Abstract
Rab GTPases and ubiquitination are critical regulators of transmembrane cargo sorting in endocytic and lysosomal targeting pathways. The endosomal protein Rabex-5 intersects these two layers of regulation by being both a guanine nucleotide exchange factor (GEF) for Rab5 and a substrate for ubiquitin (Ub) binding and conjugation. The ability of trafficking machinery components to bind ubiquitinated proteins is known to have a function in cargo sorting. Here, we demonstrate that Ub binding is essential for the recruitment of Rabex-5 from the cytosol to endosomes, independently of its GEF activity and of Rab5. We also show that monoubiquitinated Rabex-5 is enriched in the cytosol. These observations are consistent with a model whereby a cycle of Ub binding and monoubiquitination regulates the association of Rabex-5 with endosomes.
Keywords: endosomes, Rabaptin-5, Rabex-5, RabGEF1, ubiquitin
Introduction
Ubiquitination of transmembrane (TM) proteins at the plasma membrane and the TGN is a signal for endosomal sorting and delivery to the vacuole/lysosome through the multivesicular body (MVB) pathway (Dupré et al, 2004; Hicke et al, 2005; Mukhopadhyay and Riezman, 2007). Ubiquitinated TM cargoes are recognized by adaptor proteins that contain one or more of several ubiquitin (Ub)-binding domains (UBDs) and that themselves undergo Ub-binding-dependent monoubiquitination (Dupré et al, 2004; Hicke et al, 2005; Mukhopadhyay and Riezman, 2007). Another important regulatory mechanism in the endosomal system is provided by members of the Rab family of low molecular weight GTPases that are enriched in different domains of this system and coordinate cargo trafficking alongside it (Miaczynska and Zerial, 2002; Stein et al, 2003). Rab5, in particular, is enriched in clathrin-coated endocytic vesicles and endosomes and, together with its cognate guanine nucleotide exchange factor (GEF) Rabex-5 and its partner Rabaptin-5, is a key regulator of homotypic and heterotypic endosomal fusion (Stenmark et al, 1995; Horiuchi et al, 1997).
Recent evidence indicates that these two layers of regulation of endosomal trafficking, that is, cargo sorting by Ub-binding/ubiquitinated adaptors and Rab5-dependent endosome fusion, intersect. Indeed, in addition to functioning as a Rab5 GEF, Rabex-5 binds Ub and undergoes Ub binding-dependent monoubiquitination (Lee et al, 2006; Mattera et al, 2006; Penengo et al, 2006). The basis for these functions lies in the domain organization of Rabex-5 (Figure 1A), which includes (a) an A20-like Zn finger (ZnF) that has Ub ligase activity (Lee et al, 2006; Mattera et al, 2006) and binds Ub through its D58-centred surface (Lee et al, 2006; Penengo et al, 2006), (b) an inverted Ub interaction motif or ‘motif interacting with ubiquitin' (MIU) that binds Ub through its I44-centred face (Lee et al, 2006; Penengo et al, 2006), (c) a Vps9 domain that, in conjunction with an adjacent N-terminal helical bundle (HB), comprises the GEF catalytic core (Delprato et al, 2004), (d) a coiled-coil (CC) containing the Rabaptin-5-binding site (Mattera et al, 2006; Delprato and Lambright, 2007; Kalesnikoff et al, 2007; Zhu et al, 2007) and (e) a C-terminal proline-rich domain.
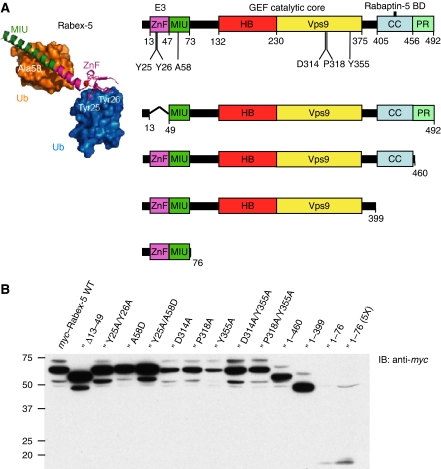
Figure 1.
Domain organization, design and expression of Rabex-5 constructs used in this study. (A) Rabex-5 domains include the A20-like Cys2/Cys2 zinc-finger (ZnF), which displays Ub protein ligase (E3) activity and binds Ub, and the motif interacting with ubiquitin (MIU) (Lee et al, 2006; Mattera et al, 2006; Penengo et al, 2006), the Vps9 module and its associated helical bundle (HB) harbouring the Rab5/Rab21 guanine nucleotide exchange factor (GEF) catalytic core (Delprato et al, 2004), the predicted amphipatic helix (CC) that binds Rabaptin-5 and acts as an autoinhibitory element of the HB-Vps9 tandem GEF activity (Mattera et al, 2006; Delprato and Lambright, 2007; Kalesnikoff et al, 2007; Zhu et al, 2007), and a C-terminal proline-rich region (PR). Residues substituted in the full-length construct (Y25 and Y26 in the ZnF, A58 in the MIU, D314, P318 and Y355 in the Vps9 domain) are indicated in the top scheme. Deletion constructs are shown underneath. The structure of the Rabex-5 ZnF and MIU domains (magenta and green ribbons, respectively) bound to two Ub molecules (blue and gold surfaces; adapted from Lee et al, 2006) is shown on the left. Tyr25/Tyr26 and Ala58 represent residues critical for Ub binding to the ZnF and MIU domains, respectively. Numbering corresponds to the bovine Rabex-5 used in this study. (B) Expression of myc-tagged Rabex-5 constructs in transiently transfected HeLa cells was assessed by SDS–PAGE of extracts and immunoblotting (IB) with anti-myc antiserum. The light bands with slower mobility (e.g. ∼73 kDa for myc–Rabex-5 WT, left lane) represent the monoubiquitinated recombinant proteins (see also Figure 4); the main bands correspond to the unmodified myc-tagged proteins, whereas the light bands with higher mobility are most likely degradation products. Notice that constructs with MIU substitutions (A58D and Y25A/A58D) do not undergo monoubiquitination. Monoubiquitination of myc–Rabex-5 Y25A/Y26A and Δ13–49 is also reduced or abolished when compared with WT. Numbers on the left indicate the position of molecular mass markers (in kilodaltons).
It has been proposed that binding of Ub to the UBD and/or coupled monoubiquitination of endocytic adaptors might regulate their activity and/or localization (Schnell and Hicke, 2003; Haglund and Dikic, 2005; Hoeller et al, 2007). Indeed, the binding of Ub to these proteins is key to the sorting of ubiquitinated TM cargo. However, many endocytic adaptors have domains that bind phosphoinositides or other proteins and, consequently, it is unclear whether Ub binding or monoubiquitination have a function in the actual recruitment of cytosolic machinery components to endosomes.
