Abstract
Subtelomeric regions are often under-represented in genome sequences of eukaryotes. One of the best known examples of the use of telomere proximity for adaptive purposes are the bloodstream expression sites (BESs) of the African trypanosome Trypanosoma brucei. To enhance our understanding of BES structure and function in host adaptation and immune evasion, the BES repertoire from the Lister 427 strain of T. brucei were independently tagged and sequenced. BESs are polymorphic in size and structure but reveal a surprisingly conserved architecture in the context of extensive recombination. Very small BESs do exist and many functioning BESs do not contain the full complement of expression site associated genes (ESAGs). The consequences of duplicated or missing ESAGs, including ESAG9, a newly named ESAG12, and additional variant surface glycoprotein genes (VSGs) were evaluated by functional assays after BESs were tagged with a drug-resistance gene. Phylogenetic analysis of constituent ESAG families suggests that BESs are sequence mosaics and that extensive recombination has shaped the evolution of the BES repertoire. This work opens important perspectives in understanding the molecular mechanisms of antigenic variation, a widely used strategy for immune evasion in pathogens, and telomere biology.
Introduction
Subtelomeres are dynamic and fast-evolving regions of eukaryotic genomes owing to their remarkable plasticity [1]–[3]. Recombination between internal repeats and chromosomal arms results in the accumulation of species-specific sequences, commonly mediating adaptation to the environment. Despite their extreme genetic diversity, common aspects of structure and function are shared across telomeres from a diverse range of organisms [4].
The ability to encode contingency functions within their subtelomeric regions has been exploited by the African Trypanosome, subspecies of which cause the debilitating disease Human African Trypanosomiasis and the related disease ‘nagana’ in livestock [reviewed in [2]. Trypanosoma brucei evades the mammalian host's immune response by periodically changing its variant surface glycoprotein (VSG) coat (reviewed in: [5]), a dense monolayer of 5×106 identical VSG dimers. The expressed VSG is located in specialised subtelomeric transcription units known as the Bloodstream Expression Sites (BES). The total number of BESs is dependent on the subspecies and strain but is believed to be about 20 for the T. brucei Lister 427 strain [6] used in the present study. To change the VSG coat, transcription is switched to an alternative VSG by recombination of a new VSG into the active BES or by transcriptional silencing of one BES with concomitant activation of a second.
Sequencing of a few BESs has revealed a diverse range of 11 polymorphic genes, called Expression Site Associated Genes (ESAGs) [7], few of which have been functionally characterised: ESAG3, ESAG5 and ESAG11 encode membrane-associated or membrane-targeted proteins [8]; ESAG4 encodes a putative transmembrane receptor with adenylate cyclase activity [9]; ESAG6 and ESAG7 encode subunits of a heterodimeric receptor that mediates uptake of host transferrin [10], [11]; ESAG8 encodes a putative nuclear DNA-binding protein [12], [13]; ESAG10 encodes a homologue of the Leishmania biopterin transporter [14] whilst ESAG9 [15] has an unknown function. As many ESAGs are members of large families and are dispersed in the genome [16]–[18], and as a low level of transcription occurs even from ‘inactive’ BESs, determining the precise function of individual ESAGs in active expression sites is challenging.
In an effort to systematically analyse the BES repertoire of a single clone, a library of transformation-associated recombination (TAR) clones was made [19]. In the present study, sequence analysis revealed that the 19 TAR clones fall into 14 distinct BES groups. Independently, 13 randomly tagged BESs were constructed and analyzed, confirming the structures and re-defining the minimal gene complement of a functional BES for survival in culture.
Patterns of sequence variation between different ESAG families were analysed revealing that expression sites are sequence mosaics - related to each other differently depending on the gene family examined - and highlighting the significant role that recombination between gene families has played in forming BES sequences. In characterising a complete BES repertoire these results provide a perspective on nucleotide diversity, but also on the mechanisms regulating their origins and diversity.
Results
Two independent approaches were undertaken. First, BESs were randomly tagged with a G418-resistance gene, and we will refer to these clones as ‘NEO-tagged’. Second, TAR-cloned BESs were sequenced, and we will refer to these clones either by their TAR clone number or by the BES to which they have been assigned.
Tagging BESs
T. brucei cell lines tagged with two drug-resistance markers were generated to select parasites that had stochastically activated an individually tagged BES from the background of highly similar BESs. The active BES1 was tagged with a Puromycin resistance gene (PUR), immediately downstream of the promoter. In the remaining silent BESs, transcription initiates at the BES promoter but is rapidly attenuated. This low level of transcriptional activity allowed the integration of a Neomycin resistance gene (NEO) immediately downstream of a second ‘silent’ BES promoter. In this way, we obtained 30 double-tagged clones. Each randomly NEO-tagged BES was typed according to its ESAG6 hypervariable region, the distance between the ESAG6 gene and the promoter, the number of BES promoters, and the sequence of the expressed VSG upon BES activation. Analysis of the 30 NEO-tagged clones and comparison to the TAR clones showed that 13 different BESs were tagged: BES1, 2, 3, 4, 5, 7, 8, 12, 13, 14, 15a and 15b (a duplicated BES, see below) and 17 (Tables 1 and 2). All tagged BES could be activated in vitro, except for BES8. Characterisation of these clones confirmed many of the apparently anomalous structures revealed by the TAR sequences (see below). The only two cloned BESs that were not tagged were BES10 and BES11 (Tables 1 and 2).
Table 1. BES overview.
| BES | TAR clone sequenced | Nomenclature of VSG genes | Deviation from conserved architecture | Drug-resistance tagged clone | NEO-tagged clone consistent with sequenced TAR clone | BES could be activated in vitro | Genome localisation: type and size (Mb) of chromosome | ||||
| 2008 | Lab-specific | MITat | Multiple copies of ESAGs? | Missing ESAGs? | Other | ||||||
| 1 (a) | 40 | 427-2 | 221 | MITat 1.2 | large-scale | - | 70 bp-repeat array (×2) | Yes | - | Yes | MBC-3.0 |
| 2 (a) | 129 | 427-9 | VO2 | MITat 1.9 | 7 (×2) | - | ingi element 3′ of 70-bp repeat | Yes | Yes | Yes | IC-0.325 |
| 3 | 2, 15 | 427-6 | 121 | MITat 1.6 | - | - | Yes | Yes | Yes | MBC-2.05 | |
| 4 | 3, 28 | 427-21 | T3 | MITat 1.21 | 7 (×3) | - | VSG ψ 3′ of 70-bp repeat | Yes | Yes | Yes | IC-0.250 |
| 5 | 98 | 427-18 | 800 | MITat 1.18 | - | - | Yes | No (c) | Yes | MBC-1.95 | |
| 7 | 65 (b), 153 | 427-3 | 224 | MITat 1.3 | 6 (×2) | 7 | ESAG3 ψ 3′ of 70-bp repeat | Yes | Yes | Yes | MBC >3.1 |
| 8 | 64 | 427-14 | - | MITat 1.14 | - | 1,2,3,4,5,6,8,9,11 | Single ESAG7 gene | Yes | No (c) | No | MBC >3.1 |
| 10 | 134 | 427-15 | - | MITat 1.15 | - | 1,2,3,4,8,11 | 23 bp trace of 70-bp repeat | No | N/A | N/A | IC-0.450 |
| 11 | 122 | 427-16 | - | MITat 1.16 | 4 (×2) | 1,2,11 | additional VSG ψ; copy of ESAG9 | No | N/A | N/A | IC-0.180 |
| 12 | 29 | 427-8 | OD1 | MITat 1.8 | - | 3,4,8 | Yes | Yes | Yes | MBC-1.7 | |
| 13 | 56 | 427-17 | JS1 | MITat 1.17 | 8 (×2) | - | Yes | Yes | Yes | IC-0.180 | |
| 14 | 10 | 427-19 | - | MITat 1.19 | - | 3,4,8 | 2 additional VSG ψs; 70 bp-repeat array (×2) | No | N/A | N/A | N/A |
| 117 (not sequenced) | 427-8 | OD1 | MITat 1.8 | N/A | N/A | N/A | Yes | Yes | Yes | MBC >3.1 | |
| 15 (two copies: 15a, 15b) | 126 | 427-11 | bR-2 | MITat 1.11 | - | - | ESAG8(×2); VSG ψ (×2); ESAG3 ψ (×2); 70 bp-repeat array (×2) | Yes (15a & 15b) | Yes | Yes | 2 MBC: 1.35, 1.7 |
| 16 | 128 (d) | 427-2 | 221 | MITat 1.2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 17 (a) | 51 (b), 59 | 427-13 | NA1 | MITat 1.13 | - | - | - | Yes | Yes | Yes | IC-0.295 |
In this paper, we propose a new simple and logical VSG nomenclature. The hyphenated name indicates the T. brucei strain, followed by a number that identifies each unique VSG. For very closely related (probably antigenically indistinguishable) members of a VSG family, the number may be followed by a lower-case letter (for example Lister 427-3). When no misunderstanding can occur between strains under discussion in a particular context, the strain information may be omitted and the VSG referred to only by its number (for example, VSG 3). ND: not determined; N/A: not applicable; IC: intermediate chromosome; MBC: Megabase chromosome. (a) BES1, 2 and 17 had been previously cloned in BACs and identified as 221 ES, VO2 ES and Bn-2 ES, respectively [27]. (b) TAR65 and TAR51 were re-classified as belonging to BES group 7 and 17, respectively, based on global alignments showing sequence identity along the length of the sequence. The only differences were found in the promoter region, the target site for recombination during construction of the library. (c) Restriction mapping of the NEO tagged BES suggests the existence of two promoters, whereas the TAR clone has a single promoter. (d) TAR clone 128 is likely to be a recombinant between expression sites belonging to BES groups 1 and 15 (see text).
