Abstract
The LEAFY (LFY) protein is a key regulator of flower development in angiosperms. Its gradually increased expression governs the sharp floral transition, and LFY subsequently controls the patterning of flower meristems by inducing the expression of floral homeotic genes. Despite a wealth of genetic data, how LFY functions at the molecular level is poorly understood. Here, we report crystal structures for the DNA-binding domain of Arabidopsis thaliana LFY bound to two target promoter elements. LFY adopts a novel seven-helix fold that binds DNA as a cooperative dimer, forming base-specific contacts in both the major and minor grooves. Cooperativity is mediated by two basic residues and plausibly accounts for LFY's effectiveness in triggering sharp developmental transitions. Our structure reveals an unexpected similarity between LFY and helix-turn-helix proteins, including homeodomain proteins known to regulate morphogenesis in higher eukaryotes. The appearance of flowering plants has been linked to the molecular evolution of LFY. Our study provides a unique framework to elucidate the molecular mechanisms underlying floral development and the evolutionary history of flowering plants.
Keywords: crystal structure, flower development, homeotic genes, LEAFY, transcriptional regulation
Introduction
Homeotic genes control developmental patterns and organ morphogenesis. In animals, they encode transcription factors of the homeodomain family, such as Hox and paired proteins, which contact DNA through one or several helix-turn-helix (HTH) motifs (Gehring et al, 1994; Underhill, 2000). In plants, most homeotic genes determining the identity of floral organs encode MADS-box transcription factors, suggesting that plants and animals have adopted distinct types of homeotic regulators (Meyerowitz, 1997; Ng and Yanofsky, 2001). In addition to organ identity genes, plants also use another class of regulators named ‘meristem identity genes', which control floral meristem versus shoot/inflorescence fate. In Arabidopsis thaliana, the meristem identity genes LEAFY (LFY) and APETALA1 (AP1) induce flower development, whereas TERMINAL FLOWER1 (TFL1) promotes inflorescence development (Blazquez et al, 2006). Mutations or ectopic expression of these genes result in complete or partial interconversions between flower and inflorescence meristems.
The LFY gene encodes a plant-specific transcription factor, which has a cardinal function in this process, regulating both the transition to flowering and the subsequent patterning of young floral meristems. During the plant vegetative growth, LFY expression increases in newly formed leaves until a certain threshold is reached. LFY then induces the expression of AP1 and CAULIFLOWER (CAL) genes and triggers the abrupt floral transition (Blazquez et al, 2006). Once the floral meristem is established, LFY governs its spatial patterning by inducing the expression of the floral homeotic ABC genes, such as AP1, AP3 or AGAMOUS (AG), which control the identity of stereotypically arranged floral organs (Coen and Meyerowitz, 1991; Lohmann and Weigel, 2002).
LFY is found in all terrestrial plants from moss to angiosperms; its sequence shows a high level of conservation throughout the plant kingdom but no apparent similarity to other proteins (Maizel et al, 2005). Unlike many plant transcription factors that evolved by gene duplication to form a multigene family (Riechmann and Ratcliffe, 2000; Shiu et al, 2005), LFY is present in single copy in most angiosperms and lfy mutants available from several species such as snapdragon, petunia, tomato or maize show, as in Arabidopsis, partial or complete flower-to-shoot conversions (Coen et al, 1990; Souer et al, 1998; Molinero-Rosales et al, 1999; Bomblies et al, 2003). In gymnosperms, a paralogous NEEDLY (NLY) clade of genes exists. No mutant is available in these species, but LFY and NLY expression patterns are also consistent with a role in reproductive organ development (reviewed in Frohlich and Chase, 2007). Because of its central role in determining floral meristem identity, and considering that NLY disappeared concomitantly with the appearance of flowers, LFY has been put at the centre of different evolutionary scenarios that rationalize the appearance of the successful angiosperm group (Albert et al, 2002; Frohlich, 2003; Frohlich and Chase, 2007; Theissen and Melzer, 2007).
LFY activates gene expression by recognizing pseudo-palindromic sequence elements (CCANTGT/G) in the promoters of its target genes, including AP1 (one site) and AG (four sites; AG-I to AG-IV) (Parcy et al, 1998; Busch et al, 1999; Lohmann et al, 2001; Lamb et al, 2002; Hong et al, 2003). LFY has two domains, a partially conserved N-terminal domain that is thought to contribute to transcriptional activation and a highly conserved C-terminal domain responsible for DNA binding (LFY-C) (Coen et al, 1990; Maizel et al, 2005). LFY functions synergistically with coregulators such as the WUSCHEL (WUS) homeodomain protein (Lenhard et al, 2001; Lohmann et al, 2001) or the UFO F-Box protein (Lee et al, 1997; Parcy et al, 1998; Chae et al, 2008).
In this study, we show that LFY binds DNA cooperatively as a dimer, a property shown to be essential to trigger developmental switches. The crystal structure of LFY-C bound to DNA reveals the molecular basis for sequence-specific recognition and cooperative binding as well as an unexpected similarity of LFY with HTH proteins such as homeodomain transcription factors. Our findings enable to formulate new hypotheses on the appearance of angiosperms in evolution.
Results and discussion
LFY-C dimerizes on DNA binding
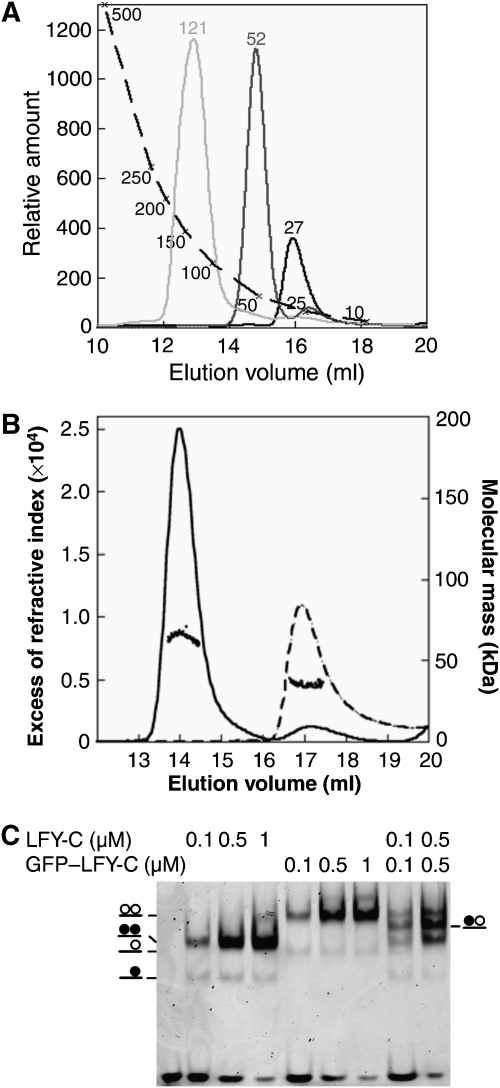
We produced the recombinant LFY DNA-binding domain (LFY-C, residues 223–424) and showed by size-exclusion chromatography (SEC) that it is monomeric in the absence of DNA (Figure 1A and B). In electrophoretic mobility shift assays (EMSAs), LFY-C recognized a DNA probe bearing an AP1 site as two distinct species: a major protein–DNA complex and a minor one of higher mobility (Figure 1C). Multi-angle laser light scattering (MALLS) coupled to SEC demonstrated that the major complex contained two LFY-C molecules per DNA duplex (Figure 1B). The homodimeric nature of LFY in this complex was confirmed by mixing untagged and GFP-tagged LFY-C and observing a single new species attributable to the formation of an LFY-C/GFP–LFY-C/DNA complex (Figure 1C). Using probes mutated in one half-site of the palindrome, we confirmed that the minor, high-mobility species corresponds to a single LFY-C monomer bound to DNA (Supplementary Figure 2).
