the proteasome that contributes to germ cell overproliferation functions in the gld-1 pathway, downstream of GLP-1/Notch signaling.
Independent functions of GLD-2 and GLD-3:
gld-2 and gld-3 function together in the gld-2 pathway to inhibit proliferation and/or promote meiotic entry (Eckmann et al. 2004). GLD-2 is a poly(A) polymerase and binds GLD-3, which is an RNA-binding Bicaudal-C family member (Wang et al. 2002; Eckmann et al. 2004). GLD-3 strongly enhances the poly(A) polymerase activity of GLD-2 (Wang et al. 2002). While gld-2 and gld-3 appear to work together in the gld-2 pathway to inhibit proliferation and/or promote meiotic entry, they also have separate functions related to other aspects of germ-line development. For example, gld-3 is involved in germ-line survival and male spermatogenesis, while gld-2 does not appear to have related functions (Kadyk and Kimble 1998; Eckmann et al. 2002). Here we show that gld-2 and gld-3 are also not equivalent in their roles in inhibiting proliferation and/or in promoting meiotic entry; when gld-2 activity is removed in pas-5(oz237); glp-1(q175) double mutants, the resulting animals are Glp, whereas the removal of gld-3 activity in these same double mutants results in a tumorous germ line. One possible explanation for this dramatic difference in phenotypes could be that eliminating one component of the GLD-2/GLD-3 complex does not completely eliminate activity; GLD-2 and/or GLD-3 may be able to partially provide the function normally provided by the complex. If this explanation is true, then a lack of gld-3 function reduces complex activity more than a lack of gld-2 activity because the gld-2(q497) pas-5(oz237); glp-1(q175) mutant has meiotic entry, whereas the triple mutant with gld-3(q730) does not. Another possible explanation for the difference in phenotypes in gld-2(q497) pas-5(oz237); glp-1(q175) and pas-5(oz237); gld-3(q730); glp-1(q175) animals is that they may have some independent mRNA targets. It has previously been suggested that GLD-2 and GLD-3 may have a number of different binding partners on the basis of their different single-mutant germ-line phenotypes (Eckmann et al. 2004). Therefore, while GLD-2 and GLD-3 likely partner to regulate certain mRNAs involved in the proliferation vs. meiotic entry decision, they may also independently bind other proteins to regulate other mRNAs involved in this same decision.
Proteasome and Notch signaling:
We have shown that decreasing proteasome activity enhances two phenotypes associated with increased Notch signaling in C. elegans. Proteasome-mediated protein degradation has previously been implicated in other systems as a regulatory mechanism controlling the level of Notch signaling. For example, dominant-negative temperature-sensitive mutations in two Drosophila proteasome subunits result in shaft-to-socket cell fate transformations and a double-socket phenotype; both of these phenotypes are associated with an increase in Notch signaling (Schweisguth 1999). The increase in Notch signaling observed in these dominant-negative proteasome subunit mutations in Drosophila is likely due, at least in part, to a decrease in degradation of the cleaved intracellular form of the Notch receptor; an activated version of the intracellular domain is stabilized in one of the dominant-negative proteasome subunit mutants (Schweisguth 1999). Proteasome-mediated degradation of the intracellular domain of the Notch receptor has also been shown or suggested to occur in other systems, including C. elegans (Mango et al. 1991; Hubbard et al. 1997; Gupta-Rossi et al. 2001; Oberg et al. 2001; Wu et al. 2001; McGill and McGlade 2003; Tsunematsu et al. 2004). This degradation is thought to depend, at least in part, on the activity of SEL-10, a component of an E3 ubiquitin ligase (Hubbard et al. 1997; Gupta-Rossi et al. 2001; Oberg et al. 2001; Wu et al. 2001; Tsunematsu et al. 2004). sel-10 was first identified in a genetic screen as a negative regulator of LIN-12/Notch signaling (Sundaram and Greenwald 1993) and has since been shown to inhibit Notch signaling in many other systems (Lai 2002). sel-10 mutants also enhance the germ-line overproliferation phenotype of a weak glp-1(gf) mutant, suggesting that GLP-1(INTRA) is also a target of proteasome-mediated degradation in the germ line (Pepper et al. 2003). Here we show that decreased proteasome activity enhances both lin-12 and glp-1 gain-of-function alleles. A likely means by which this increase in Notch signaling is achieved is through a decrease in the degradation of INTRA. However, a reduction of SEL-10-mediated INTRA degradation cannot fully explain the increase in Notch signaling seen in the pas-5(oz237) mutants. We have shown that animals lacking sel-10 activity, and that also have reduced proteasome activity due to the pas-5(oz237) allele, have a synthetic multivulval phenotype. This synthetic interaction suggests that not all proteasome-mediated inhibition