Table 2. Summary of the number of TAR clones sequenced, BES identified and BES activated.
| Number | TAR clones or BES | TAR clones or BES missing | |
| TAR clones | 19 | 40, 129, 2, 15, 3, 28, 98, 65, 153, 64, 134, 122, 29, 56, 10, 126, 128, 51, 59 | TAR128 is probably a recombination artifact |
| Unique BES | 14 | 1, 2, 3, 4, 5, 7, 8, 10, 11, 12, 13, 14, 15, 17 | TAR2 and TAR3 were duplicates of TAR15 and TAR28, respectively; TAR51 and TAR65 were re-classified |
| All BES | 15 | 1, 2, 3, 4, 5, 7, 8, 10, 11, 12, 13, 14, 15a+15b, 17 | BES15 is present in two copies in the genome |
| Tagged BES | 13 | 1, 2, 3, 4, 5, 7, 8, 12, 13, 14, 15a+15b, 17 | BES10 and BES11 were not tagged |
| Functional tagged BES | 12 | 1, 2, 3, 4, 5, 7, 12, 13, 14, 15a+15b, 17 | BES8 was tagged with NEO gene, but no G418-resistant clones could be obtained. |
Chromosomal localization of BESs
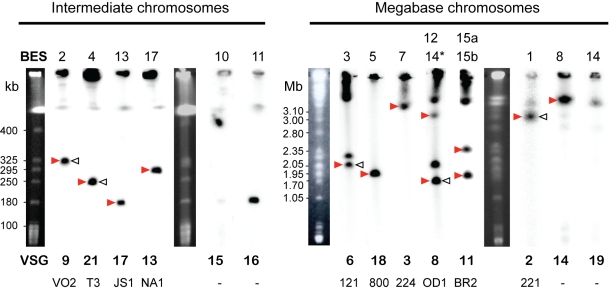
The chromosome containing each BES (indicated by arrows in Figure 1) was identified by Southern blotting of PFGE-separated chromosomal DNA with NEO and VSG probes (Figure 1). It had been shown previously that BES2 is located on an intermediate chromosome [20]. We now show that BES4, BES10, BES11, BES13 and BES17 are also present in this chromosome class and that the megabase-sized chromosomes encode all remaining BES, consistent with previous reports [21].
Figure 1. Chromosomal distribution of BES-resident VSGs.
Red arrows indicate BES randomly tagged in this study with NEO. Open arrows indicate BES activated by [58] and subsequently tagged. Names of VSG genes are indicated at the bottom, using the newly proposed strain-specific numbering (top row) and lab-specific names (bottom row). BES10 and 11 were not tagged in either study. BES14 is represented in two lanes: 14* refers to TAR117 (which was not sequenced and harbours VSG8), whereas 14$ refers to TAR10 (which was sequenced and harbours VSG9).
In some cases, the VSG probe hybridized with more than one chromosome, indicating that the genome contains multiple copies of the corresponding VSG. For instance, BES12 and BES14 contain VSG427-8, and BES15a and BES15b contain VSGbR-2/427-11, on megabase chromosomes of different sizes (Figures S1A and S1B, respectively). VSG121/427-6 is present as a non-BES copy (Dreesen O., personal communication).
Switching mechanism and frequency
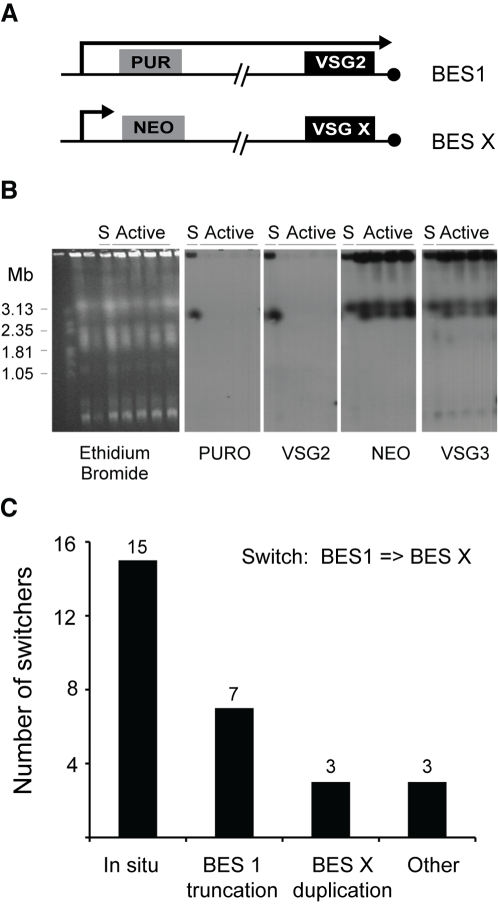
The switching assay was designed to select for trypanosomes that had undergone an in situ switch by transcriptionally silencing BES1 and activating another BES without any DNA rearrangements. To assess the efficiency of selecting in situ switchers, Puromycin- and G418-resistant clones were analyzed by PFGE. If an in situ switch had occurred, the chromosomal locations of PUR, NEO and VSG should be unchanged: PUR and VSG221/427-2 should be on chromosome VIa, whereas NEO and the newly identified VSG should be located on a different chromosome. PFGE analysis of 28 NEO-tagged clones that had activated different BES (2, 4, 5, 7, 13, 14, 15b and 17) confirmed that in situ switching was the predominant switching mechanism (15 switch events), but other switching mechanisms were also observed (Figure 2). In BES2- or BES15-tagged clones, for example, we observed that VSG221/427-2 and PUR were deleted, whereas NEO and the new VSG remained in their original chromosomal location. This suggests that activation of BES5 and BES15 was accompanied by the truncation of BES1, a phenomenon previously reported for another BES [22]. In three of the 28 cases studied, the active BES was replaced by duplicative transposition of the entire new BES. In switchers that activated BES7 (Figure 2B), BES13 or BES15b, VSG221/427-2 and PUR were lost and NEO and the new VSG genes were duplicated onto chromosome VIa. This switching mechanism has been previously detected in switchers obtained in mice [23] or by negative-selection in vitro [24]. In three cases, we observed unconventional recombination events that involved more than two chromosomes. These complicated VSG switch events that have undergone multiple DNA rearrangements have been previously documented [25].