Figure 1.
DNA-dependent dimerization of LFY-C. (A) Size-exclusion chromatography. LFY-C (40 μM, black curve), AP1 DNA (10 μM, dark grey curve), LFY-C (40 μM)+AP1 DNA (10 μM, light grey curve) were analysed. LFY-C elution at a volume corresponding to 28 kDa is consistent with the monomer size (25.7 kDa), the DNA duplex elutes earlier than expected at a volume corresponding to 52 kDa because of its elongated shape. The LFY/DNA complex elutes at a volume corresponding to 121 kDa. Molecular weights estimated from the calibration curve (dashed line) are indicated. (B) Molecular mass of LFY-C alone (dashed line) or in combination with AP1 DNA (solid line) determined by multi-angle laser light scattering and refractometry combined with size-exclusion chromatography. Elution profiles were monitored by excess refractive index (left ordinate axis). Dots show the molecular mass distribution (right ordinate axis). Average molecular mass is 64±2 kDa for the LFY-C/DNA complex (65 kDa theoretical size for a dimeric complex) and 35±1 kDa for LFY-C alone (26 kDa theoretical size for LFY-C monomer). (C) Electrophoretic mobility shift assay (EMSA) with 10 nM AP1 DNA and various LFY-C or GFP–LFY-C concentrations. Schematic complexes with LFY-C (filled circle) and GFP–LFY-C (open circle) are depicted.
Structure of the LFY DNA-binding domain bound to its DNA recognition site
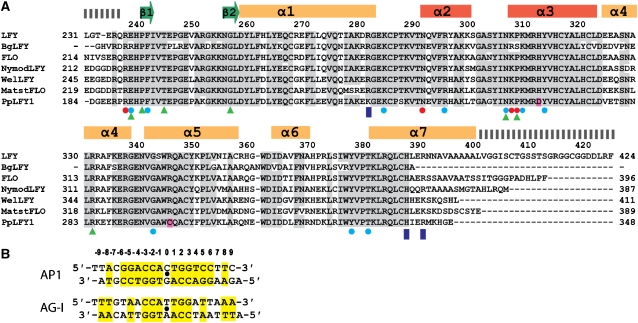
To understand how LFY specifically recognizes its DNA target sequences, we crystallized LFY-C in complex with DNA. We solved the structure of LFY-C bound to two different LFY-binding sites, AP1 and AG-I at 2.1- and 2.3-Å resolution, respectively (Figures 2, and 3A and B; Table I). The overall structure shows an LFY-C dimer bound to a pseudo-palindromic DNA duplex, where the LFY-C monomers are related by a crystallographic dyad. The DNA duplexes used for co-crystallization deviate from strict two-fold symmetry at the 5′ends and at base pairs (bp) ±9, ±7 and ±0 in the AP1 site (5′ end, bp ±7, ±6, ±4 and ±0 in the AG-I site). Nevertheless, the pseudo-dyads of the DNA duplexes coincide with the crystallographic dyad, probably as a result of the random bimodal orientation of the DNA duplex around the dyad (see Materials and methods). The resulting molecular averaging does not impair our interpretation of the protein–DNA interface. In the final 2Fo−Fc electron density map, but also in the initial solvent-flattened single isomorphous replacement with anomalous scattering (SIRAS) electron density map (Supplementary Figure 1), the sugar–phosphate backbone of the DNA is well defined and shows no evidence of conformational averaging. The density for palindromic DNA bases is clearly defined, whereas the density at non-palindromic positions is consistent with the superposition of two different base pairs. Furthermore, all residues close to the DNA are clearly defined and we do not observe any diffuse density, which suggests that each monomer undergoes only minor changes to adapt to the slightly different half-sites. Despite the differences between the AP1 and AG-I binding sites (Figure 2B), both complex structures are very similar and can be superimposed with an r.m.s. distance of 0.55 Å for 163 Cα and 19 phosphate atoms.
Figure 2.
Sequence alignments. (A) Aligned C-terminal amino-acid sequences of LFY (Arabidopsis thaliana, AAA32826), BgLFY (Brownea grandiceps, AAS79888), FLO (Antirrhinum majus, P23915), NymodLFY (Nymphea odorata, AAF77609), WelLFY (Welwitschia mirabilis, AAF23870), MatstLFY (Matteuccia struthiopteris, AAF77608) and PpLFY1 (Physcomitrella patens, BAD91043). Identical and conservatively substituted residues are depicted on a grey background. Secondary structure elements are indicated. Residues involved in interactions with DNA bases and backbone are labelled with red and blue circles, respectively. Dashed bars indicate disordered regions in the crystal, blue rectangles indicate the residues involved in dimerization. Green triangles indicate the position of Arabidopsis mutations and residues divergent in PpLFY1 are highlighted in pink. (B) Two DNA duplexes containing the LEAFY-binding sites from AP1 and AG promoters present in the LEAFY–DNA complex crystals are depicted. Base pairs related by a dyad (indicated by a black dot) are highlighted in yellow.
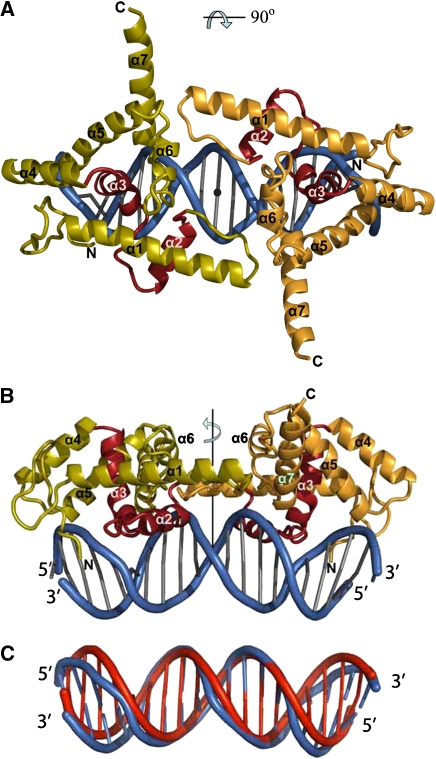
Figure 3.