In this study, we have examined the function of Ub binding and ubiquitination in membrane recruitment using Rabex-5 as a model protein. We demonstrate that Ub binding is necessary for the recruitment of Rabex-5 to early endosomes, and that this recruitment is independent of its GEF activity and of its partners, Rab5 and Rabaptin-5. We also provide evidence that monoubiquitinated Rabex-5 is enriched in the cytosol. On the basis of these results, we propose that cycles of Ub binding and monoubiquitination regulate the association of Rabex-5 with endosomes.
Results
Role of Ub binding in the recruitment of Rabex-5 to early endosomes
We asked whether the ability of Rabex-5 to interact with Ub has a function in its recruitment to early endosomes. To address this, we analysed the following myc–Rabex-5 constructs (Figure 1A): (a) Rabex-5 wild type (WT); (b) Rabex-5 Δ13–49 (deletion of the ZnF abolishing binding to the Ub D58 patch); (c) Rabex-5 A58D (lacking binding of the MIU to the Ub I44 patch) and (d) Rabex-5 Y25A/A58D (combined mutations in ZnF and MIU that completely abrogate Ub binding) (Lee et al, 2006; Penengo et al, 2006). The expression levels of these and additional constructs analysed by immunoblotting are shown in Figure 1B. Immunofluorescence microscopy showed that myc–Rabex-5 WT localized to round vesicles of ∼0.7–2.5 μm in diameter in transiently transfected HeLa cells expressing intermediate to high levels of the recombinant protein (Figure 2A). These vesicles were more concentrated in the perinuclear area and their size appeared directly proportional to the expression level of myc–Rabex-5 (Supplementary Figure S1). The vesicles contained the early endosome antigen 1 (EEA1), identifying them as early endosomes (Figure 3A–C). The EEA1-containing endosomes in myc–Rabex-5-transfected cells appeared larger than those in untransfected cells, indicating that the expression of myc–Rabex-5 causes enlargement of endosomes (Figure 3A–C). This enlargement is reminiscent of the morphological changes observed on expression of proteins that form a tight complex with Rabex-5, such as Rabaptin-5 (Stenmark et al, 1995) or Rabaptin-5β (Gournier et al, 1998).
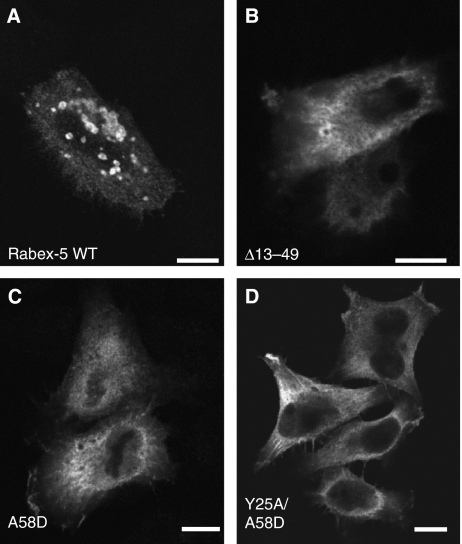
Figure 2.
Mutations impairing Ub binding alter the localization of Rabex-5. HeLa cells were transiently transfected and fixed with 4% formaldehyde ∼24 h after transfection. Fixed cells were incubated with mouse anti-myc and Alexa 568-conjugated anti-mouse antisera. The substitutions in the ZnF and MIU domains interfere with Ub binding to Rabex-5 (Lee et al, 2006; Mattera et al, 2006; Penengo et al, 2006). myc–Rabex-5 wild type (WT) (A) is predominantly recruited to large (∼2 μm) vesicles that are positive for the early endosomal marker EEA1 (see Figure 3); in contrast, expression of comparable levels of myc–Rabex-5 Ub-binding mutants, such as Δ13–49 (deletion of the ZnF) (B), A58D (C) and Y25A/A58D (D), results in predominantly cytosolic accumulation (some threads and membrane ruffles are also visible). Enlarged vesicles containing the recombinant proteins were observed in 99, 2, 1 and 1% of cells expressing intermediate to high levels of myc–Rabex-5 WT or the Δ13–49, A58D and Y25A/A58D mutants, respectively (n=396, 643, 534 and 372, respectively). Approximately 8–12% of cells transfected with either Rabex-5 A58D or Rabex-5 Y25A/A58D also exhibited large artificial aggregates containing the recombinant proteins that were clearly discernable from the vesicles containing myc–Rabex-5 WT; this aggregation was less frequent (0–3%) in cells transfected with the ΔZnF or Y25A/Y26A mutants. Scale bars=10 μm.
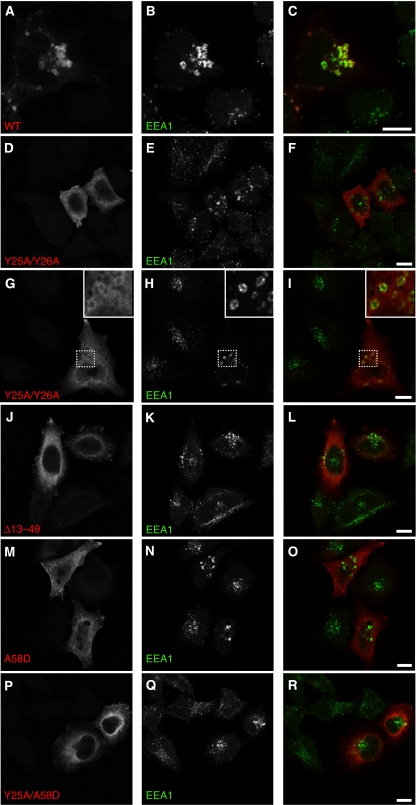
Figure 3.
Recruitment of myc–Rabex-5 WT but not Ub-binding mutants to early endosomes. HeLa cells were transfected and fixed as described in the legend to Figure 2. Fixed cells were incubated with rabbit anti-myc and mouse anti-EEA1 antisera, followed by incubation with Alexa 568-conjugated anti-rabbit and Alexa 488-conjugated anti-mouse antisera. Rabex-5 WT (A–C) is recruited to early endosomes, whereas the Rabex-5 Ub-binding mutants (D–R) exhibit a predominantly cytosolic distribution. Although expression of all constructs results in enlargement of early endosomes as compared with untransfected cells, only myc–Rabex-5 WT is recruited to these structures. The enlargement of early endosomes in cells transfected with myc–Rabex-5 WT was greater than in cells expressing the Ub-binding mutants. In some instances, particularly for Rabex-5 Y25A/Y26A (G–I, insets), it was possible to observe decoration of enlarged early endosomes by the Ub-binding mutants amidst their predominantly cytosolic distribution. Insets in (G–I) represent a × 3 magnification of areas in the dotted squares. (C, F, I, L, O, R) Merged images of the panels at their left. Scale bars=10 μm.