Figure 2. Types of switching mechanisms observed for tagged BES clones in vitro.
A. NEO-tagged clones have PUR in BES1 and NEO in an unknown BES. B. Example of a BES duplication. PFGE analysis of a BES7-tagged clone (17.9), when this BES is silent (S) and after activation. NEO and VSG3 genes were duplicated onto the chromosome that originally contained PUR and VSG221/427-2. C. Quantification of the frequency of the different types of mechanisms. “Other” refers to recombination events that involved more than 2 chromosomes.
In our cell-lines, the frequency of switching between BES1 and a second BES can be measured as the ratio between the number of G418-resistant clones and the total number of starting cells. Switching frequency was measured for clones in which the NEO gene was in BES4, 5, 13, 15b and 17. Consistent with previous reports, the average frequency was around 2±3×10−6 switchers/cell, with BES15 showing the lowest switching frequency (8.3×10−8 switchers/cell) [20], [26]. However, it should be noted that these measurements were based on a small number of independent clones (usually 3). A larger-scale study would be necessary to assess if the differences of switching frequency are statistically significant and how those correlate with the DNA sequence of the activated BES.
Sequencing and assembly of TAR clones
Nineteen TAR clones [19] were sequenced, manually finished and analysed. This set represents 14 unique BESs, with TAR clones from BESs 9 and 16 having been re-classified (Table 2). For each BES group, at least one clone was sequenced (Figure 3, Table 1), choosing a larger sized clone wherever possible. Additional clones were sequenced for BES3 and BES4 to verify the sequence and clone integrity. Global alignments showed that TAR clones 2 and 15 (BES3) as well as 3 and 28 (BES4) are 100% identical at the DNA level with the exception of an ESAG7 triplication in TAR28. In addition, TAR clones from BES1 (TAR40), BES2 (TAR129) and BES17 (TAR59) were compared to the same BES previously sequenced from a BAC library [27] and were 99%,100% and 99% identical, respectively, indicating that TAR cloning produced stable clones.
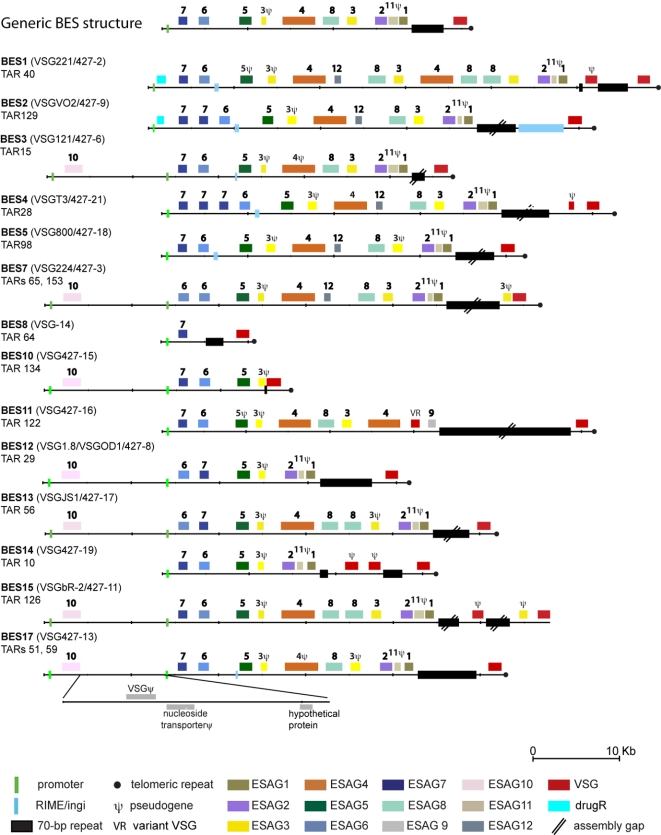
Figure 3. Overview of T. brucei Lister 427 BES.
BES are drawn to scale and have been aligned at their 5′-most ESAG7 or ESAG6 sequence. The inset shows the regions conserved between the dual promoters present in some of the clones. The VSG indicated in front of the BES label refers to the telomere-proximal VSG.
BES architecture
Eighteen of the TAR clones contain full-length BESs, ending in telomeric hexamer repeats, and the 14 BES types show a striking conservation in overall structure and order of ESAGs (Figure 3). The 5′ end of each TAR clone contains the BES promoter that was used as the recombination target during cloning. Half of the BESs have a second promoter approximately 13 kb upstream from the first [28] which accounts for the differences in clone sizes observed in some of the BES groups. The region between the two promoters encodes ESAG10 and a variety of pseudogenes that are commonly found in T. brucei subtelomeric regions (Figure 3). The presence of two promoters in some BES was confirmed by restriction mapping of the NEO-tagged clones. BES5 and BES8 were the only two discrepancies: NEO-tagged clones suggested the presence of 2 promoters, whereas TAR clones only had one, presumably because the downstream promoter was the target of the TAR cloning (Figure S3B). Downstream of the second promoter, the majority of sites encode polymorphic variants of ESAG7, ESAG6, ESAG5, ESAG3, ESAG4, ESAG8, ESAG3, ESAG2, ESAG11 and ESAG1, in this conserved order. The only exception was an inversion between ESAG7 and ESAG6 in two BES groups (TAR29/BES12 and TAR56/BES13).
Nine BESs encode at least one copy of each ESAG (excluding ESAG10). We also observed frequent duplication of ESAGs, including a large-scale duplication in BES1 (TAR40), an ESAG7 triplication in BES4/TAR28 and three cases of divergent additional copies of ESAG8 (BES1, BES13 and BES15).
The terminal VSGs are flanked upstream by tracts of 70-bp repeats, whose lengths vary in our assemblies between 0.2–7.1 kb, although this repeat region was only manually finished in 10 clones (Figure 3). The distance between the VSG and the telomeric repeats could be assessed in 17 TAR clones and ranged from 200–1590 nucleotides.
As described above, the analysis of the NEO-tagged clones was consistent with the analysis of TAR clones except in one case. Although VSG221/427-2 is a single-copy gene (Figure 1), we found it in both BES1/TAR40 and BES16/TAR128. The presence of VSG221/427-2 in BES1 has been firmly established from sequencing one BAC and two TAR clones. The promoter-proximal region (approx 40 kb) of TAR128 is identical to TAR126. Taken this data together, we conclude that TAR128 may be an artefact that resulted from recombination between BES1 and BES15. PCR analysis of additional TAR clones of BES16 failed to detect VSG221/427-2 (data not shown), further suggesting that TAR128 is not representative of BES16.
Non-functional and novel BES genes
A defining feature shared by most BESs is the strict conservation of the 5′ copy of ESAG3 and ESAG11 as pseudogenes (Figure 3). The predominance of the latter is corroborated by a previous study describing the characterisation of this gene family [7]. In addition to these two pseudogenes, single frameshifts cause the presumed inactivation of ESAG4 and ESAG5 in some BESs. An analysis of ESAG processing signals gave further indication that some of the ESAGs may not localise correctly within the cell, including a number of ESAG4 genes which lack sequences for surface targeting.
In five BESs (BESs 1, 2, 4, 5, 7) we identified a novel highly conserved putative protein coding sequence with a predicted signal peptide that we have designated ESAG12 (Figure 3). No functional clues could be discerned from sequence similarity or protein domain search results. A single homologue is found in the TREU 927 genome (Tb927.7.7510), although this is likely to be an under-representation due to the lack of subtelomeric sequence in the current TREU 927 genome assembly [18].