of Notch signaling is achieved through sel-10. The target of this sel-10-independent protein degradation could be a positive regulator of Notch signaling or a core component of Notch signaling, including INTRA. Indeed, there is evidence that an elimination of SEL-10 activity does not completely abolish proteasome-mediated degradation of INTRA. This evidence comes from the gain-of-function phenotype of the glp-1(q35) allele. As mentioned, glp-1(q35) results in a truncated GLP-1 protein, removing the PEST domain that is involved in targeting INTRA for degradation (Mango et al. 1991). The multivulval phenotype is presumably due to INTRA not being degraded by the proteasome, resulting in an increase in Notch signaling (Mango et al. 1991; Westlund et al. 1997). In contrast, sel-10 mutant animals, which should also have a decrease in the degradation of INTRA, do not display a multivulval phenotype in an otherwise wild-type background (Sundaram and Greenwald 1993; Hubbard et al. 1997). Therefore, glp-1(q35) animals presumably have a greater increase in Notch signaling than sel-10 animals, suggesting that some INTRA is still being targeted for degradation in sel 10 mutants. Loss of sel-10 activity does enhance both lin-12(gf) and glp-1(gf) alleles, suggesting that sel-10 mutants do have an increase in Notch signaling, presumably due to increased INTRA stability (Sundaram and Greenwald 1993; Hubbard et al. 1997; Pepper et al. 2003); however, the increase in Notch signaling in sel-10 mutants is not as great as the increase in Notch signaling in glp-1(q35) animals. The synthetic multivulval phenotype in pas-5(oz237); sel-10 mutants strongly supports the model that a lack of sel-10 activity does not abolish proteasome-mediated inhibition of Notch signaling. As mentioned, this inhibition could be accomplished through sel-10-independent degradation of INTRA, or it could involve another regulator or component of Notch signaling. sel-10-independent proteasome-mediated degradation of INTRA has previously been described (McGill and McGlade 2003). Mammalian Notch is ubiquitinated by the E3-ubiquitin ligase Itch, in association with Numb, targeting the Notch intracellular domain for proteasome-mediated degradation that does not require the PEST domain (McGill and McGlade 2003). C. elegans LIN-12(INTRA) can also be degraded in a mechanism independent of sel-10; however, this is thought to be mediated by the lysosome, not by the proteasome (Shaye and Greenwald 2005). Therefore, the sel-10-independent increase in Notch signaling that is achieved in pas-5(oz237) mutants may identify a novel level of regulation of Notch signaling, at least in C. elegans, that is accomplished through activity of the proteasome.
Proteasomal regulation of the GLD-1 pathway:
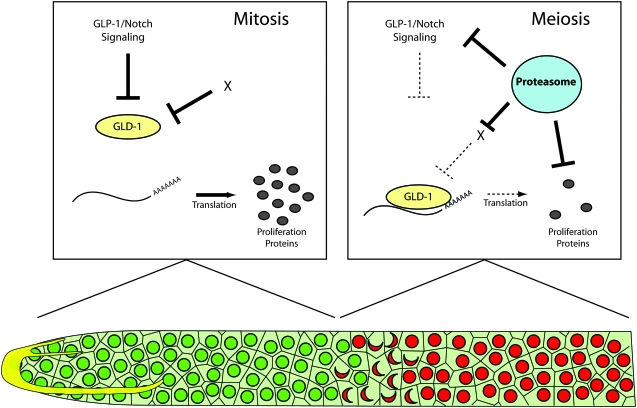
We have shown that stabilization of INTRA is not sufficient for the synthetic tumorous phenotype with gld-2 pathway genes; neither gld-2(q497); glp-1(q35) nor gld-3(q730); glp-1(q35) animals show overproliferation (Table 4). Additionally, since pas-5(oz237); gld-3(q730); glp-1(q175) animals do show some overproliferation, at least one factor that is degraded less efficiently in a pas-5(oz237) mutant and contributes to overproliferation functions downstream or parallel to the GLP-1/Notch signaling pathway. Our synthetic tumor tests show that pas-5(oz237) is synthetic tumorous with gld-2 pathway genes, but not with gld-1 pathway genes, suggesting that the proteasome interacts with the gld-1 pathway. The proteins in the gld-1 pathway that are normally degraded by the proteasome to regulate the proliferation vs. meiotic entry decision cannot be the protein products of either of the two known gld-1 pathway genes, gld-1 or nos-3, because stabilization of these proteins would increase the activity of the gld-1 pathway, whereas it is a decrease in gld-1 pathway activity that contributes to overproliferation. As mentioned above, GLD-1 is a translational inhibitor, likely inhibiting the translation of genes necessary for proliferation (Jan et al. 1999; Lee and Schedl 2001, 2004; Xu et al. 2001; Marin and Evans 2003; Lakiza et al. 2005). A proteasome target(s) that promotes proliferation may also be an mRNA target of GLD-1, and together the proteasome and GLD-1 inhibit the expression or stability of the protein(s) that is necessary for proliferation. In this model, germ cells in the distal end of the gonad express proteins that contribute to the proliferative state of the cells (Figure 6). As cells move proximally and switch from proliferation to meiotic prophase, GLD-1 inhibits further translation of the proteins. However, some proliferation proteins are likely to persist in these cells as they move from the mitotic region of the gonad to the region where they enter meiotic prophase. To allow for entry into meiotic prophase, these proliferation-promoting proteins could then be targeted for degradation by the proteasome (Figure 6).