Structure of the LFY-C dimer bound to DNA. (A, B) Two orthogonal views of the LFY-C dimer (residues 237–399) bound to DNA. Monomers are coloured in olive and orange with the helix-turn-helix (HTH, helices α2 and α3) motif in red. The DNA duplex is depicted in blue. Figures 3, 4B, 5A and 6 were produced with program Pymol (Delano, 2002). (C) Superposition of the DNA duplex found in the LEAFY–DNA complex (blue) with regular B-form DNA (red).
Table 1.
Structure determination of the LEAFY–DNA complex
| Data statistics | |||||
|---|---|---|---|---|---|
| Data set | Resolution (Å)a | Reflections measured/unique | Rmeas (%)b | I/σ | Completeness (%) |
| AP1/LFY: space group P6522, unit cell dimensions a=b=98.8 Å, c=177.4 Å | |||||
| Native | 20–2.1 (2.2–2.1) | 503 443/29 859 | 4.8 (54.3) | 38.5 (5.0) | 97.4 (83.9) |
| EMTS | 20–2.4 (2.5–2.4) | 283 416/20 463 | 11.1 (50.2) | 15.5 (4.7) | 99.6 (99.3) |
| AG-I/LFY: space group P6522, unit cell dimensions a=b=98.4 Å, c=176.4 Å | |||||
| Native | 20–2.3 (2.4–2.3) | 328 932/23 456 | 7.9 (75.1) | 26.7 (3.4) | 99.8 (100.0) |
| Phasing statistics for the EMTS derivative (SIRAS) | |||||
| Wavelength (Å) | 0.934 | ||||
| Phasing powerc | 1.68 | ||||
| Figure of merit | 0.468 | ||||
| Rcullisd | 0.674 | ||||
| Number of mercury sites | 4 | ||||
| Refinement statistics | |||||
| LFY/AP1 | LFY/AG-I | ||||
| Resolutiona | 19.1–2.1 (2.2–2.1) | 20.0–2.3 (2.4–2.3) | |||
| Total number of non-hydrogen protein atoms | 1352 | 1332 | |||
| Total number of non-hydrogen DNA atoms | 526 | 567 | |||
| Number of water molecules | 148 | 105 | |||
| R-factor (%)a | 21.0 (25.0) for 26 896 reflections | 22.1 (29.0) for 22 559 reflections | |||
| Rfree (%)a,e | 23.7 (26.4) for 1521 reflections | 24.9 (30.7) for 1183 reflections | |||
| r.m.s. deviations | |||||
| Bond lengths (Å) | 0.009 | 0.008 | |||
| Bond angles (deg) | 1.22 | 1.21 | |||
| Average temperature factors (Å2) | |||||
| Protein | 41.1 | 39.8 | |||
| DNA | 48.1 | 46.4 | |||
| Solvent | 42.8 | 48.5 | |||
| r.m.s.d. of covalently linked atoms (Å2) | 2.55 | 3.45 | |||
| Residues in Ramachandran plotf | |||||
| Most favoured regions (%) | 93.2 (136) | 95.2 (139) | |||
| Additionally allowed regions (%) | 6.8 (10) | 4.8 (7) | |||
| Generously allowed regions (%) | 0 (0) | 0 (0) | |||
| Disallowed region (%) | 0 (0) | 0 (0) | |||
| aValues for the highest resolution range are given in parenthesis. | |||||
| bRmeas is a redundancy independent R-factor as defined in Diederichs and Karplus (1997). | |||||
| cPhasing power is the mean value of the heavy-atom structure factor amplitudes divided by the mean lack of closure. | |||||
| dRcullis is the mean lack of closure divided by the mean isomorphous difference. | |||||
| eRfree was calculated from a subset of 5% of the data. | |||||
| fNumber of residues are given in parentheses. | |||||
LFY-C (with residues 237–399 ordered in the crystal structure) adopts a compact fold that interacts principally with a single DNA half-site (Figures 3A and B, and 4A and B). The fold is defined by two short β-strands followed by seven helices connected by short loops (Figure 3A and B). The absence of any extended hydrophobic patches at its surface suggests that LFY-C represents an autonomous DNA-binding domain without a large interface to its N-terminal domain. Helices α2 and α3 define a HTH motif (Aravind et al, 2005), with helix α3 occupying the major groove and mediating most of the DNA contacts. The DNA in the complex adopts a B-DNA-like conformation exhibiting an overall bend of about 20° (Figure 3C), which can be localized to two kinks of about 10° at base pairs ±2/±3. Both ends of the DNA duplex are AT rich and the minor grooves are narrower compared with classical B-DNA. Narrowing of the minor groove is slightly more pronounced in the AG-I duplex than in AP1.
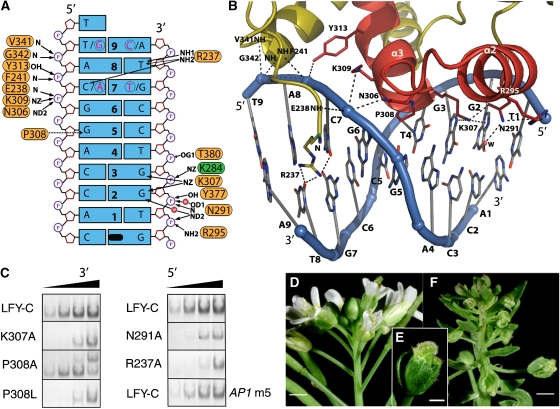
Figure 4.
DNA recognition by LEAFY. (A) Protein–DNA interactions in one AP1 half-site. Dyad-related base pairs 7 and 9 from the other half-site are shown in pink and encircled. Polar and hydrophobic interactions are shown with solid and dashed arrows, respectively. K284 belongs to the other monomer and is depicted in green. The pseudo-dyad coinciding with the crystallographic dyad is depicted in black. (B) Ribbon diagram of one LEAFY monomer bound to its AP1 half-site. The protein is coloured in olive except for the HTH motif shown in red. Polar interactions are indicated by dashed lines. For clarity, only side chains in contact with DNA are shown. (C) Effect of selected mutations on LFY-C DNA-binding affinity to AP1 DNA. EMSAs were performed with wild-type and mutant LFY-C (100–250–750–2000 nM from left to right). Only dimeric complexes are shown except for P308A that gave rise to an unknown higher complex. AP1 m5 mutant DNA contains base pair C:G instead of A:T at position±8 (see Supplementary Table 3 for full DNA sequences). Phenotype of the wild-type Arabidopsis inflorescence (D) and lfy-28 (P308L) mutant inflorescence (F) and flower (E). Scale bar is 1 mm on (D, F) and 0.5 mm on (E).