Some BES deviate from the overall conserved structure
Several BESs diverge from this conserved structure either by encoding additional sequences or because they represent truncated sites, lacking members of individual ESAG families. Most BES harbour only the VSG between the 70 bp-repeat array and the telomere (Figure 3). BESs 2, 4, 7 and 15 are exceptions to this rule, showing additional sequences (ingi element, VSG or ESAG3 pseudogene). A further three sites encode duplicated regions of 70-bp repeats (BES1, 14 and 15), in each instance sandwiching VSG pseudogenes (see below). Five of the BES groups do not contain one or more of the expected ESAG repertoire. Amongst this set are:
BES7 (TARs 65,153) lacks ESAG7, but contains tandem copies of ESAG6, with different hypervariable regions, with ESAG6a resembling an ESAG6/ESAG7 mosaic. This BES could be activated in vitro at the same frequency as other BESs. In order to investigate whether any DNA rearrangements have donated a copy of ESAG7 to the newly activated BES7, DNA from cells with an active or silent BES7 were compared by PCR and restriction mapping (Figure S2). No differences were observed, suggesting that a BES lacking ESAG7 can indeed be activated, confirming previous observations [29].
BES8 (TAR64) is unusually small and only contains a single copy of ESAG7 followed by the 70-bp repeat region and the VSG (Figure 3). Long-range analysis of chromosomal DNA from a BES8-tagged clone digested with Apa I supported this finding, with the NEO and VSG427-14-hybridizing restriction fragment being only ∼20 kb in length (Figure S3A). Genotyping of the NEO-tagged clone confirmed the presence of ESAG7 and absence of ESAG6 (data not shown). Attempts to activate this BES in two independent clones were unsuccessful, suggesting that this mini-BES may not be functional.
BES10 (TAR134) lacks the central portion (ESAG 4-8-3-2-11-1). TAR134 also encodes only a 23-bp fraction of a 70-bp repeat. This site was not among those that were tagged with a drug resistance marker, so its potential for activation could not be assessed.
BES14 (TAR10), also lacks the ESAG4-8-3 gene cluster and has two VSG pseudogenes sandwiched between 70-bp repeat arrays. An inconsistency was found between the NEO-tagged clone and TAR sequence. The NEO-tagged clone did not harbour the VSG predicted by the TAR clone. Instead, the NEO tagged cell line expressed VSG427-8 upon activation, rather than VSG427-19, as predicted by the TAR sequence. We confirmed by PFGE that the BES activation did not involve gene rearrangements (data not shown), indicating that VSG427-8 was at the BES14 locus prior to activation. Further examination of the TAR clones in the BES14 set revealed TAR117 to correspond to the NEO-tagged clone. An interesting consequence of these results is that two BESs (BES12 and BES14) contain the VSG427-8 gene (Figures 1 and S1A).
In BES11 (TAR122), the ESAG2-11-1 cluster has been replaced by a VSG-related gene (VR, Figure 3) and a full-length copy of ESAG9, the only copy found to date in a T. brucei BES. The VR gene is believed to be a VSG that has already evolved a new function and, like other VRs, has no 70-bp repeat sequences in its upstream flank [30]. Similarly to BES10, this site was not tagged with a drug selectable marker and therefore, activation in vitro could not be assessed.
BES12 (TAR29) has a deletion of the ESAG4-8-3 cluster, but successful tagging and activation demonstrated that this site was functional in vitro (Table 1).
Investigating the forces driving BES diversity
In order to investigate the role of recombination in the evolution of bloodstream expressions sites, we examined changes in phylogenetic relationships along the BESs using two methods.
Likelihood comparisons of optimal and constrained trees
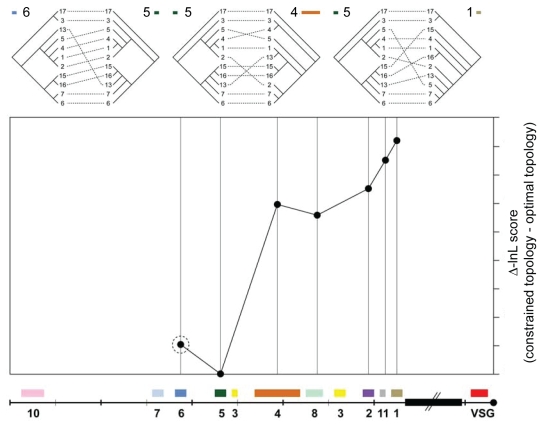
There were obvious discrepancies between the optimal phylogenies for different ESAG loci, suggesting that the phylogenetic signal varied along the BESs. This was manifest in both the different shapes of ESAG trees and comparisons of their log-likelihood scores (Figures 4 and S4). Comparing the likelihood score of an optimal topology for any given ESAG with the score obtained for a tree constrained with the signal of another ESAG locus, typically indicated a decrease in likelihood and therefore a decreased probability of shared histories. Indeed, the drop in likelihood was generally proportional with physical distance, such that topologies from different ends of the expression sites were most dissimilar (Figures 4 and S4). Although topological differences between the phylogenies of neighbouring ESAGs were observed, these were non-significant and fell within the margins of systematic error. In fact, comparisons for all genes (Figure S4) showed that ESAGs fell into three clusters with regard to phylogenetic signal: ESAGs 7, 6 and 5, ESAGs 4 and 8, and ESAGs 2, 1 and 11 with compatible phylogenetic signals within, but not between, these clusters.
Figure 4. Quantifying the difference in phylogenetic signal along the bloodstream expression site.
A. Three tanglegrams relate the ESAG5 phylogeny with those of ESAG6 (close match), ESAG4 (moderate incongruence) and ESAG1 (severe incongruence). Dashed lines link corresponding expression sites in each tree. Incongruence between trees increases from left to right. B. Comparison of likelihood scores between optimal and constrained tree topologies for ESAG5. The phylogenies of six other ESAG loci (indicated by a cartoon of the expression site) were used to constrain the estimation of the ESAG5 tree; the difference in likelihood score between each of these constrained trees and the optimal ESAG5 tree is plotted along the expression site. Non-significant differences in likelihood are denoted by a dashed circle as evident when constraining the ESAG5 topology with the ESAG6 topology; however, enforcing the topologies of central loci (ESAGs 4 and 8) caused a moderate decrease in likelihood, whilst constraining with ESAGs 2, 11 or 1 caused a larger decrease.
Recombination analyses
To examine the fine scale structure of recombination within the BESs, a genetic algorithm-based tool (GARD) [31] was used to infer the numbers and locations of recombination breakpoints. Although this method relies on phylogenetic incompatibility to deduce recombination, GARD differs from the constrained tree analyses described above in two important respects: (i) the locations of putative breakpoints are inferred rather than fixed a priori, allowing us to characterize recombination within individual loci, and (ii) GARD is able to detect gene conversion events that affect branch lengths but not tree topology. When applied to the nucleotide alignments of each ESAG locus and to five intergenic regions, this method identified a large number of recombination tracts with distinct phylogenetic histories (Tables S1 and S2). Mean recombination tract lengths ranged from 90 bp/tract for ESAG1 to 687 bp/tract for ESAG10, and mean tract lengths were similar in both coding and non-coding sequences. The GARD analyses demonstrate that phylogenetic breakpoints occur throughout the expression sites, consistent with frequent genetic exchange.
Phylogenetic inference of recombination can be misled by substitution rate variation, as could occur if different genes or regions within a gene are subject to different selection pressures. Therefore, a summary statistic-based approach, the Pairwise Homoplasy Index (Φ) Test [32], which has been shown to be robust to substitution rate variation, was applied to each alignment to validate the GARD analyses, and to each of the individual recombination tracts inferred by GARD to identify any further recombination that might exist. With the exception of ESAG10, which is highly conserved, significant values of Φ were obtained for each ESAG alignment, consistent with recombination within these loci (Table S2). In addition, the Φ values were significant in at least some of the GARD-inferred tracts within all ESAG loci except ESAG8 and ESAG10, and in 30% of individual tracts overall. These results indicate that there are additional breakpoints not detected by GARD, and support the conclusion that there is extensive recombination throughout the entire BES.