Figure 6.—
Model of proteasome activity in regulating the proliferation vs. meiotic entry decision. In this model, proliferative cells in the mitotic zone (green) have active GLP-1/Notch signaling. This signaling inhibits the activity of GLD-1 by limiting its expression in the distal end (Hansen and Schedl 2006), which allows for specific mRNAs to be translated and produce proteins necessary for proliferation (blue). As part of one model, an unknown factor (X) may also inhibit GLD-1 activity in a manner independent of GLP-1/Notch signaling (see discussion). As cells move proximally, GLP-1/Notch signaling is reduced due to the distance from the LAG-2 ligand expressed in the DTC (yellow), and cells enter meiotic prophase (red). The proteasome also degrades a core component(s) or positive regulator of GLP-1/Notch signaling, reducing the level of signaling further. Reduced GLP-1/Notch signaling allows GLD-1 to bind target mRNAs, preventing further production of proliferation proteins. The proteasome could interact with the GLD-1 pathway by degrading a negative regulator of GLD-1 activity (factor X) and/or by degrading proliferation proteins that were translated while cells were still in mitosis, but persist as cells enter meiotic prophase. In either situation, proliferation protein levels are reduced, which allows cells to enter meiotic prophase (red). Dashed lines represent reduced activity.
An alternative or additional mechanism by which the proteasome could interact with the gld-1 pathway to regulate the proliferation vs. meiotic entry decision is through degradation of a negative regulator of GLD-1 activity (Figure 6). In this model, an unknown negative regulator (X) inhibits GLD-1 activity, allowing cells to remain proliferative. This inhibition of GLD-1 activity would have to occur independently of GLP-1/Notch signaling. As cells enter meiosis, the proteasome degrades the negative regulator, which allows GLD-1 to repress the translation of proteins necessary for proliferation; the cells are then able to enter meiotic prophase.
Proteasome-mediated regulation of a developmental decision:
The degradation of proteins by the proteasome is increasingly being realized as an important means of regulating many cellular processes and developmental decisions (Groll et al. 2005; Bowerman and Kurz 2006; Naujokat and Saric 2007). Indeed, the proteasome is involved in regulating so many fundamental processes, such as the cell cycle (Glotzer et al. 1991; Hershko et al. 1991), that it is sometimes difficult to tease apart specific functions. Our identification of a partial loss-of-function allele of a subunit of the 20S proteasome, which only partially reduces proteasome activity, has allowed us to identify a role for the proteasome in regulating the proliferation vs. meiotic entry decision in the C. elegans germ line. The proteasome has also been implicated in regulating the differentiation of mammalian stem cells (Naujokat and Saric 2007). The next step in understanding how protein degradation regulates the proliferation vs. meiotic entry decision in the C. elegans germ line, which will also likely help in our understanding of how protein degradation controls stem cell differentiation in general, will be to identify the proteins that are degraded by the proteasome and the E3 ubiquitin ligases that target them for destruction.
Acknowledgments
We thank Laura Wilson Berry and Tim Schedl who isolated the oz237 allele under National Institutes of Health (NIH) grant GM63310. We are grateful to Malcolm Williamson who helped with the initial mapping of the oz237 allele. We also thank Pavel Pasierbek and Joseph Loidl for anti-REC-8 antibodies and Monique Zetka for anti-HIM-3 antibodies. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. A.K. was partially funded by an undergraduate student research award salary support grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). D.H. is a scholar with the Alberta Heritage Foundation for Medical Research. This work was funded with a Discovery grant from NSERC to D.H.