DNA recognition in the major and minor grooves
Sequence-specific contacts between LFY and the DNA involve both the minor and major grooves. Base-specific contacts in the major groove are formed by Asn291 and Lys307 in helices α2 and α3, which together specify the two invariant guanines at positions ±2 and ±3 (Figure 4A and B). Mutating either of these residues into alanine resulted in considerably lower DNA-binding affinity (Figure 4C), whereas previous studies showed loss of binding when the corresponding base pairs were mutated (Parcy et al, 1998; Busch et al, 1999). The Arabidopsis lfy-20 mutation (N306D) adjacent to Lys307 also leads to a reduced DNA-binding affinity (Supplementary Figure 3) and a weak lfy phenotype in planta (Weigel et al, 1992; Maizel et al, 2005), presumably because the negatively charged aspartate interacts unfavourably with the DNA backbone (Figure 4A and B).
Base-specific recognition in the minor groove is mediated by Arg237, which is the first ordered N-terminal residue in the crystal structure. At the AP1 site, its side chain points towards A:T base pairs ±8 and contacts the exocyclic O2 of thymine 8 and also the O2 of cytosine 7 in one half-site, or the O2 of cytosine-9 in the other half-site (Figure 4A and B; Supplementary Figure 4). In the AG-I site, T:A base pair 8 is replaced by A:T, and in the LFY/AG-I complex, the Arg237 side chain adopts a different conformation, which allows it to recognize the thymine of the opposite strand (Supplementary Figure 4). The importance of this interaction is underscored by the presence of A:T or T:A base pairs at position 8 in all 12 confirmed half-sites (Parcy et al, 1998; Busch et al, 1999; Lohmann et al, 2001; Lamb et al, 2002; Hong et al, 2003). The consensus LFY-binding site is therefore more accurately defined as T/ANNNNCCANTGT/GNNNNT/A (with the centre of the pseudo-palindrome underlined). The Arg237 side chain is inserted into an AT-rich narrow minor groove (Figure 4A and B), similar to that observed in the Hox homeodomain–Exd–DNA complex, where the narrow minor grove was shown to enhance the electrostatic interaction between DNA backbone and arginine side chain (Joshi et al, 2007). The R237A mutation led to a strongly reduced affinity of LFY-C for AP1 (Figure 4C). In contrast, changing the adenine 8 into a cytosine in AP1 reduced only moderately the LFY-C-binding affinity (Figure 4C; AP1 m5), presumably because the arginine side chain can contact the adjacent base (Supplementary Figure 4). Finally, next to Arg237, the two lfy mutations (lfy-4 (E238K) and lfy-5 (P240L)) result in decreased in vitro binding affinities (Supplementary Figure 3) and lead to a mutant phenotype in planta (Weigel et al, 1992).
An unusual contact with DNA is mediated by Pro308 that points between the guanines in base pairs ±5 and ±6, which results in a pronounced propeller twist for base pair ±5 and local bending of the DNA at this position (Figure 4B). The mutant lfy-28 (P308L) is impaired in DNA binding and gives rise to an intermediate to strong phenotype in planta (Figure 4C–F), as a likely consequence of a steric clash of the leucine side chain with the guanine bases. In contrast, a small side chain such as alanine perfectly fits in this protein–DNA interface and, indeed, the mutant protein P308A showed a wild-type-binding affinity (Figure 4C). Pro308 is not strictly conserved and is substituted by serine in some Brownea species (Figure 2A). This substitution probably modifies DNA binding, because serine can form direct hydrogen bonds to DNA bases at positions ±4 and +5 and it replaces P308, which locally distorts DNA. However, the conformational flexibility of serine and its ability to function as hydrogen bond donor or acceptor makes it difficult to predict the preferred binding specificity. Moreover, P308S is systematically associated with the K307R substitution, affecting the base-contacting residue Lys307 (Figure 2A). LFY proteins from Brownea species might therefore recognize significantly different DNA target sites.
Similar to most protein–DNA co-crystal structures, not all bases in the consensus LFY site T/ANNNNCCANTGT/G NNNNT/A are specified through direct interactions with the protein. Additional specificity presumably arises from sequence-dependent deformability of the DNA, sometimes referred to as ‘indirect readout'. Dinucleotide steps CA/TG at bp±1/bp±2 are part of the consensus LFY site and are particularly flexible, which might facilitate the observed kink of the DNA at base pairs ±2/±3. However, these particular sequences are not critically required as they are not conserved in the AP3-I binding site (Lamb et al, 2002).
Not all LFY mutations directly affect DNA contacts. Mutations lfy-3 (T244M) and lfy-9 (R331K) (Weigel et al, 1992) disturb two interacting amino acids, both of which contribute to a polar network that connects N-terminal residues with helices α1 and α4. Similarly, two other residues (His312 and Arg345) interact in a typical planar stacking. His312 and Arg345 are conserved except in the LFY protein from Physcomitrella patens (PpLFY1) where they are substituted by aspartate and cysteine, respectively (Maizel et al, 2005). His312 forms part of helix α3 and is located just one helical turn above Pro308 at the N-terminal end of helix α3. In addition, the preceding residue Lys307 directly contacts the guanine in base pairs ±2 (Figure 4A). The loss of the His312/Arg345 stacking interaction in the moss PpLFY1 likely affects the orientation of helix α3, explaining the altered DNA-binding properties of this orthologue, whereas reverting the aspartate into histidine restores the binding activity of PpLFY1 to canonical LFY-binding sites (Maizel et al, 2005).