Evidence of molecular adaptation within ESAG families
The role of selection in the evolution of bloodstream expression sites was examined by estimating the ratio of the non-synonymous and synonymous substitution rates (ω) within each ESAG family. A likelihood ratio test was used to assess whether the distribution of ω across codons is best explained by a model (M1a) which assumes that all sites are evolving neutrally or under purifying selection, or by a model (M2a) which includes an additional category of sites under positive selection (i.e., ω>1). As indicated by the higher likelihood scores of model M2a (Table S3), these tests provide significant evidence of adaptive evolution within ESAGs 4, 5, 6, and 7, and are nearly significant (p<0.1) for ESAG1 and ESAG3b. In contrast, there is no evidence of adaptive evolution in ESAGs 2, 8 and 10. Selective pressures at individual codons were inferred using a Bayesian approach with model M2a used as a prior distribution on ω. As expected, the majority of sites appear to be subject to strong purifying selection (ω≪0.5), while the numbers of positively-selected sites corroborate the gene-wide analyses, with ESAGs 4–7 displaying widespread adaptation (Table S4). Taken together, these results indicate that all ESAGs are under strong purifying selection, presumably to maintain functionality, and that most ESAGs contain at least some sites which have undergone adaptive evolution over the course of BES differentiation.
Discussion
The elucidation of the BES repertoire of a single T. brucei strain has produced a global view of BES structure and evolution. We have sequenced 19 out of 182 clones of BESs from T. brucei 427, chosen to represent as many different BESs as possible based on the preliminary analysis of the original set [19] and including duplicates. Sequence analysis identified 14 distinct BESs, one of which exists in two copies, giving a total number of 15 BESs. Have we found all of the BESs in T. brucei 427? Assuming that the promoter sequence is conserved in all BESs, we estimated the chance of missing any BESs in the entire library by simulating samples of 182 randomly sampled clones from populations with different numbers of underlying BESs. In such a simulation, no sites are ‘missed’ if there were 19 or fewer BESs. Based on all of the available data the subset chosen for sequencing appears to cover most, if not all, BESs. Thirteen of these 15 BES were tagged with the Neomycin resistance gene in 30 random integration events. Assuming equal probability of tagging any end we expect to miss at least 2 out of 15 BESs in 30 tagging events 65% of the time (simulation of 1000 random samples of 30 tagging events). No BESs were tagged that were absent from the collection of TAR clones and we therefore are confident that the library contains the entire T. brucei Lister 427 BES repertoire. We will however only be able to address the question as to whether the size of the BES repertoire of T. brucei Lister 427 is shared by other Trypanosoma brucei isolates once a large number of strains are analyzed. The genome strain T. brucei TREU 927 has fewer BESs based on hybridization analysis [33] and a similar set of T. brucei 927 BES TAR clones with only 5–8 promoter and ESAG6 region sequence types have been identified (Becker and Louis, unpublished). In addition to having fewer BESs, T. brucei 927 also has smaller VSG basic copy arrays compared with T. brucei 427, though the genome strain T. brucei TREU 927 has one of the smallest genomes analysed among T. brucei isolates [34]. This makes it likely that the relatively large number of BESs that we observe here with T. brucei 427 does indeed fall within the range of BES copy numbers that may be observed among different isolates.
The validity of the TAR cloning is demonstrated by the identical sequences obtained from two independent TAR clones of the same BES, by the comparison to previously published sequences and by the congruence with the data obtained from the NEO-tagged BES. Given both the number of TAR clones from known BESs and TAR cloned DNA that does not exist in the genome, we get a maximum estimate of 15% of aberrant TAR clones in the original library. In fact, only one of the 19 TAR clones selected for sequencing (TAR128) is inconsistent with other data and could be a recombinant between two BESs generated during TAR cloning. The other clones originally classified with this clone in BES group 16 appear not to be recombinant and could be sequenced in the future.
The sequencing effort has also highlighted the need to return to the original TAR clone set to account for misclassifications based on the hypervariable region of ESAG6 and the promoter sequence. Given that the promoter sequence was the target for recombination, the actual cloning procedure may have introduced variability in this region not present in the genome. As a consequence, two TAR clones were re-assigned in this study to different BES groups, giving fewer groupings than the original classification into 19 groups. It is clear that correct and complete characterisation of the telomeres requires a combination of saturation cloning and sequencing of well-chosen clones.
Characterisation of parasites with activated NEO-tagged BESs revealed that, as expected, in situ switching was the most frequent event. Interestingly, in three out of 28 switchers, we observed a switching mechanism, in which the previously active BES (BES1) was replaced by a duplication of the newly activated BES (7, 13 or 15b). Most likely recombination between the two subtelomeric regions occurs upstream of the drug-resistance genes, possibly at the promoter region, the 50 bp-repeat array, or in an even more internal chromosomal location. It is unclear which of the two BES copies is the active one. Nevertheless, our observations confirm what has been previously reported that an entire BES can be duplicated and transposed to a different locus [23], [24], which is reminiscent of break-induced replication in yeast. In fact, when a double-strand break is induced in subtelomeric regions of S. cerevisiae, that end is often replaced by a duplicated copy of another end (reviewed in [35]). Such mechanism could explain the existence of two copies of BES15 and it may also have contributed to the large number of BES in T. brucei 427 relative to other isolates.
The BESs presented here show considerable diversity in the ESAG complement, though there is an overall conservation of order that agrees with the generic BES structure previously proposed [27] (top diagram in Figure 3). There are some clear exceptions within the overall conservation, ranging from a small BES containing only ESAG7 and a VSG, to a BES containing ESAG duplications and triplications. The BES-tagging data presented here redefine the requirement for a minimal BES, which appears only to entail functional copies of ESAGs 1, 2 and 6, although ESAG1 is not essential within an active BES [16], [17] and ESAG2 is absent from the BES encoding SRA, the T. b. rhodesiense gene able to confer resistance to human serum in infections [36]. There appears to be no requirement for ESAG7, as previously postulated [27]. The smallest BES we came across as part of this study only encodes ESAG7 in addition to VSG (BES8). We were unfortunately unable to show activation of this site in vitro but it may yet indicate that none of the ESAGs need to be expressed from the active BES. However, the absolute requirement of ESAGs in an expression site is difficult to test because low-level transcription of ESAG6 and ESAG7 and indeed other ESAGs may occur from ‘silent’ sites [37], [38]. Even if these truncated BES are not functional, they may still contribute sequences via gene conversion to other BESs.
One surprising finding was the presence of additional sequences downstream of the 70-bp repeat in some BESs, as well as the number of BESs containing additional VSG pseudogenes. The presence of pseudogenes towards the downstream end of the BES is not entirely unexpected though, as this area is a recombinational hotspot, continually accepting incoming copies of VSG cassettes in order to mediate antigenic variation. As the recombination processes involved may not be constrained by the high stringency normally imposed by mismatch repair [39], it has to be expected that there will be errors, such as a duplicated cassette being copied into another cassette, rather than replacing it. Most of the VSGs in the silent archive are full-length pseudogenes, at least some of which contribute ‘healthy’ sequence to novel mosaic expressed VSGs [30]. It has been speculated that silent BESs might safely foster the stepwise assembly of mosaics, in contrast to the active BES, from which there is an absolute requirement for expression of an intact VSG [40], [41]. The present findings are compatible with this proposal, but not conclusive. The VSG pseudogenes could be copies of silent VSG pseudogenes, or partially assembled mosaics, or genes that have degenerated in situ through lack of requirement for expression. Work is underway to address the question of mosaic VSG assembly and expression.
The repertoire of BESs gives some indication of their proliferation among telomeres and their macroevolution. The driving forces that have shaped this repertoire are segmental duplications and deletions, recombination events typical of subtelomeres. However, gene content does not always coincide with the relationships among ESAG genes; for instance, BES13, 15 and 16 all possess a tandem duplication of ESAG8, but are not closely related based on their ESAG8 nucleotide sequences. The incongruence between gene trees for the different ESAGs confirms that BESs have not diversified through ‘orthodox’ patterns of duplication and divergence. Rather, each BES is a mosaic and the product of frequent recombination between telomeres; this may occur through reciprocal crossover events between homologous strands at mitosis, or through non-reciprocal gene conversion. Recombination is frequently non-homologous, as we observe both segmental deletions and duplications, (for instance the substantial duplications and triplications within BES1), and variations in length, with some BESs greatly reduced in size (e.g. BES8 and BES10).