References
- Austin, J., and J. Kimble, 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51 589–599. [DOI] [PubMed] [Google Scholar]
- Austin, J., and J. Kimble, 1989. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell 58 565–571. [DOI] [PubMed] [Google Scholar]
- Bowerman, B., and T. Kurz, 2006. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development 133 773–784. [DOI] [PubMed] [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden, S. L., E. R. Troemel, T. C. Evans and J. Kimble, 1994. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120 2901. [DOI] [PubMed] [Google Scholar]
- Crittenden, S. L., D. S. Bernstein, J. L. Bachorik, B. E. Thompson and M. Gallegos, 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417 660. [DOI] [PubMed] [Google Scholar]
- Eckmann, C. R., B. Kraemer, M. Wickens and J. Kimble, 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3 697–710. [DOI] [PubMed] [Google Scholar]
- Eckmann, C., S. L. Crittenden, N. Suh and J. Kimble, 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, R., M. K. Barton, J. Kimble and T. Schedl, 1995. a gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, R., E. Maine and T. Schedl, 1995. b Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 325–330. [DOI] [PubMed] [Google Scholar]
- Glotzer, M., A. W. Murray and M. W. Kirschner, 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349 132–138. [DOI] [PubMed] [Google Scholar]
- Greenwald, I., P. W. Sternberg and H. R. Horvitz, 1983. The lin-12 locus specifies cell fate in Caenorhabditis elegans. Cell 34 435–444. [DOI] [PubMed] [Google Scholar]
- Groll, M., M. Bochtler, H. Brandstetter, T. Clausen and R. Huber, 2005. Molecular machines for protein degradation. Chembiochem 6 222–256. [DOI] [PubMed] [Google Scholar]
- Gupta, B. P., and P. W. Sternberg, 2002. Tissue-specific regulation of the LIM homeobox gene lin-11 during development of the Caenorhabditis elegans egg-laying system. Dev. Biol. 247 102–115. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi, N., O. Le Bail, H. Gonen, C. Brou, F. Logeat et al., 2001. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276 34371–34378. [DOI] [PubMed] [Google Scholar]
- Hansen, D., and T. Schedl, 2006. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr. Top. Dev. Biol. 76 185–215. [DOI] [PubMed] [Google Scholar]
- Hansen, D., E. J. Hubbard and T. Schedl, 2004. a Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268 342. [DOI] [PubMed] [Google Scholar]
- Hansen, D., L. Wilson-Berry, T. Dang and T. Schedl, 2004. b Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131 93–104. [DOI] [PubMed] [Google Scholar]
- Henderson, S. T., D. Gao, E. J. Lambie and J. Kimble, 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120 2913–2924. [DOI] [PubMed] [Google Scholar]
- Hershko, A., D. Ganoth, J. Pehrson, R. E. Palazzo and L. H. Cohen, 1991. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J. Biol. Chem. 266 16376–16379. [PubMed] [Google Scholar]
- Hubbard, E. J., G. Wu, J. Kitajewski and I. Greenwald, 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11 3182–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, S., H. T. Schwartz, H. R. Horvitz and B. Conradt, 2004. The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc. Natl. Acad. Sci. USA 101 12549–12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski, J., and K. Kornfeld, 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, E., C. K. Motzny, L. E. Graves and E. B. Goodwin, 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. R., and T. Schedl, 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9 1491–1504. [DOI] [PubMed] [Google Scholar]
- Jones, A. R., R. Francis and T. Schedl, 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180 165. [DOI] [PubMed] [Google Scholar]
- Kadyk, L. C., and J. Kimble, 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kahn, N. W., S. L. Rea, S. Moyle, A. Kell and T. E. Johnson, 2008. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 409 205–213. [DOI] [PubMed] [Google Scholar]
- Kimble, J., and S. L. Crittenden, 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23 405–433. [DOI] [PubMed] [Google Scholar]
- Kimble, J. E., and J. G. White, 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81 208–219. [DOI] [PubMed] [Google Scholar]
- Koepp, D. M., L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu et al., 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294 173–177. [DOI] [PubMed] [Google Scholar]
- Kraemer, B., S. Crittenden, M. Gallegos, G. Moulder, R. Barstead et al., 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9 1009–1018. [DOI] [PubMed] [Google Scholar]
- Kramer, J. M., R. P. French, E. C. Park and J. J. Johnson, 1990. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol. Cell. Biol. 10 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E. C., 2002. Protein degradation: four E3s for the notch pathway. Curr. Biol. 12 R74–R78. [DOI] [PubMed] [Google Scholar]
- Lakiza, O., L. Frater, Y. Yoo, E. Villavicencio, D. Walterhouse et al., 2005. STAR proteins quaking-6 and GLD-1 regulate translation of the homologues GLI1 and tra-1 through a conserved RNA 3′UTR-based mechanism. Dev. Biol. 287 98–110. [DOI] [PubMed] [Google Scholar]
- Lamont, L. B., S. L. Crittenden, D. Bernstein, M. Wickens and J. Kimble, 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7 697–707. [DOI] [PubMed] [Google Scholar]