Structural basis for cooperative DNA binding
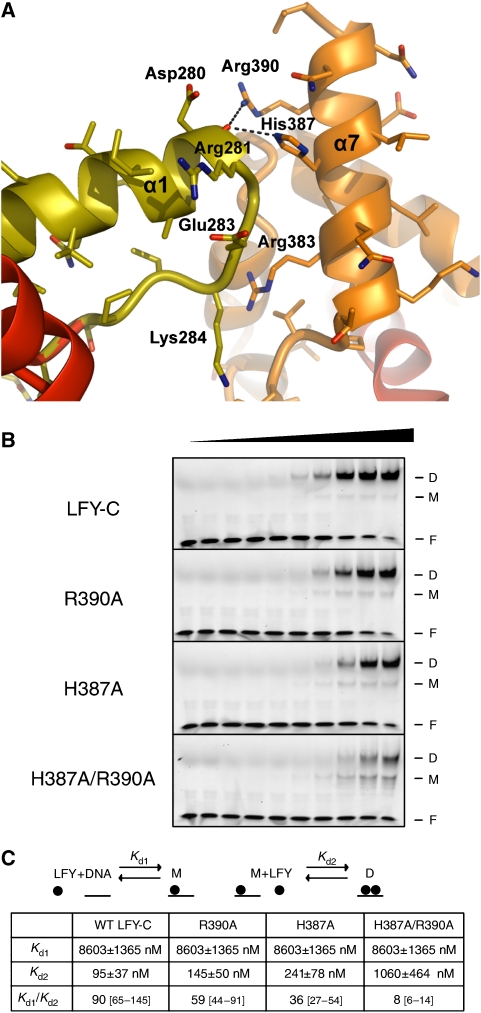
The structure of the LFY-C/DNA complex also reveals important monomer–monomer interactions governing its DNA-binding mode (Figure 5A). Our EMSA analysis shows that LFY-C binds DNA in a cooperative manner: the monomeric complex is present only in minor amounts as compared with the dimeric complex, even at low LFY-C concentrations (Figure 5B) and binding of the second monomer occurs with a 90-fold higher affinity than binding of the first one (Figure 5C; Supplementary Figure 5). This type of cooperative binding can result either from DNA conformability, where binding of one monomer favours the binding of the second monomer, or from protein–protein interactions between DNA-bound monomers (Senear et al, 1998; Schumacher et al, 2002; Panne et al, 2004). Our structure suggests the latter. The LFY dimer comprises a small interface of 420 Å2 buried surface area formed by loop α12 and helix α7 in which the two residues His387 and Arg390 form hydrogen bonds with the backbone carbonyl of Asp280 (Figure 5A). We validated the importance of these contacts by mutagenesis: cooperativity of binding is moderately affected in the H387A or R390A single mutants but more strongly reduced in a H387A/R390A double mutant (Figure 5B and C; Supplementary Figure 5). Therefore, the small monomer–monomer interface (with a major contribution of His387 and Arg390) rather than DNA conformability is responsible for the cooperative binding. Whether the N-terminal domain of LFY also participates in dimerization, in the presence or absence of DNA, will require additional experiments.
Figure 5.
The LFY-C dimer interface mediates cooperative binding. (A) The dimer interface is viewed perpendicular to the DNA axis. Polar contacts between the two monomers (in orange and olive) are shown with dashed lines. (B) EMSA with increasing concentrations (0, 10, 20, 50, 100, 200, 500, 1000, 2000 and 3000 nM from left to right) of LFY-C wild-type, R390A mutant, H387A mutant, and H387A/R390A double mutant and 50 μM AP1 DNA. Free DNA (F), monomeric (M) and dimeric (D) complexes are indicated. (C) Estimation of dissociation constants for wild-type LFY-C and three mutant versions (H387A, R390A and H387A/R390A). Binding of LFY-C to AP1 DNA was modelled as two equilibrium reactions as detailed in Supplementary data: (1) Binding of a first LFY-C monomer to AP1 DNA, leading to the formation of the monomeric complex (M) and characterized by the Kd1 dissociation constant; (2) binding of a second LFY-C monomer to M, leading to the formation of the dimeric complex (D) and characterized by Kd2. EMSA signals from (B) were quantified and the corresponding experimental values were fitted with theoretical equations describing the two equilibria. The errors and intervals between square brackets indicated correspond to the 95% confidence interval. An elevated Kd1/Kd2 ratio reflects a high level of cooperativity, whereas a ratio of 1 would indicate an absence of binding cooperativity. The single mutations resulted in a weak decrease of cooperativity, whereas the H387A/R390A double mutation strongly decreased the cooperativity.
A better understanding of LFY's DNA-binding mode also provides insight into its molecular switch function. DNA-binding cooperativity, as well as dimerization, allows transcription factors to work at lower concentrations and to enhance the sigmoidality of their response curves. When combined with feedback loops, it has been shown essential for threshold-dependent genetic switches (Burz et al, 1998; Cherry and Adler, 2000). LFY is involved in a positive autoregulation loop through activation of the homologous AP1 and CAL genes, that in turn activate LFY expression (Bowman et al, 1993; Liljegren et al, 1999). LFY-binding cooperativity combined with the AP1/CAL feedback loop therefore provides a plausible explanation for the threshold-dependent floral switch triggered by LFY.
Many transcription factors bind DNA as homodimers but also form heterodimers, thereby extending their spectrum of recognized DNA target sequences (Klemm et al, 1998; Garvie and Wolberger, 2001). LFY has been shown to activate the AG organ identity gene synergistically with the homeodomain protein WUS (Lohmann et al, 2001). As adjacent WUS- and LFY-binding sites are present on the AG regulatory sequence, it has been suggested that LFY and WUS could bind simultaneously (Lohmann et al, 2001; Hong et al, 2003). Preliminary model building indicates that LFY homodimers cannot be accommodated with WUS at adjacent LFY- and WUS-binding sites. This observation raises the intriguing possibility that they might either compete for the same binding sites or more likely could form LFY–WUS heterodimers.
LFY shows similarities with HTH proteins
The nature and origin of LFY had so far remained elusive: LFY-C's primary sequence shows unusually strong sequence conservation within its family but has no apparent similarity to any described transcription factor. The crystal structure of LFY-C bound to DNA reveals a seven-helix domain with many residues involved in protein–DNA interactions, tightly constrained packing interactions in the hydrophobic core and protein–protein interactions with the other monomer. Presumably, these observed tight structural and functional constraints on many residues spread over the entire DNA-binding domain explain the high level of sequence conservation within LFY-C.
The LFY-C structure contains an unpredicted HTH motif formed by helices α2 and α3 as part of the overall fold. HTH motifs are present in a wide variety of DNA-binding proteins throughout the three kingdoms of life. They are typically found in a bundle of 3–6 α-helices or combined with β-sheets (winged HTH/fork head domain), which provide a stabilizing hydrophobic core (Weigel and Jackle, 1990; Aravind et al, 2005). Comparison of LFY-C against the Protein Data Bank using program DALI (Holm and Sander, 1993) detects similarity of relative short α-helical segments (∼60 amino-acid residues) with different α-helical proteins including HTH proteins (maximal Dali Z-score 3.0, pairs with Z<2.0 are structurally dissimilar). A search comprising only the first three N-terminal helices, including the HTH motif, mainly showed similarity to different HTH proteins with slightly higher scores (maximal Dali Z-score: 4.5). When considering just the three helices α1, α2 and α3, LFY aligns well with other three-helix bundle HTH proteins, including the homeodomain protein engrailed (r.m.s.d.40Cα=2.9 Å), the paired domain (r.m.s.d.44Cα=3.5 Å) and the Tc3 transposase (r.m.s.d.30Cα=2.4 Å). LFY and partitioning protein KorB (r.m.s.d.71Cα=3.7 Å) share some similarity beyond the typical DNA/RNA-binding three-helical bundle core (Russell and Barton, 1992; Khare et al, 2004), where five of the seven LFY-C helices, including the HTH motif, roughly superimpose with KorB helices. However, LFY cannot be easily assigned to any of the described classes of HTH proteins (Aravind et al, 2005) and it therefore represents a new variant of multi-helical bundle proteins.