This process is consistent with previous observations of ‘telomere exchange’ [42], though the prevalence of mosaics demonstrates that whole telomeres are not typically exchanged. Instead, the data suggest that clusters of contiguous ESAGs share common histories, and therefore have not frequently been split by recombination. There appears to be a physical limit on the frequency of recombination breakpoints, which seldom occur in between ESAGs 7, 6 and 5, ESAGs 4 and 8 or between ESAGs2, 1 and 11 (Figure 4, Table S2). However, the transposition of an isolated ESAG7 copy into an active expression site (without ESAG6 and 5) has been reported under in vitro drug and serum selection pressure [38]. This provides experimental evidence that ESAGs can and do move between sites, with the loss of a particular allele from the active BES; our data qualify this by showing that ESAG7 usually transpose in unison with ESAGs6 and 5 (otherwise they would not share a common phylogenetic history).
Given that frequent breakpoints coincide with the position of ESAG3 loci between linkage blocks, these could play a role in orientating expression sites prior to the exchange of intervening regions. This could also explain why ESAG3a, b and c sequences do not cluster by position (data not shown). It is therefore conceivable that the conserved, repetitive, higher order structure of expression sites may be maintained as a template for recombination, either to enhance the function of ESAGs, or of the VSG switching mechanism. Alternatively, the overall structure could be a secondary, non-adaptive result of the VSG switching events.
The majority of ESAGs within expression sites are unusual in that they are part of larger gene families present at chromosome internal locations within the genome but ‘behave’ like a tandem gene array or highly linked loci [43]. Internal and telomeric gene copies seem to evolve in very different ways. Internal copies of some families, for example ESAG4 and 5 are more diverse and retain orthology with corresponding loci in the related species. Telomeric ESAG copies in contrast have relatively low levels of variation and therefore always form a clade in ESAG family phylogenies (data not shown). This indicates that frequent recombination between telomeric copies leads to homogenisation and concerted evolution of ESAG families and both GARD and PHI analyses have shown that most ESAG alignments contain chimaeras (Figures 4 and S5, Table S1). In addition, localised diversification has clearly occurred in some ESAG families such as ESAG7 and 6, where the most diversifying codons can be mapped to the exposed surface of the molecule, including the hypervariable region. [11] (M. Carrington, University of Cambridge, personal communication).
We also postulate that a by-product of the recombination between ESAG families is the prevalence of some ESAGs as apparent pseudogenes. Like VSGs, ESAG copies may be routinely corrupted but continue to provide genetic resources for functional copies, since they can be rescued by subsequent gene conversion events. Whether ESAG evolution is adaptive, or simply a secondary consequence of sharing the expression site environment, remains to be seen.
The conclusion that both the whole BES and individual ESAGs are mosaics raises the question as to how it is possible for two contiguous markers, such as ESAG1 and 2, to display parallel evolutionary histories, when both gene families are being homogenised by frequent gene conversions. This should effectively randomise the relationships of each ESAG family relative to each other. Essentially, we could be observing recombination on two different scales. Entire BESs recombine, perhaps as a secondary effect of the VSG switching mechanism, but this is limited by spatial proximity. At a finer scale, individual ESAGs recombine with paralogs in other expression sites, which restrains the normal process of divergence, tending to homogenise the entire family. Our observation that BESs retain phylogenetic signals, i.e. a pattern of divergence, demonstrates that substitutions affecting ESAG sequences are not entirely homogenised by recombination. Therefore, for phylogenetic signals to persist, the substitution rate, perhaps augmented by positive selection, must limit the extent of homogenisation through recombination in a dynamic equilibrium. Neighbouring ESAGs that are not separated by recombination share this equilibrium and therefore, phylogenetic signal.
This work opens important perspectives in understanding the molecular mechanisms of antigenic variation and telomere biology. The fact that the short BES8 and BES10 could not be activated in vitro opens the possibility of using these sites to identify the minimum requirements to make a functional BES. The role of telomeres in BES transcription has been extensively studied by several groups (reviewed in [44]). Truncated BES may be a useful tool to study telomere biology in T. brucei, since the distance between the promoter and the telomere is much shorter. Finally, BES sequences and overall architecture will aid the identification of the mediators involved in VSG switching. Such studies may reveal mechanisms that are relevant to telomere biology and antigenic variation in other important pathogens.
Materials and Methods
Trypanosoma brucei
The BES TAR library was constructed from T. brucei Lister 427 strain clone 221a [45], in which a hygromycin-resistance gene had been previously introduced immediately downstream of the promoter of the active VSG221/427-2 expression site and a neomycin-resistance gene downstream of the promoter of the silent VSGVO2 expression site [22]. BES-tagged cell lines were also derived from wild-type T. brucei 221a. Bloodstream-form trypanosomes were grown in HMI-9 medium [46]. Stable transfections were performed using the BTX Electroporator [47].
Isolation of TAR clones and preparation of sequencing libraries
Yeast artificial chromosomes (YACs) containing BESs, TAR-cloned using the conserved VSG BES promoter as a recombination targets, have been previously described [19]. A representative clone was chosen from each group, selecting a larger sized clone wherever possible. Clones were isolated from yeast transformants through separation of the YAC DNA from endogenous chromosomes by CHEF (contour clamped homogeneous electric field) gel electrophoresis [48]. The recovered DNA was fragmented by sonication and end-repaired with mung bean nuclease prior to preparative agarose gel electrophoresis. DNA fragments of 2–4 kb and 1.4–2 kb were purified and cloned into pUC18 plasmid vector.
Sequence determination, assembly and annotation
Nineteen TAR clones were sequenced by random sequencing of small insert libraries using dye-terminator chemistry on an ABI 3730 sequencing machine. Sequence reads were assembled using Phrap [www.phrap.org; P. Green, unpublished)]. Manual base calling and finishing was carried out using Gap4 software (http://www.mrc-lmb.cam.ac.uk/pubseq/manual/gap4_unix_1.html). Gaps and low quality regions of the sequence were resolved by primer walking and targeted polymerase chain reactions (PCR). A number of gaps, all exclusively located in the 70-bp repeat regions, remain in the assemblies and are indicated by 100 N's (Figure 3).
Each clone was annotated using the Artemis software [49]. Protein coding sequences were predicted as previously described and analysed to assign putative functions as previously described [18]. The full annotation of 19 expression sites can be viewed and searched via GeneDB (http://www.genedb.org/genedb/tbrucei427/) and sequences have been submitted to EMBL with the following accession numbers: FM162566 - FM162583.
Current VSG nomenclature reflects multiple naming schemes. We therefore propose a new systematic nomenclature based on sequential numbering with reference to the T. brucei Lister 427 strain (Table 1).
Sequence alignments
The Needle program, an implementation of Needleman-Wunsch algorithm in the EMBOSS [50] software package was used to perform global alignments using default parameters. The region of the imperfect 70-bp repeats was excluded from each alignment. Local alignments were performed using ClustalW (http://www.ebi.ac.uk/clustalw/index.html) with default parameters
Generation of T. brucei clones with tagged BES
BES-tagged clones were generated by introducing a Puromycin resistance gene (PUR) downstream of the promoter of a wild-type VSG221/427-2 expressing cell line. NEO was subsequently integrated downstream of the promoters of random BESs conferring resistance to G418. Clones obtained from this transfection were named BF-LF17.x or BF-LF18.x. depending on the presence or absence of the 50-bp repeat array upstream of BES1 promoter (Figueiredo et al., unpublished) (see also Data S1).
BES switching assay
To prevent premature growth of in situ switchers, NEO-tagged clones were grown under Puromycin selection. After 5 days, cells were subcloned by limiting dilution in the absence of any drug to allow independent switching in each subclone. 2.4×107 cells of 2–5 subclones were subsequently diluted in HMI-9 containing 100 µg/mL G418, and distributed in a 24-well plate. After 6–8 days, the number of G418-resistant clones was counted. Switching frequency was calculated by dividing the number of G418-resistant clones by the total number of plated cells. One or two clones from each plate were tested for Puromycin sensitivity by diluting 105 cells in 5 mL of HMI-9 containing Puromycin at 1 µg/mL. Cell growth was scored after 2 days. Western Blotting was performed to confirm that G418-resistant cells no longer expressed VSG221/427-2.