Significantly, expression of comparable levels of myc–Rabex-5 Ub-binding mutants (Figure 1B) resulted in a predominantly cytosolic distribution (along with filaments and membrane ruffles in some cells) in contrast to the WT protein, which was vesicle-associated (Figure 2A–D). Expression of the Ub-binding mutants still caused some enlargement of EEA1-labelled endosomes (Figure 3D–R), albeit to a lesser extent than the expression of myc–Rabex-5 WT (Figure 3A–C). In some instances, particularly for Rabex-5 Y25A/Y26A, it was possible to discern faint decoration of enlarged early endosomes by Rabex-5 Ub-binding mutants amidst their predominantly cytosolic distribution (Figure 3G–I, insets), suggesting a residual or transient association of these mutants with endosomes. Consistent with the confocal microscopy, subcellular fractionation showed that the majority of myc–Rabex-5 WT was membrane-bound, whereas the myc–Rabex-5 Y25A/Y26A and A58D mutants were mostly cytosolic (Figure 4A). These observations indicated that (a) decreased binding of Rabex-5 to Ub impairs its recruitment to early endosomes; (b) lack of recruitment of the myc–Rabex-5 Ub-binding mutants to early endosomes is not caused by the disruption of this compartment as EEA1-positive endosomes are visible in cells transfected with these constructs.
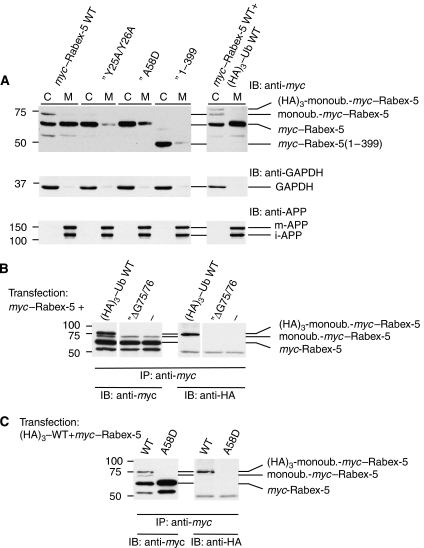
Figure 4.
Subcellular fractionation of myc–Rabex-5 constructs and ubiquitination assays. (A) HeLa cells were transfected with the indicated constructs and subjected to nitrogen cavitation ∼24 h after transfection. Postnuclear (900 g) supernatants were centrifuged at 300 000 g to yield cytosolic (C) and crude membrane (M) fractions that were analysed by SDS–PAGE and immunoblotting (IB). Cytosol and membrane samples loaded onto gels represent 0.75% of the total of each fraction. The mobilities of (HA)3-monoubiquitinated myc–Rabex-5, monoubiquitinated myc–Rabex-5 (see legend of B, C) and myc–Rabex-5 are shown on the right of the upper blots. Note that both (HA)3-monoubiquitinated myc–Rabex-5 (cells co-transfected with myc–Rabex-5 and (HA)3–Ub in blot shown on right) and monoubiquitinated myc–Rabex-5 (blots on left and right) are enriched in the cytosolic fraction. GAPDH and amyloid precursor protein (APP) were used as markers of the cytosolic and membrane fractions, respectively (middle and lower blots). The two bands detected with anti-APP correspond to the mature N- plus O-glycosylated (m-APP) and immature N-glycosylated (i-APP) forms, respectively (Chyung and Selkoe, 2003). (B, C) HeLa cells were transfected with myc–Rabex-5 with or without the indicated (HA)3–Ub constructs. Cells were lysed ∼24 h after transfection and the lysates were subjected to immunoprecipitation (IP) with mouse anti-myc followed by SDS–PAGE and IB with rabbit anti-myc (left) or mouse anti-HA (right) antisera. The mobilities of (HA)3-monoubiquitinated myc–Rabex-5, monoubiquitinated myc–Rabex-5 and myc–Rabex-5 are shown on the right of the blot. Identification of the second band from top (∼73 kDa) as myc–Rabex-5 monoubiquitinated with endogenous Ub is based on (a) its immunoreactivity with anti-myc but not anti-HA (B, C); (b) the presence of this band in immunoprecipitates of cells transfected only with myc–Rabex-5 (no co-transfection with (HA)3–Ub) or co-transfected with unconjugatable (HA)3–UbΔG75/76 (B) (demonstrating it does not originate from proteolysis of (HA)3-mono-ubiquitinated-Rabex-5); (c) the absence of this band, along with that corresponding to (HA)3-monoubiquitinated-Rabex-5 (∼76 kDa) in immunoprecipitated extracts of cells transfected with Rabex-5 A58D, a mutant that does not undergo monoubiquitination (Mattera et al, 2006; Penengo et al, 2006) (C) and (d) the similar enrichment of both the 76- and 73-kDa species in the cytosolic fraction (A, blot on right). Monoubiquitination of myc–Rabex-5 by endogenous ubiquitin is not inhibited by the co-expression of (HA)3–UbΔG75/G76 Ub (anti-myc blot in B), consistent with the requirement for Ub glycine 76 in the formation of an Ub adenylate during the first step of Ub protein conjugation (Pickart et al, 1994). The lower band above the 50 kDa standard in the anti-myc blot is a myc–Rabex-5 cleavage product, whereas the anti-HA immunoreactivity at ∼50 kDa corresponds to the IgG heavy chain of the mouse anti-myc used for IP.
Both endogenous and recombinant Rabex-5 are monoubiquitinated in HeLa cells (Mattera et al, 2006; Penengo et al, 2006). Transfection of HeLa cells with myc–Rabex-5 and SDS–PAGE of cell lysates or anti-myc immunoprecipitates followed by anti-myc immunoblotting resulted in the detection of myc–Rabex-5 (∼64 kDa) along with a more slowly migrating band at ∼73 kDa (Figure 4A–C, left lanes). A similar analysis of cells co-transfected with myc–Rabex-5 and (HA)3-tagged Ub showed an additional band corresponding to (HA)3-monoubiquitinated-Rabex-5 (∼76 kDa) (Figure 4B and C, left lanes). Both the (HA)3-monoubiquitinated-Rabex-5 (∼76 kDa) and the ∼73 kDa bands are not detected in immunoprecipitates from cells transfected with myc–Rabex-5 A58D (a mutant that does not undergo Ub conjugation) (Figure 4C). On the basis of this observation and on the detection of the ∼73 kDa band in myc–Rabex-5-transfected cells independently of co-transfection with (HA)3–Ub (Figure 4A and B), indicating that it does not represent proteolysis of (HA)3-monoubiquitinated Rabex-5, we conclude that this species corresponds to myc–Rabex-5 conjugated to endogenous Ub. Significantly, the blots with cytosolic and membrane fractions revealed that, whereas most of myc–Rabex-5 WT was membrane bound, monoubiquitinated-myc–Rabex-5 WT (conjugated to either endogenous or (HA)3-tagged Ub) was predominantly cytosolic (Figure 4A, left and right blots). The relative enrichment of monoubiquitinated-Rabex-5 in the cytosol when compared with the unmodified protein indicates that it is the ability to bind Ub, as opposed to its covalent modification with Ub, that drives membrane association of Rabex-5.