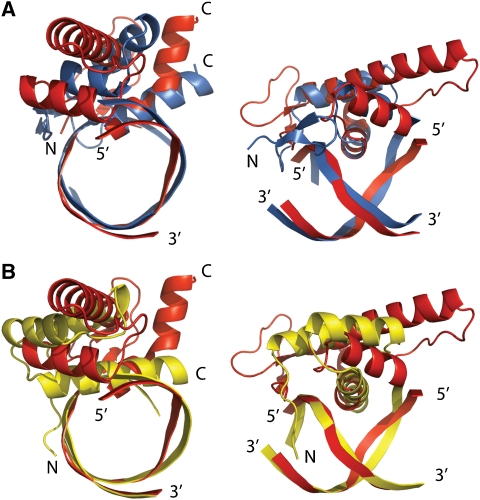
The DNA recognition mode of LFY is similar to those observed for the paired domain, Tc3A transposase, Hin recombinase and λ repressor (van Pouderoyen et al, 1997; Xu et al, 1999). The axis of the recognition helix α3 in the HTH of these proteins is oriented parallel to the edges of the nucleotide bases. Only the N terminus of the recognition helix is inserted into the major groove of the DNA, whereas the short helix α2 has a supporting function. In contrast, in homeodomain proteins, the long probe helix α3 runs more parallel to the neighbouring DNA phosphate backbone, and mainly the central part of helix α3 contacts the DNA (Figure 6). Similarity between LFY and the paired domain also includes a small two-stranded β-sheet, which precedes the three-helix bundle, and N-terminal residues, which are inserted into the minor groove. However, the minor groove contacting residues are located at the most N-terminal end of LFY-C, whereas in the paired domain they protrude from the loop connecting the two short N-terminal β strands.
Figure 6.
Comparison of LFY-C with paired and homeodomain DNA binding. (A) Two orthogonal views of LFY-C helices α1–α3 bound to their DNA target site (red) superimposed with the three-helical bundle core of the N-terminal subdomain of the paired domain of Drosophila Prd (blue, PDB-id: 1pdn). (B) Superposition with the homeodomain of Drosophila engrailed bound to DNA (yellow, PDB-id: 1hdd), where the centre of recognition helix α3 inserts into the major groove.
Sequence similarities are too weak to suggest a precise evolutionary origin for LFY, although structural resemblances indicate that it might derive from ancestral HTH proteins, including paired and homeodomain proteins (Rosinski and Atchley, 1999; Breitling and Gerber, 2000; Aravind et al, 2005). Until now, most plant homeotic genes were found to encode MADS box transcription factors, whereas plant homeodomain proteins rather control meristem homoeostasis and cell division (Meyerowitz, 1997; Ng and Yanofsky, 2001). Our study reveals that the LFY master regulator, which determines flower meristem fate and controls the expression of floral organ identity genes, shares structural similarity with other HTH proteins, indicating that this universal DNA-binding motif has also been adopted in plants to trigger major developmental switches.
Prospects regarding the appearance of angiosperms
The LFY-C structure combined with more than 200 LFY sequences from all types of terrestrial plants offers a unique opportunity to detect key residues in evolution. Some charged LFY-C surface residues (such as Lys253 or Lys254) are strictly conserved, suggesting that they might participate in interactions with other proteins. Other residues are conserved in all angiosperms but not in the non-flowering plants. For example, R390, identified as one of the residues mediating interaction between monomers and cooperative binding, has been conserved in angiosperm LFY proteins, whereas most LFY from non-flowering plants, such as gymnosperms and ferns, show a lysine at this position. This amino-acid change presumably weakens the interaction between monomers and thereby reduces the DNA-binding affinity. The acquisition of R390 might therefore have been important for flower evolution. Because LFY stands at the very centre of the network regulating flower development, it has been proposed that modifications of the LFY gene contributed to the appearance of floral structures in evolution (Albert et al, 2002; Frohlich, 2003; Frohlich and Chase, 2007; Theissen and Melzer, 2007). The availability of the LFY-C crystal structure provides a unique framework for generating plausible hypotheses that relate the appearance of angiosperms to specific events during the molecular evolution of LFY. The ‘functional synthesis' approach that combines phylogeny, biochemical and structural analyses with functional assays in vivo (Dean and Thornton, 2007) can now be applied to LFY to try to solve one of the most puzzling enigmas of plant biology: the origin of flowers.
Materials and methods
Plant material
The lfy-28 mutant allele of A. thaliana (accession Landsberg erecta) was kindly provided by D Weigel (Max Planck Institute, Tübingen, Germany) and originally isolated by J Fletcher (PGEC, Albany). lfy-28 mutant had been back-crossed twice with the wild type, and individuals showing a mutant phenotype were selected from segregating populations. Plants were grown at 25°C in long days (16 h light).
Plasmid constructions
Expression plasmids. LFY-C (residues 223–424 from A. thaliana LFY cDNA) was amplified from pIL-8 (obtained from D Weigel) with Pfu Turbo Polymerase (Stratagene, France) and primers oFP1242 (5′CTCTCGAGCCCGGGCTAGAAACGCAAGTCGTCGCC3′) and oFP1244 (5′CTCTCGAGCCCGGGCTATCCGGTACAGCTAATACCGCC3′), subcloned into pCR-TOPO-BluntII (Invitrogen, Cergy Pontoise, France) and shuttled to pETM-11 (Dummler et al, 2005) as NcoI/XhoI fragment to yield the pCH28 expression vector. pETM-11 contains an N-terminal 6 × His tag followed by a tobacco etch virus (TEV) cleavage site.
LFY–GFP plasmid. A GFP fragment was amplified from pBS-GLFY plasmid obtained from X Wu (Wu et al, 2003) using primers oETH1001 5′CCCACTACTGAGAATCTTTATTTTCAGGGCCAGTT CAGTAAAGGAGAAGAAC3′ and oETH1002 5′CCCCAAACCACTACCTCCGTTGCCGTTATCCTGTT TGTATAGTTCATCCAT3′. The amplified fragment was subsequently used as a megaprimer to amplify plasmid pCH28 and yield pETH8 (6His–TEV–GFP–LFY-C).
Expression plasmids for mutant LFY-C. pCH45 (K307A), pCH46 (N291A), pCH47 (R237A), pCH48 (P308A), pCH49 (D280K), pCH50 (H387A/R390A), pCH54 (H387A), pEDW127 (R390A), pCH55 (lfy-28, P308L), pETH21 (lfy-4, E238K), pETH23 (lfy-20, N306D) and pCH56 (lfy-5, P240L) were derived from pCH28 using the megaprimer strategy with appropriate primers (Kirsch and Joly, 1998). All plasmids were verified by sequencing.