Cloning the ESAG6 hypervariable region
The ESAG6 hypervariable region of each NEO-tagged BES was PCR amplified using a NEO-specific primer and one of two ESAG6-specific primers (5′-TAAAGAGAGTTGTTCACTCAC-3′ or 5′-TGTTCACTCACTCTCTTTGAC-3′) and the products gel-purified and sequenced. From clones with two consecutive ESAG6 genes, the ESAG6 hypervariable region from the most telomeric ESAG6 gene, was reamplified from the largest PCR fragment by nested-PCR with primers: 5′-CACTAATGATCAGCTTTACG-3′ and 5′-GACTCTTTTACACGTGAATC-3′. The resulting 1-kb fragment was purified and sequenced.
Cloning the expressed VSG
Total RNA was extracted from ∼108 G418-resistant cells with RNA STAT-60 reagent (TEL-TEST, Inc.), following the manufacturer's instructions. cDNA was synthesized using a PolyT-primer and the StrataScript™ First Strand Synthesis System (Stratagene). VSG cDNA was amplified with primers that bind to the spliced leader (5′-GACTAGTTTCTGTACTATAT-3′) and to a conserved sequence found in the 3′UTR of all VSG (5′-GTGTTAAAATATATC-3′). Using the spliced leader primer ensured that the amplified product originated only from trans-spliced RNA and not from a DNA contaminant. PCR products were subsequently cloned in pGEM-T Easy (Promega) and sequenced.
Chromosome separation by CHEF
T. brucei chromosomes were separated using a CHEF electrophoresis apparatus (CHEF-DR III, Bio-Rad), loading DNA from approximately 2×107 cells per lane. Electrophoresis conditions for the panel separating T. brucei intermediate chromosomes were: 25 s pulse time, 6 V/cm, for 20 h, in 0.5×TBE, at 14°C, using a gel of 1% high-strength agarose (Helena Biosciences). T. brucei megabase chromosomes were separated using 1400–700 s linear ramped pulse, 2.5 V/cm, for 144 h in 1×TBE at 14°C in a gel with 1.2% high-strength agarose.
Long-range Southern blotting
Agarose-embedded chromosome plugs were digested with 40 units of Apa I or Apa I and Xma I (New England Biolabs), as previously described [51]. Restriction products were separated on a 1.0% agarose gel in 0.5×Tris-Borate-EDTA buffer (TBE) in the Stratagene Rotating Agarose Gel Electrophoresis variant of Pulsed Field Gel Electrophoresis (PFGE) at 12°C and with a rotation angle of 120°. The running program for Apa I restriction fragments consisted of a 5 to 15 sec linear ramp at a constant 155 V for 18 h. For Apa I and Xma I restriction fragments the program was 1 to 4 sec linear ramp at a constant 150 V for 18 h. After alkaline transfer and hybridization with a radioactive probe, the Hybond-N+ Nylon membranes (Amersham Biosciences) were typically washed at 65°C as follows: twice for 20 min with 2×Sodium Chloride/Sodium Citrate (SSC), 0.1% SDS and twice for 20 min with 0.5×SSC, 0.1%SDS.
Comparison of phylogenetic signal among ESAGs
ClustalX was used to align homologs of 7 ESAG loci from 11 different expression sites [52]. The maximum likelihood phylogenetic tree topology was estimated for each locus using the program PHYML [53], with a GTR+Γ model and parameters estimated from the data. By constraining the resulting trees with the optimal topology of each other locus in turn a further six tree topologies were produced for each locus. The differences in likelihood value between the optimal tree and each constrained tree were calculated and the significance evaluated using the Shimodaira-Hasegawa test [54] thus quantifying the difference in phylogenetic signal between different loci along the expression site. Such likelihood comparisons require that two trees have equal taxon sets. For this reason a consistent gene complement from each expression site was used and ESAG10 and ESAG7 were therefore excluded. ESAG3 was also not used because it is present in three positions along the expression site and does not cluster by position; hence, its phylogenetic signal was not amenable to positional comparisons.
Characterization of recombination
Sequence alignments of each ESAG family and selected intergenic regions were generated with ClustalX as described above, manually edited and gap-stripped. Each alignment was tested for evidence of recombination using two approaches. (i) The likelihood-based method implemented in the program GARD ([31]; http://www.datamonkey.org/GARD) was used to identify putative recombination and gene conversion breakpoints, employing a nucleotide substitution model specified by the HyPhy software package ([55]; http://www.hyphy.org). (ii) The program Phi [32]; http://www.mcb.mcgill.ca/trevor) was used to assess the significance of the pairwise homoplasy index (Φw) with a window size of 100 bases and 10,000 permutations per test.
Adaptive evolution of ESAG sequences
ESAG protein-coding sequences were analysed for adaptive evolution by estimating the relative rates of non-synonymous and synonymous substitutions within each ESAG family. Positive selection is indicated by a non-synonymous/synonymous rate ratio (ω) greater than 1. The method was modified for recombinant sequences by allowing the genealogy to vary across the alignment while sharing the parameters of the codon substitution model between tracts [56]. The dimensions and genealogies of putative recombination tracts were provided by the GARD analyses described previously. HyPhy was used to obtain maximum likelihood estimates for two nested models of the distribution of ω. A likelihood ratio test for positive selection was performed with a χ2-distribution on two degrees of freedom by comparing twice the log-likelihood difference between a nearly-neutral model (M1a, in which ω cannot exceed 1) and a selection model (M2a) that included an additional category for ω>1 [57]. Evidence for adaptive evolution at individual codons was tested using an empirical Bayes method [57]: model M2a, with the maximum likelihood estimated parameters, was taken as a prior distribution for ω, and Bayes' formula was used to calculate the posterior distribution of ω at each codon, given the sequence data.
Supporting Information
PFGE Southern blotting.
(2.68 MB EPS)
Absence of ESAG7 from BES7.
(1.82 MB EPS)
Long-range restriction mapping of NEO-tagged clones.
(5.15 MB EPS)
Quantifying the difference in phylogenetic signal along the bloodstream expression site.
(1.97 MB EPS)
GARD analysis and interpretation using ESAG2 sequences as an example.
(5.49 MB EPS)
Locations of breakpoints inferred by GARD analysis of ESAG alignments.
(0.38 MB EPS)
Diversity and recombination within BES coding and intergenic sequences.
(0.45 MB EPS)
Non-synonymous substitution rates for ESAGs.
(0.35 MB EPS)
Positively selected codons in ESAG proteins.