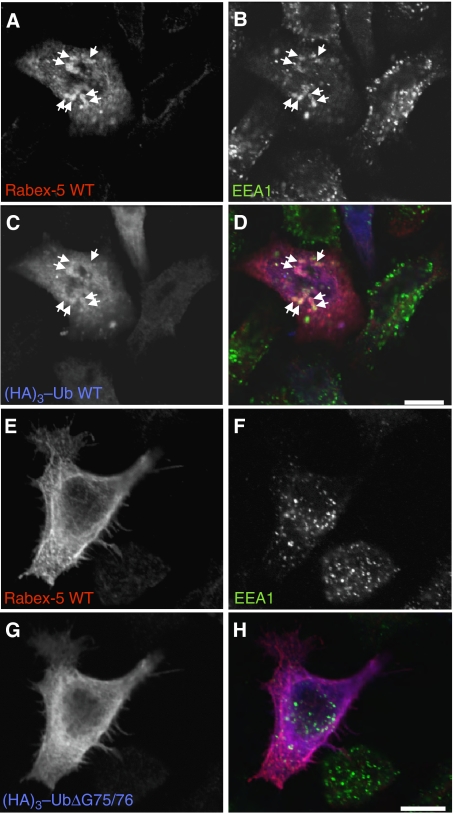
Interestingly, in cells co-transfected with myc–Rabex-5 and (HA)3–Ub, the anti-HA immunoreactivity was localized diffusely as well as to myc–Rabex-5-containing enlarged early endosomes, consistent with the binding of myc–Rabex-5 to ubiquitinated partners on early endosomes (Figure 5A–D). To obtain further evidence for a role of Ub binding in Rabex-5 recruitment to endosomes, we co-expressed myc–Rabex-5 with (HA)3–UbΔG75/G76, a Ub mutant that can bind UBD but cannot undergo covalent conjugation (Stang et al, 2004; Figure 4B). This mutant thus behaves as a competitive inhibitor of Ub binding (Stang et al, 2004). Indeed, we observed that the expression of this Ub mutant decreased localization of myc–Rabex-5 to early endosomes (Figure 5E–H). These results suggest that Rabex-5 is recruited to early endosomes due to its interaction with ubiquitinated partners.
Figure 5.
Colocalization of myc–Rabex-5 and (HA)3–Ub on early endosomes; interference of Rabex-5 recruitment to early endosomes by (HA)3–UbΔG75/76. Cells were co-transfected with myc–Rabex-5 and (HA)3–Ub or (HA)3–UbΔG75/76 and fixed 24 h after transfection. Fixed cells were incubated with chicken anti-myc, mouse anti-EEA1 and rabbit anti-HA, followed by incubation with Alexa 594-conjugated anti-chicken, Alexa 488-conjugated anti-mouse and Alexa 647-conjugated anti-rabbit antisera. Arrows in A–D show colocalization of myc–Rabex-5 with (HA)3–Ub WT on early endosomes. We observed this phenotype in 90 and 28% of cells co-transfected with (HA)3–Ub or (HA)3–UbΔG75/76, respectively (n=104 and 421 cells, respectively, exhibiting medium–high expression levels of myc–Rabex-5). The phenotype in >70% of cells co-transfected with (HA)3–UbΔG75/76 is shown in (E–H). (D, H) Merged images of the preceding panels. Scale bars=10 μm.
The C-terminal CC is also required for the recruitment of Rabex-5 to endosomes
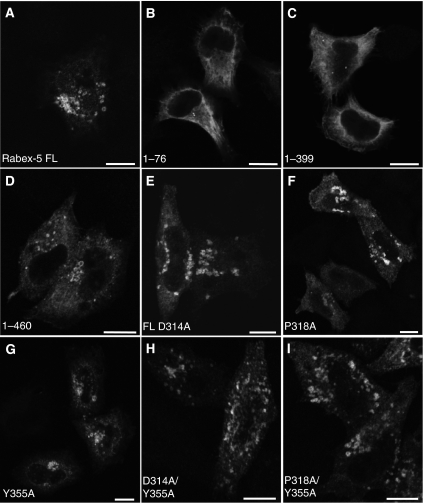
We next asked whether Rabex-5 domains other than the ZnF and MIU are also important for its recruitment to early endosomes. Expression of myc–Rabex-5(1–76) (having only ZnF and MIU domains) in HeLa cells resulted in diffuse cytoplasmic staining along with a few small puncta (Figure 6B) that did not colocalize with endosomal markers (not shown). Given the low expression of this construct (Figure 1B), we examined the distribution of myc–Rabex-5(1–399), which includes the Vps9 domain with its associated HB and the ZnF-MIU tandem, and of myc–Rabex-5(1–460), a construct also including the C-terminal CC that contains the Rabaptin-5-binding site (these two constructs exhibited expression levels similar to the WT; Figure 1B). myc–Rabex-5(1–399) also exhibited a predominantly cytosolic distribution with occasional small puncta (Figure 6C), consistent with the subcellular fractionation of this construct (Figure 4A). In contrast, myc–Rabex-5(1–460) was recruited to early endosomes (Figure 6D). These findings indicate that (a) binding of Rabex-5 to ubiquitinated partners is necessary (Figures 2, 3, 4 and 5) but not sufficient (Figure 6B and C) for recruitment to endosomes; (b) the Rabex-5 HB-Vps9 tandem, comprising the Rabex-5 GEF catalytic core, is also insufficient for this recruitment (Figure 6C) and (c) in addition to the ZnF and MIU domains, the Rabex-5 C-terminal CC is required for endosomal recruitment (Figure 6C and D).
Figure 6.
Roles of the Vps9 domain (residues 230–375), C-terminal coiled-coil (residues 405–456) and GEF activity in the recruitment of Rabex-5 to early endosomes. HeLa cells were transiently transfected with the indicated constructs (A–D) (see Figure 1A for schematic representation of the constructs). Cells were fixed and stained as described in the legend to Figure 2. The substitutions in Vps9 residues (i.e. D314, P318 and Y355; E–I) impair the Rab5/Rab21 GEF activity of Rabex-5 (Delprato et al, 2004). All substitutions in the Vps9 module were introduced in the context of the Rabex-5 FL construct. Scale bars=10 μm.
Rabex-5 GEF activity is dispensable for recruitment to endosomes
The results described in the preceding paragraph indicated that the Rabex-5 GEF domain is not sufficient for Rabex-5 recruitment to early endosomes. To determine whether Rabex-5 GEF activity is necessary, we used GEF-inactive mutants of Rabex-5 generated by alanine substitution of residues D314, P318 and Y355 in the myc–Rabex-5 full-length construct. These residues cluster near the N termini of the αV4 and αV6 helices in the Rabex-5 Vps9 domain, are highly conserved in other Vps9 domains and their substitution by alanine severely impairs Rabex-5 GEF activity towards Rab5 and Rab21 in vitro (Delprato et al, 2004). Interestingly, single alanine substitutions in residues D314, P318 and Y355 in myc–Rabex-5 did not affect its recruitment to early endosomes (Figure 6E–G). Although these single substitutions decrease the Rabex-5 kcat/Km measured in vitro by two orders of magnitude (Delprato et al, 2004), we wondered whether a small residual activity could still drive GEF-dependent changes leading to the recruitment of Rabex-5 to early endosomes. However, combined substitution of D314/Y355 or P318/Y355 was also inconsequential for the recruitment of myc–Rabex-5 to early endosomes (Figure 6H and I), demonstrating that its GEF activity is not required for this function.