Protein expression, purification and crystallization
Wild-type and mutant LFY-C domains were expressed using Escherichia coli strain RosettaBlue(DE3)pLysS (Novagen, Strasbourg, France). After induction by 0.5 mM IPTG, cells were grown overnight at 22°C. For cell lysis, the pellet of 1 l culture was sonicated in 30 ml lysis buffer A (500 mM NaCl, 20 mM Tris–HCl pH 8, 5 mM imidazole, 5% glycerol, 5 mM Tris(2-carboxyethyl)phosphine hydrochloride), one protease inhibitor cocktail tablet Complete EDTA-free (Roche, Meylan, France) and centrifuged for 40 min at 30 000 g. The clear supernatant was incubated for about 1 h with 1 ml Ni-NTA resin (Qiagen, Courtaboeuf, France). The resin was transferred into a column, washed with 20 column volumes (CVs) of buffer A, buffer A+50 mM imidazole (10 CV) and eluted with buffer A+380 mM imidazole. The fractions containing the protein were pooled and applied to a Hi-load Superdex-200 16/60 prep grade column (GE Healthcare, Orsay, France) equilibrated with 200 mM NaCl, 20 mM Tris–HCl pH 8, 5 mM dithiothreitol (DTT) to eliminate aggregated proteins by SEC. Protein concentration was estimated using the Bradford assay (Bradford, 1976).
For crystallographic experiments, after elution on the metal-affinity column, the histidine tag was cleaved at 4°C overnight with TEV protease (0.01% w/w, 16 h, 4°C) during the dialysis step against buffer B (500 mM NaCl, 20 mM Tris pH 7.5, 5 mM DTT). The TEV protease, the histidine tag and the uncleaved protein were removed by repassing the dialysed sample over the Ni-affinity column. The protein was separated from the remaining DNA contamination using the anion-exchange column MonoQ HR10/10 (GE Healthcare) pre-equilibrated in buffer B. Pure protein was recovered in the flow-through, whereas DNA remained bound to the resin. Aggregated protein was removed by SEC with Superdex S75GL column (GE Healthcare) in 200 mM NaCl, 10 mM Tris pH 7.5 and 5 mM DTT. The protein concentration was adjusted to 7.5 mg/ml. DNA oligonucleotides were chemically synthesized and purified by anion-exchange chromatography following established procedures (Cramer and Muller, 1997).
EMSAs
Single-stranded oligonucleotides, 5′-labelled with tetra-methylcarboxy-rhodamine (Sigma, Saint Quentin Fallavier, France), were annealed to non-fluorescent complementary oligonucleotides in annealing buffer (10 mM Tris pH 7.5, 150 mM NaCl and 1 mM EDTA). The sequences of oligonucleotides used are indicated in Supplementary Table 1. Binding reactions were performed in 20 μl binding buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.5, 1% glycerol, 0.25 mM EDTA, 2 mM MgCl2 and 1 mM DTT) supplemented with 28 ng/μl fish sperm DNA (Roche) and 10 nM double-stranded DNA probe or 140 ng/μl fish sperm DNA for 50 nM DNA probe (Figure 5). Binding reactions were loaded onto native 6% polyacrylamide gels 0.5 × TBE (45 mM Tris, 45 mM boric acid and 1 mM EDTA pH 8) and electrophoresed at 90 V for 80 min at 4°C. Gels were scanned on a Typhoon 9400 scanner (Molecular Dynamics, Sunnyvale, CA; excitation light 532 nm, emission filter 580 BP 30) and signals were quantified using ImageQuant software (Molecular Dynamics). Estimations of Kd1 and Kd2 (Figure 5; Supplementary Figure 5) were based on the quantifications of binding experiments shown in Figure 5B. The binding model equations used to calculate these Kd values are explained in detail in Supplementary data.
SEC
The molecular size of LFY-C/AP1 complex was determined using a Superdex-200 10/300GL column (GE Healthcare), equilibrated with buffer containing 150 mM NaCl, 16 mM Tris–HCl pH 7.5, 0.6 mM EDTA and 1 mM DTT, and calibrated with low and high molecular weight protein standards (gel filtration calibration kit; GE Healthcare). Our samples (LFY-C 40 μM, AP1 WT 10 μM and LFY-C 40 μM+AP1 WT 10 μM) were analysed in the same buffer as protein standards, and molecular size is deduced from the standard curve.
Analytical SEC and MALLS-SEC
Separation by SEC was carried out with a S200 Superdex column (GE Healthcare). The column was equilibrated in 20 mM Tris–HCl, 150 mM NaCl buffer at pH 7.5. Separations were performed at 20°C with a flow rate of 0.6 ml min−1. Protein solution (50 μl) at a concentration of 5 mg ml−1 was injected. The elution was monitored by using a DAWN-EOS detector with a laser emitting at 690 nm for online MALLS measurement (Wyatt Technology Corp., Santa Barbara, CA), and with a RI2000 detector for online refractive index measurements (Schambeck SFD). Molecular mass calculation was performed as described using the ASTRA software (Gerard et al, 2007).
Crystallization
For co-crystallization using the hanging drop method, protein and DNA duplexes were mixed in a molar ratio of 2:1. The best crystals were obtained at 4°C with 20-mer oligonucleotides bearing complementary A:T overhangs and with 10% PEG 400, 100 mM KCl, 10 mM CaCl2, 50 mM HEPES (NaOH) pH 7.0 as reservoir solution. Single crystals grew to a maximal size of 300 × 300 × 500 μm3 and were stepwise transferred to reservoir solution containing 30% (v/v) glycerol for cryo-protection. For preparation of the mercury derivative, the crystals were soaked in the reservoir solution supplemented with 0.1 mM ethylmercury thiosalicylate (EMTS) for 2 h.