(0.32 MB EPS)
Acknowledgments
We thank the core sequencing and informatics teams at the Wellcome Trust Sanger Institute for their assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The Wellcome Trust supports the Sanger Institute Pathogen Genomics group. LMF, MK and GAMC are supported in part by Grant Number R01AI021729 from the National Institute of Allergy And Infectious Diseases (NIAID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 2.Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- 3.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- 5.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Redpath MB, Windle H, Nolan D, Pays E, Voorheis HP, et al. ESAG11, a new VSG expression site-associated gene from Trypanosoma brucei. Mol Biochem Parasitol. 2000;111:223–228. doi: 10.1016/s0166-6851(00)00305-4. [DOI] [PubMed] [Google Scholar]
- 8.Pays E, Lips S, Nolan D, Vanhamme L, Perez-Morga D. The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts. Mol Biochem Parasitol. 2001;114:1–16. doi: 10.1016/s0166-6851(01)00242-0. [DOI] [PubMed] [Google Scholar]
- 9.Paindavoine P, Rolin S, Van Assel S, Geuskens M, Jauniaux JC, et al. A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol Cell Biol. 1992;12:1218–1225. doi: 10.1128/mcb.12.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligtenberg MJ, Bitter W, Kieft R, Steverding D, Janssen H, et al. Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. Embo J. 1994;13:2565–2573. doi: 10.1002/j.1460-2075.1994.tb06546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmon D, Hanocq-Quertier J, Paturiaux-Hanocq F, Pays A, Tebabi P, et al. Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. Embo J. 1997;16:7272–7278. doi: 10.1093/emboj/16.24.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoek M, Engstler M, Cross GA. Expression-site-associated gene 8 (ESAG8) of Trypanosoma brucei is apparently essential and accumulates in the nucleolus. J Cell Sci. 2000;113(Pt 22):3959–3968. doi: 10.1242/jcs.113.22.3959. [DOI] [PubMed] [Google Scholar]
- 13.Hoek M, Zanders T, Cross GAM. Trypanosoma brucei expression-site-associated-gene 8 protein interacts with a Pumilio family protein. Mol Biochem Parasitol. 2002;120:269–283. doi: 10.1016/s0166-6851(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 14.Gottesdiener KM. A new VSG expression site-associated gene (ESAG) in the promoter region of Trypanosoma brucei encodes a protein with 10 potential transmembrane domains. Mol Biochem Parasitol. 1994;63:143–151. doi: 10.1016/0166-6851(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 15.Florent IC, Raibaud A, Eisen H. A family of genes related to a new expression site-associated gene in Trypanosoma equiperdum. Mol Cell Biol. 1991;11:2180–2188. doi: 10.1128/mcb.11.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carruthers VB, Navarro M, Cross GA. Targeted disruption of expression site-associated gene-1 in bloodstream-form Trypanosoma brucei. Mol Biochem Parasitol. 1996;81:65–79. doi: 10.1016/0166-6851(96)02672-2. [DOI] [PubMed] [Google Scholar]
- 17.Morgan RW, El-Sayed NM, Kepa JK, Pedram M, Donelson JE. Differential expression of the expression site-associated gene I family in African trypanosomes. J Biol Chem. 1996;271:9771–9777. doi: 10.1074/jbc.271.16.9771. [DOI] [PubMed] [Google Scholar]
- 18.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 19.Becker M, Aitcheson N, Byles E, Wickstead B, Louis E, et al. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 21.Melville SE, Leech V, Navarro M, Cross GAM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol Biochem Parasitol. 2000;111:261–273. doi: 10.1016/s0166-6851(00)00316-9. [DOI] [PubMed] [Google Scholar]
- 22.Rudenko G, Chaves I, Dirks-Mulder A, Borst P. Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol Biochem Parasitol. 1998;95:97–109. doi: 10.1016/s0166-6851(98)00099-1. [DOI] [PubMed] [Google Scholar]
- 23.Pays E, Delauw MF, Van Assel S, Laurent M, Vervoort T, et al. Modifications of a Trypanosoma b. brucei antigen gene repertoire by different DNA recombinational mechanisms. Cell. 1983;35:721–731. doi: 10.1016/0092-8674(83)90105-8. [DOI] [PubMed] [Google Scholar]
- 24.Cross M, Taylor MC, Borst P. Frequent loss of the active site during variant surface glycoprotein expression site switching in vitro in Trypanosoma brucei. Molecular & Cellular Biology. 1998;18:198–205. doi: 10.1128/mcb.18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myler PJ, Aline RF, Jr, Scholler JK, Stuart KD. Multiple events associated with antigenic switching in Trypanosoma brucei. Mol Biochem Parasitol. 1988;29:227–241. doi: 10.1016/0166-6851(88)90078-3. [DOI] [PubMed] [Google Scholar]
- 26.Lamont GS, Tucker RS, Cross GAM. Analysis of antigen switching rates in Trypanosoma brucei. Parasitology. 1986;92:355–367. doi: 10.1017/s003118200006412x. [DOI] [PubMed] [Google Scholar]
- 27.Berriman M, Hall N, Sheader K, Bringaud F, Tiwari B, et al. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol. 2002;122:131–140. doi: 10.1016/s0166-6851(02)00092-0. [DOI] [PubMed] [Google Scholar]
- 28.Gottesdiener KM, Goriparthi L, Masucci JP, Van der Ploeg LH. A proposed mechanism for promoter-associated DNA rearrangement events at a variant surface glycoprotein gene expression site. Mol Cell Biol. 1992;12:4784–4795. doi: 10.1128/mcb.12.10.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansorge I, Steverding D, Melville S, Hartmann C, Clayton C. Transcription of ‘inactive’ expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol Biochem Parasitol. 1999;101:81–94. doi: 10.1016/s0166-6851(99)00060-2. [DOI] [PubMed] [Google Scholar]
- 30.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 32.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Molecular & Biochemical Parasitology. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- 34.Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haber JE. Chromosome breakage and repair. Genetics. 2006;173:1181–1185. doi: 10.1093/genetics/173.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 37.Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Van Xong H, et al. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 38.van Luenen HG, Kieft R, Mussmann R, Engstler M, ter Riet B, et al. Trypanosomes change their transferrin receptor expression to allow effective uptake of host transferrin. Mol Microbiol. 2005;58:151–165. doi: 10.1111/j.1365-2958.2005.04831.x. [DOI] [PubMed] [Google Scholar]
- 39.Barnes RL, McCulloch R. Trypanosoma brucei homologous recombination is dependent on substrate length and homology, though displays a differential dependence on mismatch repair as substrate length decreases. Nucleic Acids Res. 2007;35:3478–3493. doi: 10.1093/nar/gkm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barry JD, Marcello L, Morrison LJ, Read AF, Lythgoe K, et al. What the genome sequence is revealing about trypanosome antigenic variation. Biochem Soc Trans. 2005;33:986–989. doi: 10.1042/BST20050986. [DOI] [PubMed] [Google Scholar]
- 41.Sheader K, Vaughan S, Minchin J, Hughes K, Gull K, et al. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci U S A. 2005;102:8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudenko G, McCulloch R, Dirks-Mulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- 43.Jackson AP. Tandem gene arrays in Trypanosoma brucei: comparative phylogenomic analysis of duplicate sequence variation. BMC Evol Biol. 2007;7:54. doi: 10.1186/1471-2148-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dreesen O, Li B, Cross GA. Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol. 2007;5:70–75. doi: 10.1038/nrmicro1577. [DOI] [PubMed] [Google Scholar]
- 45.Johnson JG, Cross GA. Selective cleavage of variant surface glycoproteins from Trypanosoma brucei. Biochem J. 1979;178:689–697. doi: 10.1042/bj1780689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 47.Wirtz E, Hartmann C, Clayton C. Gene expression mediated by bacteriophage T3 and T7 RNA polymerases in transgenic trypanosomes. Nucleic Acids Res. 1994;22:3887–3894. doi: 10.1093/nar/22.19.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leech V, Quail MA, Melville SE. Separation, digestion, and cloning of intact parasite chromosomes embedded in agarose. Methods Mol Biol. 2004;270:335–352. doi: 10.1385/1-59259-793-9:335. [DOI] [PubMed] [Google Scholar]
- 49.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 50.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 51.Dreesen O, Cross GA. Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol Cell Biol. 2006;26:4911–4919. doi: 10.1128/MCB.00212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 55.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 56.Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- 57.Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aitcheson N, Talbot S, Shapiro J, Hughes K, Adkin C, et al. VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection. Mol Microbiol. 2005;57:1608–1622. doi: 10.1111/j.1365-2958.2005.04795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PFGE Southern blotting.
(2.68 MB EPS)
Absence of ESAG7 from BES7.
(1.82 MB EPS)
Long-range restriction mapping of NEO-tagged clones.
(5.15 MB EPS)
Quantifying the difference in phylogenetic signal along the bloodstream expression site.
(1.97 MB EPS)
GARD analysis and interpretation using ESAG2 sequences as an example.
(5.49 MB EPS)
Locations of breakpoints inferred by GARD analysis of ESAG alignments.
(0.38 MB EPS)
Diversity and recombination within BES coding and intergenic sequences.
(0.45 MB EPS)
Non-synonymous substitution rates for ESAGs.
(0.35 MB EPS)
Positively selected codons in ESAG proteins.
(0.32 MB EPS)