Rab5 is not required for the recruitment of Rabex-5 to endosomes
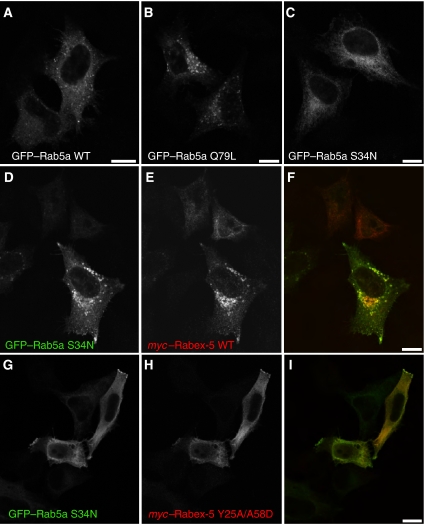
We next analysed the function of Rab5 in Rabex-5 recruitment to early endosomes using two independent approaches. The first consisted of examining the effect of co-transfection with a dominant-negative Rab5 mutant (S34N) on the association of myc–Rabex-5 with early endosomes. Recombinant Rab5a WT, dominant-active ‘GTP-locked' Rab5a Q79L or dominant-negative ‘GDP-locked' Rab5a S34N localize to small vesicles, large vesicles (enlarged early endosomes) or cytosol, respectively (Figure 7A–C). Importantly, co-expression of myc–Rabex-5 and GFP–Rab5a S34N did not prevent the recruitment of myc–Rabex-5 to early endosomes but resulted in stabilization of Rab5a S34N on the Rabex-5-containing early endosomes (Figure 7D–F). This is consistent with a recent report showing colocalization of recombinant Rabex-5 and Rab5 S34N in BHK cells (Zhu et al, 2007). In contrast, co-expression of Ub-binding/conjugation-deficient mutants of Rabex-5, such as Rabex-5 Y25A/A58D, did not affect the cytosolic distribution of GFP–Rab5a S34N (Figure 7G–I). The colocalization of myc–Rabex-5 WT and GFP–Rab5a S34N (Figure 7D–F) is consistent with the high affinity of GEFs for the GDP-bound forms of monomeric GTPases and argues against a requirement for ‘activatable' Rab5 in the recruitment of myc–Rabex-5 to early endosomes. Rather, it is Rabex-5 that recruits GFP–Rab5a S34N to endosomes.
Figure 7.
Co-expression of Rab5a S34N does not inhibit the recruitment of myc–Rabex-5 to early endosomes. (A–C) HeLa cells were transfected with GFP–Rab5a WT, GFP–Rab5a Q79L (constitutively active, ‘GTP-locked') or GFP–Rab5a S34N (dominant negative, displaying higher affinity for GDP than for GTP) (Li and Stahl, 1993; Stenmark et al, 1994). Transfected cells were fixed and incubated with rabbit anti-GFP followed by Alexa 488-conjugated anti-rabbit antiserum. (D–I) HeLa cells were co-transfected with GFP–Rab5a S34N and myc–Rabex-5 WT (D–F) or myc–Rabex-5 Y25A/A58D (G–I), fixed and incubated with rabbit anti-GFP and mouse anti-myc followed by incubation with Alexa 488-conjugated anti-rabbit and Alexa 568-conjugated anti-mouse antisera. (F, I) Merged images of the panels at their left. Scale bars=10 μm.
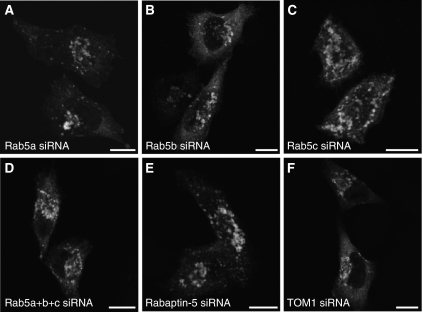
To independently test the requirement for Rab5 in the recruitment of Rabex-5 to early endosomes, we used small interfering RNAs (siRNA) specific for each of the three Rab5 isoforms in human cells (Rab5a, Rab5b and Rab5c) (Supplementary Figure S2). Treatment of HeLa cells for 72 h with each of the specific siRNAs resulted only in the depletion of the targeted Rab5 isoform, whereas combined treatment abrogated the expression of all three isoforms (Supplementary Figure S2). Importantly, either individual removal or combined depletion of all Rab5 isoforms did not significantly affect the recruitment of myc–Rabex-5 to early endosomes (Figure 8A–D). These experiments thus demonstrated that the recruitment of Rabex-5 to early endosomes is independent of Rab5.
Figure 8.
siRNA-mediated depletion of Rab5a, Rab5b, Rab5c or Rabaptin-5 does not affect the recruitment of Rabex-5 to early endosomes. HeLa cells were transfected with the indicated siRNA (A–F). Approximately 36 h after start of treatment, cells were trypsinized, plated on glass coverslips and transfected (∼12 h after plating) with myc–Rabex-5. Cells were fixed ∼24 after transfection with myc–Rabex-5 (72 h cumulative treatment with siRNAs) and stained for myc–Rabex-5 as indicated in the legend to Figure 2. myc–Rabex-5 recruitment to early endosomes was observed in 98, 96, 96, 85, 98 and 97% of cells treated with siRNAs for Rab5a, Rab5b, Rab5c, Rab5a+b+c, Rabaptin-5 or TOM1 (negative control), respectively, (n=221, 354, 334, 155, 302 and 352, respectively; cells with intermediate–high levels of myc–Rabex-5 were counted). A relatively lower transfection efficiency with myc–Rabex-5 (∼50% of the average in the other experimental groups) was observed in cells treated with Rab5a+b+c siRNA. Scale bars=10 μm.
Endosomal recruitment of Rabex-5 is independent of Rabaptin-5 and of Rabex-5 self-association
Given that the Rabaptin-5-binding site lies within the Rabex-5 C-terminal CC, and that this region is also necessary for the recruitment of Rabex-5 to early endosomes (Figure 6C and D), we tested whether Rabaptin-5 is required for recruitment. Interestingly, depletion of Rabaptin-5 (Supplementary Figure S2) did not affect the recruitment of myc–Rabex-5 to early endosomes (Figure 8E and F). CCs are often important for the stabilization of homo- and heterodimers (Burkhard et al, 2001). We thus hypothesized that Rabex-5 might undergo self-association driven by its C-terminal CC, and that this may be required for endosomal recruitment. We tested this hypothesis using the yeast two-hybrid system. The results showed that Rabex-5 indeed undergoes self-association, but this interaction is driven by the ZnF (1–49 fragment) as opposed to the C-terminal CC (400–460 fragment) (Supplementary Figure S3). This indicates that the role of the C-terminal CC in the recruitment of Rabex-5 to early endosomes must be related to interactions with factors other than Rabaptin-5 or Rabex-5 itself.