X-ray structure determination
The crystals of the LFY-C/AP1/DNA complex belong to space group P6522 (a=b=98.8 Å, c=177.4 Å), diffracted up to 2.1 Å resolution and contain half a complex per asymmetric unit. Crystals of the LFY-C/AG-I/DNA complex are isomorphous but diffracted slightly weaker (Table I). Diffraction data collected at ESRF beamlines ID14-1, ID29 and ID23-2 were processed using program XDS (Kabsch, 1993). The structure of the LFY-C/AP1/DNA complex was solved using the SIRAS method with EMTS as derivative. The quality of native and derivative data sets is summarized in Table I. Mercury sites were located using program SOLVE (Terwilliger and Berendzen, 1999) and phases were calculated with program SHARP (de la Fortelle and Bricogne, 1997). The experimental electron density map (Supplementary Figure 1) allowed us to automatically build the initial model using program ARP/wARP (Perrakis et al, 2001) followed by manually adjusting some side chain conformations with program COOT (Emsley and Cowtan, 2004) and refinement with program Refmac5 including a TLS refinement with seven groups (Murshudov et al, 1997) and later with program Phenix (Adams et al, 2002). In space group P6522, the two monomers bound to the pseudo-palindromic DNA duplex are related by a crystallographic dyad. In the crystal, the pseudo-dyad of the DNA coincides with the crystallographic dyad, although the DNA duplexes deviate from strict two-fold symmetry at base pairs 0, ±7, ±9 and the overhanging 5′-end, in the AP1 site and at base pairs 0, ±4, ±6 and ±7 and the overhanging 5′-end in the AG-I site. To confirm our space group assignment and the underlying assumption that the DNA duplexes used for co-crystallization are randomly distributed in two orientations, the data were reprocessed in the lower symmetry space group P65 lacking the dyad, which did not significantly change the Rmeas values. Subsequently, models of the LFY-C dimer bound to the 20-mer DNA duplex were built in space group P65 for the AP1 and AG-I sites and refined in two independent orientations yielding very similar final Rcryst and Rfree values compared with the refinement in space group P6522. In both orientations (and for both target sites), the final Fo−Fc electron density maps showed pairs of difference Fourier peaks (∼7σ) of similar height at the non-palindromic bases, indicating that a unique orientation of the DNA duplexes does not correctly describe the situation in the crystals. Finally, simulated-annealing omit maps in space group P65 where the non-palindromic bases were omitted showed averaged densities for the omitted bases in both complexes, further confirming the assigned space group P6522.
To account for the two orientations of the DNA in the crystal during the refinement, two nucleotides with 50% occupancy were introduced at the non-palindromic positions. The final model of the LFY-C/AP1 complex at 2.1-Å resolution (Rcrystal=21.0%; Rfree=23.7%) comprises residues 237–399 of the LFY DNA-binding domain, whereas the poorly conserved 25 C-terminal residues are disordered. For the refinement of the LFY/AG-I complex, the AP1 DNA sequence in the LFY/AP1 complex was replaced with the AG-I sequence. Multiple rounds of refinement (including TLS refinement with seven groups) using program Refmac5 (Murshudov et al, 1997) and Phenix (Adams et al, 2002) yielded a model with Rcryst of 22.1% and Rfree of 24.9% using data between 20 and 2.3 Å resolution. The atomic coordinates and structure factors for the LFY/AP1 and LFY/AG-I complexes have been deposited with the Protein Data Bank under accession codes 2vy1, r2vy1sf and 2vy2, r2vy2sf, respectively.
Supplementary Material
Supplementary data
Acknowledgments
We thank D Weigel for providing materials and advice, X Wu for material, L Blanchoin, C Guérin, R Dumas, M Jamin, C Ebel and G Schoehn for help with protein expression and characterization, L Blanchoin, A Maizel, M Blazquez, E Dorcey and C Petosa for critical reading of the paper, R Russell for structure comparisons, the EMBL/ESRF Joint Structural Biology Group for access and support at the ESRF beamlines and the crystallization facility of the Partnership for Structural Biology for support. Funding was provided by ATIP (CNRS) to FP and RB, ATIP+ (CNRS) and ANR BLAN-0211 to FP, Region Rhône-Alpes/Cluster 9 to CH, Programme Emergence of the Region Rhône-Alpes to DP.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Albert VA, Oppenheimer DG, Lindqvist C (2002) Pleiotropy, redundancy and the evolution of flowers. Trends Plant Sci 7: 297–301 [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Ferrandiz C, Madueno F, Parcy F (2006) How floral meristems are built. Plant Mol Biol 60: 855–870 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J (2003) Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breitling R, Gerber JK (2000) Origin of the paired domain. Dev Genes Evol 210: 644–650 [DOI] [PubMed] [Google Scholar]
- Burz DS, Rivera-Pomar R, Jackle H, Hanes SD (1998) Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J 17: 5998–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Chae E, Tan QK, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Cherry JL, Adler FR (2000) How to make a biological switch. J Theor Biol 203: 117–133 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Cramer P, Muller CW (1997) Engineering of diffraction-quality crystals of the NF-kappaB P52 homodimer:DNA complex. FEBS Lett 405: 373–377 [DOI] [PubMed] [Google Scholar]
- de la Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW (2007) Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet 8: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific [Google Scholar]
- Diederichs K, Karplus P (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol 4: 269–275 [DOI] [PubMed] [Google Scholar]
- Dummler A, Lawrence AM, de Marco A (2005) Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb Cell Fact 4: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Frohlich MW (2003) An evolutionary scenario for the origin of flowers. Nat Rev Genet 4: 559–566 [DOI] [PubMed] [Google Scholar]
- Frohlich MW, Chase MW (2007) After a dozen years of progress the origin of angiosperms is still a great mystery. Nature 450: 1184–1189 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C (2001) Recognition of specific DNA sequences. Mol Cell 8: 937–946 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T (1994) Homeodomain proteins. Annu Rev Biochem 63: 487–526 [DOI] [PubMed] [Google Scholar]
- Gerard FC, Ribeiro Ede A Jr, Albertini AA, Gutsche I, Zaccai G, Ruigrok RW, Jamin M (2007) Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46: 10328–10338 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS (2007) Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch WJ (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Khare D, Ziegelin G, Lanka E, Heinemann U (2004) Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat Struct Mol Biol 11: 656–663 [DOI] [PubMed] [Google Scholar]
- Kirsch RD, Joly E (1998) An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res 26: 1848–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR (1998) Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol 16: 569–592 [DOI] [PubMed] [Google Scholar]
- Lamb RS, Hill TA, Tan QK, Irish VF (2002) Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129: 2079–2086 [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Weigel D (2002) Building beauty: the genetic control of floral patterning. Dev Cell 2: 135–142 [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260–263 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM (1997) Plants and the logic of development. Genetics 145: 5–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gomez P, Capel J, Lozano R (1999) FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J 20: 685–693 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC (2004) Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J 23: 4384–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Bush MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS (2001) ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr 57: 1445–1450 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR (1999) Molecular evolution of helix-turn-helix proteins. J Mol Evol 49: 301–309 [DOI] [PubMed] [Google Scholar]
- Russell RB, Barton GJ (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins 14: 309–323 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG (2002) Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J 21: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear DF, Ross JB, Laue TM (1998) Analysis of protein and DNA-mediated contributions to cooperative assembly of protein–DNA complexes. Methods 16: 3–20 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Shih MC, Li WH (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R (1998) Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125: 733–742 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Melzer R (2007) Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot (London) 100: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DA (2000) Genetic and biochemical diversity in the Pax gene family. Biochem Cell Biol 78: 629–638 [PubMed] [Google Scholar]
- van Pouderoyen G, Ketting RF, Perrakis A, Plasterk RH, Sixma TK (1997) Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J 16: 6044–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jackle H (1990) The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63: 455–456 [DOI] [PubMed] [Google Scholar]
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D (2003) Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO (1999) Crystal structure of the human Pax6 paired domain–DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev 13: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data