- Lee, M. H., and T. Schedl, 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. H., and T. Schedl, 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango, S. E., E. M. Maine and J. Kimble, 1991. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature 352 811–815. [DOI] [PubMed] [Google Scholar]
- Mao, J. H., J. Perez-Losada, D. Wu, R. Delrosario, R. Tsunematsu et al., 2004. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432 775–779. [DOI] [PubMed] [Google Scholar]
- Marin, V. A., and T. C. Evans, 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130 2623–2632. [DOI] [PubMed] [Google Scholar]
- McGill, M. A., and C. J. McGlade, 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278 23196–23203. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg, K. H., D. W. Bell, D. C. Wahrer, D. A. Haber and I. K. Hariharan, 2001. Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413 311–316. [DOI] [PubMed] [Google Scholar]
- Nateri, A. S., L. Riera-Sans, C. Da Costa and A. Behrens, 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303 1374–1378. [DOI] [PubMed] [Google Scholar]
- Naujokat, C., and T. Saric, 2007. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem Cells 25 2408–2418. [DOI] [PubMed] [Google Scholar]
- Oberg, C., J. Li, A. Pauley, E. Wolf, M. Gurney et al., 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276 35847–35853. [DOI] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer et al., 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A. S., D. J. Killian and E. J. Hubbard, 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien, P. W., S. Cameron and H. R. Horvitz, 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412 198–202. [DOI] [PubMed] [Google Scholar]
- Rogers, S., R. Wells and M. Rechsteiner, 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234 364–368. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F., 1999. Dominant-negative mutation in the beta2 and beta6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage. Proc. Natl. Acad. Sci. USA 96 11382–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye, D. D., and I. Greenwald, 2005. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 132 5081–5092. [DOI] [PubMed] [Google Scholar]
- Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt et al., 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413 316–322. [DOI] [PubMed] [Google Scholar]
- Suh, N., B. Jedamzik, C. R. Eckmann, M. Wickens and J. Kimble, 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103 15108–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988. Methods, pp. 587–606 in The Nematode Caenorhabditis elegans, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sundaram, M. V., and I. Greenwald, 1993. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics 135 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz, D., 1988. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature 332 281–284. [DOI] [PubMed] [Google Scholar]
- Tax, F. E., J. J. Yeargers and J. H. Thomas, 1994. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368 150–154. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395 854. [DOI] [PubMed] [Google Scholar]
- Tsunematsu, R., K. Nakayama, Y. Oike, M. Nishiyama, N. Ishida et al., 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279 9417–9423. [DOI] [PubMed] [Google Scholar]
- Tulina, N., and E. Matunis, 2001. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294 2546. [DOI] [PubMed] [Google Scholar]
- Wagner, E., and J. Lykke-Andersen, 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115 3033–3038. [DOI] [PubMed] [Google Scholar]
- Wang, L., C. R. Eckmann, L. C. Kadyk, M. Wickens and J. Kimble, 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419 312–316. [DOI] [PubMed] [Google Scholar]
- Welcker, M., A. Orian, J. Jin, J. E. Grim, J. W. Harper et al., 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101 9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C., and I. Greenwald, 1999. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans. J. Cell Biol. 145 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlund, B., L. W. Berry and T. Schedl, 1997. Regulation of germline proliferation in Caenorhabditis elegans. Adv. Dev. Biol. 5 43–80. [Google Scholar]
- Wharton, R. P., and G. Struhl, 1991. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67 955–967. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]
- Wu, G., S. Lyapina, I. Das, J. Li, M. Gurney et al., 2001. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21 7403–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, T., and A. C. Spradling, 2000. A niche maintaining germ line stem cells in the Drosophila ovary. Science 290 328. [DOI] [PubMed] [Google Scholar]
- Xu, L., J. Paulsen, Y. Yoo, E. B. Goodwin and S. Strome, 2001. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics 159 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada, M., S. Hatakeyama, T. Kamura, M. Nishiyama, R. Tsunematsu et al., 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem, J., and I. Greenwald, 1989. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell 58 553–563. [DOI] [PubMed] [Google Scholar]
- Yoo, A. S., and I. Greenwald, 2005. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science 310 1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka, M. C., I. Kawasaki, S. Strome and F. Muller, 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]