Discussion
Here, we demonstrate that Ub binding is critical for the recruitment of Rabex-5 to early endosomes, and that this recruitment is independent of Rabex-5 GEF activity and of its partners Rab5 and Rabaptin-5. Moreover, we show that monoubiquitinated Rabex-5 is enriched in the cytosol. On the basis of these findings, we propose that a cycle of Ub binding and monoubiquitination regulates Rabex-5 association with endosomes (Figure 9).
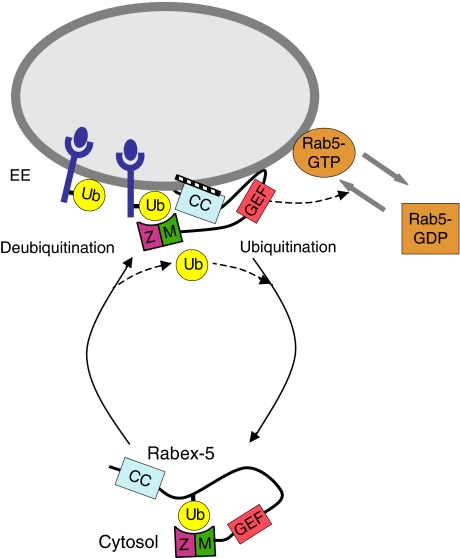
Figure 9.
Proposed model for regulation of Rabex-5 association to early endosomes by Ub binding and monoubiquitination. The scheme depicts the proposed recruitment of Rabex-5 to early endosomes by virtue of its ability to bind ubiquitinated cargo through the ZnF (Z) and MIU (M) domains, and by the interaction of the C-terminal coiled-coil (CC) with as yet unidentified factors. The activation of Rab5 catalysed by the GEF activity of the HB-Vps9 tandem catalytic core (denoted as GEF) is also shown. We hypothesize that cargo conveyance followed by UBD-dependent monoubiquitination results in dissociation of Rabex-5 from endosomes to the cytosol, where the covalently linked Ub moiety in cis (position shown is arbitrary) may bind intramolecularly to the ZnF/MIU domains. Intermolecular interactions in Rabex-5 oligomers where the Ub binds to ZnF/MIU domains in trans are also possible. Deubiquitination of Rabex-5, presumably by endosomal deubiquitinating enzymes, releases this interaction allowing resumption of the cycle. The model is consistent with the enrichment of non-ubiquitinated Rabex-5 and monoubiquitinated Rabex-5 in the membrane and cytosolic fractions, respectively, and is thought to apply to Rabex-5 in dynamic equilibrium with the endosomal compartment.
Ub binding and CC domains are required for the recruitment of Rabex-5 to early endosomes
Although necessary, interaction with Ub is not sufficient for endosomal recruitment of Rabex-5: the C-terminal CC (which includes the Rabaptin-5-binding site) is also required. This requirement, however, is unrelated to its binding to Rabaptin-5.
We hypothesized that the C-terminal CC could mediate self-association of Rabex-5. Although we found that Rabex-5 self-associates, this association depends on an N-terminal fragment including the ZnF domain and not on the C-terminal CC. This is consistent with the function of this type of ZnF in the self-association of the founding member of the family, A20 (Klinkenberg et al, 2001). Our findings indicate that the requirement for the C-terminal CC in endosomal recruitment of Rabex-5 most likely reflects its interaction with as yet unidentified factors other than Rabaptin-5. Of interest, the C-terminal fragment of mouse Rabex-5 (residues 380–491), which includes the Rabaptin-5-binding site, interacts with H-Ras (Tam et al, 2004). This suggests that the Rabex-5 C-terminal CC is a platform for interaction with multiple partners that may be required, in addition to Ub binding, for its stabilization on early endosomes. The importance of Ub binding and the C-terminal CC in the recruitment of Rabex-5 to early endosomes demonstrated here does not exclude that other domains may also contribute to this association (Zhu et al, 2007).
Recruitment of Rabex-5 to early endosomes is independent of its GEF activity and of Rab5
The observed association of Rabex-5 with early endosomes independently of its role as a Rabaptin-5-activatable Rab5 GEF indicates that recruitment of Rabex-5 precedes Rab5 activation. This is consistent with the recruitment of in vitro transcribed/translated free Rabex-5 (i.e. not complexed with Rabaptin-5) to early endosomal membranes stripped of Rab5 by treatment with Rab GDP dissociation inhibitor (Lippé et al, 2001). It is also noteworthy that Rabex-5 GEF activity and Rab5 are also unnecessary for enlargement of early endosomes in HeLa cells transfected with myc–Rabex-5. This suggests that Rabex-5 overexpression may interfere with the sorting of ubiquitinated cargo, leading to compartment expansion or, alternatively, that Rabex-5 may promote endosomal fusion independently of Rab5 and its effectors (e.g. by linking endosomes through Ub binding and CC domains).
Role of Ub binding in the recruitment of trafficking machinery components to endosomes
Ubiquitination of TM proteins at the plasma membrane and TGN constitutes a signal for delivery to the vacuole/lysosome through endosomes and the MVB pathway (Dupré et al, 2004; Hicke et al, 2005; Mukhopadhyay and Riezman, 2007). Ub moieties conjugated to the TM cargoes are recognized by cytosolic/peripheral components of the trafficking machinery that contain UBD and that themselves undergo ubiquitination (Dupré et al, 2004; Hicke et al, 2005; Mukhopadhyay and Riezman, 2007). Examples of such components involved in endocytosis of ubiquitinated epidermal growth factor receptor (EGFR) are the adaptor proteins EGFR substrate (Eps) 15 and 15R, Eps15-interacting proteins (epsins) 1 and 2, and the suppressors of T-cell receptor signalling (Sts) 1 and 2. Subsequent delivery of ubiquitinated TM cargoes to internal vesicles of MVBs is carried out by the ESCRT complexes (Hurley and Emr, 2006; Williams and Urbé, 2007). Some of the subunits of these complexes also contain UBD (e.g. Hrs and STAM in ESCRT-0, TSG101 in ESCRT-I, Eap45 in ESCRT-II) (Hurley and Emr, 2006) and undergo UBD-dependent ubiquitination (e.g. Hrs, STAM, TSG101) (Katz et al, 2002; Polo et al, 2002; Amit et al, 2004; Komada and Kitamura, 2005). Recruitment of these proteins to endocytic structures and endosomes, however, is primarily mediated by binding to membrane phosphoinositides (e.g. phosphatidylinositol 4,5-bisphosphate for epsins (Horvath et al, 2007) and phosphatidylinositol 3-phosphate for Hrs and Eap45 (Hurley and Emr, 2006; Teo et al, 2006)) or to other proteins (e.g. AP-2 and clathrin for epsins (Brett et al, 2002; Horvath et al, 2007), clathrin for Hrs, Hrs for STAM and TSG101), independently of Ub binding/ubiquitination (Dupré et al, 2004; Komada and Kitamura, 2005). Therefore, the ability of these proteins to bind Ub is not thought to be required for recruitment to endosomes but for other functions such as cargo sorting. In contrast, we show here that Ub binding and ubiquitination are critical determinants of Rabex-5 association to and dissociation from membranes. To our knowledge, this represents the first demonstration that Ub binding is essential for the membrane recruitment of a component of the trafficking machinery from the cytosol to endosomes. This conclusion is consistent with the agonist- and time-dependent colocalization of EGFR and Rabex-5 in endosomes, and with the Rabex-5 UBD-dependent co-immunoprecipitation of these proteins (Penengo et al, 2006).
A cycle of Ub binding and monoubiquitination regulates Rabex-5 association with endosomes
On the basis of our findings, we propose that Ub binding and monoubiquitination regulate the association of Rabex-5 with endosomes (Figure 9). According to this model, Rabex-5 is recruited to early endosomes by interaction of its UBD with TM ubiquitinated cargo (or ubiquitinated adaptors bound to this cargo). Conveyance of cargo, followed by UBD-dependent monoubiquitination, results in dissociation of Rabex-5 from membranes and localization to the cytosol in a conformation where its UBD may bind the Ub moiety covalently linked to this molecule. Deubiquitination of Rabex-5, presumably by endosomal deubiquitinating enzymes such as AMSH and UBPY (Clague and Urbé, 2006), may release this intramolecular interaction allowing resumption of the cycle. In this model, Rabex-5 is first recruited to endosomes by virtue of its ability to interact with ubiquitinated proteins and other as yet unidentified factors (i.e. those interacting with the C-terminal CC). It is then that Rabex-5 can activate Rab-5, leading to enhanced homotypic and heterotypic fusion. This integration of cargo recognition with endosome fusion is thus an example of the coordinated nature of these two fundamental processes in intracellular protein trafficking.
Materials and methods
DNA constructs
Constructs based on pCI-neo (Promega, Madison, WI) encoding full-length myc-tagged bovine Rabex-5 and its A58D and Δ13–49 mutants were previously described (Mattera et al, 2006). pCI-neo-myc-Rabex-5(1–399) and (1–460) were generated by introducing stop codons in the full-length construct by site-directed mutagenesis (SDM) (QuikChange™ kit; Stratagene, La Jolla, CA). The same approach was used to generate pCI-neo-myc-Rabex-5(1–76) from the internal deletion construct pCI-neo-myc-Rabex-5(1–76^400–492) (previously generated by SDM of pCI-neo-myc-Rabex-5 full length). The Y25A/Y26A, Y25A/A58D, D314A, P318A, Y355A, D314A/Y355A and P318A/Y355A substitutions were also generated by SDM of the pCI-neo-myc-Rabex-5 full-length template. The pCI-neo-myc-(HA)3-human-UbΔG75–76 was obtained by SDM of the previously described full-length construct (Mattera et al, 2006).
The WT, Q79L and S34N forms of dog-Rab5a were cloned in the KpnI and BamHI sites of pEGFP-C1, whereas the constructs based on the pReceiver-M06 vector for the expression of N-terminal HA-tagged human-Rab5a, -Rab5b and -Rab5c were purchased from GeneCopoeia (Germantown, MD).
The pGAD424 and pGBT9 (Clontech, Mountain View, CA) yeast two-hybrid constructs for the expression of full-length or fragments of bovine Rabex-5 and human Rabaptin-5 were previously described (Mattera et al, 2003, 2006), with the exception of (a) pGBT9-Rabex-5(1–49), obtained by SDM of pGBT9-Rabex-5 and (b) pGBT9-Rabex-5(1–76^400–460), obtained by ligating a PstI fragment excised from the corresponding pGAD424 construct into the large fragment obtained after PstI digestion of pGBT9-Rabex-5.
Antibodies
For immunofluorescence microscopy, we used mouse anti-myc (9E10; Covance Research Products, Berkeley, CA), rabbit anti-myc (gift from RS Hegde, CBMP, NICHD), chicken anti-myc (Molecular Probes, Eugene, OR), mouse anti-EEA1 (BD Biosciences, San Jose, CA), mouse anti-HA (HA.11; Covance Research Products), rabbit anti-HA (Affinity BioReagents, Golden, CO) and rabbit anti-GFP (Molecular Probes). For immunoblotting of lysates or subcellular fractions from cells transfected with pCI-neo-myc-Rabex-5 constructs, we used rabbit anti-myc (Cell Signaling, Danvers, MA), goat anti-GAPDH (Santa Cruz Biotech, Santa Cruz, CA) and a rabbit antiserum against the C terminus of amyloid precursor protein (Zymed-Invitrogen Immunodetection, Carlsbad, CA). Cell lysates from ubiquitination assays were immunoprecipitated with mouse anti-myc (Invitrogen, Carlsbad, CA) and immunoblotted with rabbit anti-myc (Cell Signaling) and mouse anti-HA (Covance Research Products). For immunoblotting of siRNA-treated cells, we used rabbit antisera to Rab5 (pan Rab5), Rab5a, Rab5b and Rab5c from Santa Cruz Biotech. The isoform specificity was confirmed using HeLa cells transfected with HA-tagged constructs of Rab5a, Rab5b and Rab5c (Supplementary Figure S2). In the siRNA experiments, we also used mouse anti-Rabaptin-5 and anti-Rabex-5 (BD Biosciences), rabbit anti-TOM1 against the human TOM1 C-terminal sequence RKKTQEKDDDMLFAL (custom-designed, generated by Covance Research Products) and mouse anti-α-tubulin (Sigma-Aldrich, St Louis, MO).
Cell transfection and immunofluorescence microscopy
HeLa cells (American Type Culture Collection, Manassas, VA) were cultured under a humidified atmosphere (95:5 air/CO2) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (supplemented DMEM). For transfections, we used a ratio of 3 μg of total DNA per well of a six-well plate (with cells at 30–50% confluency plated on 12-mm coverslips using Fugene™ (Roche Applied Science, Indianapolis, IN). Transfected cells were fixed with 4% formaldehyde ∼20–24 h after transfection. Fixed cells were incubated with the indicated primary and secondary antisera and imaged with an Olympus FluoView confocal microscope ( × 60 oil immersion lens).
Subcellular fractionation
Cells cultured on 100-mm dishes (three dishes per construct) were transfected at a ratio of 10 μg of total plasmid DNA per dish at 30–50% confluency using Fugene™ reagent. Approximately 20–24 h after transfection, cells were washed twice with ice-cold phosphate-buffered saline and resuspended in 3 ml of 50 mM HEPES pH 7.4, 5 mM NaCl, 100 mM KCl, 3 mM MgCl2, 1 mM EGTA supplemented with protease inhibitors (EDTA-free, Complete™; Roche Applied Science) (cavitation buffer). Resuspended cells were subjected to nitrogen cavitation for 20 min at 3450 kPa (500 l.b.f./in.2) and 4°C using a cell disruption bomb (Parr Instrument Co., Moline, IL), followed by centrifugation for 10 min at 900 g and 4°C. The postnuclear supernatants were further centrifuged for 30 min at 300 000 g. Both the supernatants (cytosol) and the pellets (membrane fraction, resuspended in 600 μl of cavitation buffer) were stored at −80°C.
Ubiquitination assays
HeLa cells cultured on 100-mm dishes were transfected with Fugene™ reagent using a ratio of 10 μg of total plasmid DNA (7.5 μg of pCI-neo-myc-Rabex-5 and 2.5 μg of pCI-neo-(HA)3-Ub constructs) and incubated in supplemented DMEM. Cells were lysed ∼20–24 h after transfection in 50 mM Tris–HCl pH 7.4, 75 mM NaCl, 0.4% (v/v) Triton X-100 supplemented with the above described protease inhibitors. Lysates were incubated, centrifuged and subjected to immunoprecipitation, SDS–PAGE and immunoblotting as described previously (Mattera et al, 2006).
RNA interference
ON-TARGETplus™ siRNA (pooled sets of four) from Dharmacon (Chicago, IL) was used for RNA interference in HeLa cells against human Rab5a (LQ-004009-00), Rab5b (LQ-004010-00), Rab5c (LQ-004011-00) and Rabaptin-5 (LQ-017645-01). The Rabaptin-5-specific pool does not target Rabaptin-5β, a related protein that shares 42% sequence identity with Rabaptin-5 but is ∼15 times less abundant in HeLa cells (Gournier et al, 1998). The custom-designed siRNA for TOM1, sense sequence r(CCUGUGUCAAGAACUGCGG)dTT, was from Qiagen (Valencia, CA). Cells grown on six-well plates were transfected with 200 nM total siRNA using Oligofectamine™ (Invitrogen). Approximately 38 h after siRNA treatment, cells were split and re-plated on 12-mm coverslips and six-well plates. Cells re-plated on coverslips were transfected after ∼48 h of siRNA treatment with pCI-neo-myc-Rabex-5 and fixed 24 h later with 4% formaldehyde (72 and 24 h of siRNA and plasmid DNA transfection, respectively). Cells re-plated on six-well plates were lysed 72 h after siRNA transfection for immunoblotting purposes.
Supplementary Material
Supplementary Figures S1–S3
Acknowledgments
We thank P Burgos, G Mardones and R Rojas for helpful discussions and advice, and H Tsai for expert technical assistance. This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
References
- Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U, Yarden Y (2004) Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev 18: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10: 797–809 [DOI] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82–88 [DOI] [PubMed] [Google Scholar]
- Chyung JH, Selkoe DJ (2003) Inhibition of receptor-mediated endocytosis demonstrates generation of amyloid beta-protein at the cell surface. J Biol Chem 278: 51035–51043 [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbé S (2006) Endocytosis: the DUB version. Trends Cell Biol 16: 551–559 [DOI] [PubMed] [Google Scholar]
- Delprato A, Lambright DG (2007) Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol 14: 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A, Merithew E, Lambright DG (2004) Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118: 607–617 [DOI] [PubMed] [Google Scholar]
- Dupré S, Urban-Grimal D, Haguenauer-Tsapis R (2004) Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta 1695: 89–111 [DOI] [PubMed] [Google Scholar]
- Gournier H, Stenmark H, Rybin V, Lippé R, Zerial M (1998) Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J 17: 1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I (2005) Ubiquitylation and cell signaling. EMBO J 24: 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dötsch V, Dikic I (2007) E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell 26: 891–898 [DOI] [PubMed] [Google Scholar]
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90: 1149–1159 [DOI] [PubMed] [Google Scholar]
- Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ (2007) Epsin: inducing membrane curvature. Int J Biochem Cell Biol 39: 1765–1770 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35: 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalesnikoff J, Rios EJ, Chen CC, Barbieri MA, Tsai M, Tam SY, Galli SJ (2007) Roles of RabGEF1/Rabex-5 domains in regulating Fc epsilon RI surface expression and Fc epsilon RI-dependent responses in mast cells. Blood 109: 5308–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K, Yarden Y (2002) Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3: 740–751 [DOI] [PubMed] [Google Scholar]
- Klinkenberg M, Van Huffel S, Heyninck K, Beyaert R (2001) Functional redundancy of the zinc fingers of A20 for inhibition of NF-kappaB activation and protein–protein interactions. FEBS Lett 498: 93–97 [DOI] [PubMed] [Google Scholar]
- Komada M, Kitamura N (2005) The Hrs/STAM complex in the downregulation of receptor tyrosine kinases. J Biochem 137: 1–8 [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol 13: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Stahl PD (1993) Structure–function relationship of the small GTPase rab5. J Biol Chem 268: 24475–24480 [PubMed] [Google Scholar]
- Lippé R, Miaczynska M, Rybin V, Runge A, Zerial M (2001) Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell 12: 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS (2003) Divalent interaction of the GGAs with the Rabaptin-5–Rabex-5 complex. EMBO J 22: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Tsai YC, Weissman AM, Bonifacino JS (2006) The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem 281: 6874–6883 [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Zerial M (2002) Mosaic organization of the endocytic pathway. Exp Cell Res 272: 8–14 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Kasperek EM, Beal R, Kim A (1994) Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1). J Biol Chem 269: 7115–7123 [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, Raiborg C, Stenmark H, Madshus IH (2004) Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol Biol Cell 15: 3591–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MP, Dong J, Wandinger-Ness A (2003) Rab proteins and endocytic trafficking: potential targets for therapeutic intervention. Adv Drug Deliv Rev 55: 1421–1437 [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lütcke A, Gruenberg J, Zerial M (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13: 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Vitale G, Ullrich O, Zerial M (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83: 423–432 [DOI] [PubMed] [Google Scholar]
- Tam SY, Tsai M, Snouwaert JN, Kalesnikoff J, Scherrer D, Nakae S, Chatterjea D, Bouley DM, Galli SJ (2004) RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat Immunol 5: 844–852 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2006) ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125: 99–111 [DOI] [PubMed] [Google Scholar]
- Williams RL, Urbé S (2007) The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 5: 355–368 [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhu G, Liu J, Liang Z, Zhang XC, Li G (2007) Rabaptin-5-independent membrane targeting and Rab5 activation by Rabex-5 in the cell. Mol Biol Cell 18: 4119–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S3