The liver plays an important role in adjusting metabolic processes to daily feeding–fasting cycles. This role is manifested by the circadian expression of many liver genes involved in the metabolism of lipids, proteins, carbohydrates, and xenobiotics (1–4). Moreover, the disruption or mutation of the essential clock genes Bmal1 and Clock results in various metabolic disorders (for review see ref. 5). Although these studies clearly demonstrate that BMAL1 and CLOCK are important regulators of metabolism, they do not address the question of whether the phenotypes have been caused by the loss of these genes or the loss of rhythms. However, in this issue of PNAS, Weitz and coworkers (6) present compelling evidence for the physiological importance of a functional liver clock. Using the Cre-loxP recombination strategy in mice, these authors inactivated Bmal1 either in all cells (Bmal1−/−) or specifically in hepatocytes (L-Bmal1−/−). Animals with a liver-specific Bmal1 disruption suffer from hypoglycemia specifically during the inactivity phase. Yet mice deficient for BMAL1 in all cells do not show overt problems with their resting blood sugar levels. Hence, in L-Bmal1−/− mice, the lack of liver rhythms rather than that of BMAL1 must have been the cause of impaired glucose homeostasis.
The circadian timing system has a hierarchical structure in that a master pacemaker in the brain's suprachiasmatic nucleus (SCN) synchronizes slave oscillators in nearly all body cells (1). Central and peripheral clocks have a similar molecular make-up, and both tick in a self-sustained and cell-autonomous fashion (7–9). The molecular clockwork relies on a negative feedback of gene expression (10). Two cryptochrome (CRY1 and CRY2) and two period proteins (PER1 and PER2) constitute the central part of the molecular oscillators. The genes encoding these proteins are activated by heterodimers of the transcription factors BMAL1 and CLOCK (or its paralog NPAS2). As a consequence, PER and CRY proteins accumulate in the cell and form heterotypic complexes, and once these have reached a critical threshold concentration (and/or activity) they annul the transactivation potential of BMAL1-CLOCK/NPAS2 heterodimers and thereby shut off transcription of their own genes. This results in a decrease of PER and CRY proteins below the level required for autorepression and a new daily PER–CRY accumulation cycle can ensue. The robustness of this transcription/translation feedback loop is augmented by cycles of posttranslational modifications that affect the activity and stability of PER and CRY proteins (10–12).
Owing to their intercellular connections, SCN neuron clocks maintain phase coherence indefinitely in vivo and for weeks in brain tissue explants (7). In contrast, hepatocyte oscillators or fibroblasts do not talk to each other and rapidly desynchronize in SCN-lesioned animals (13) or in tissue culture (9, 14). Hence, peripheral slave oscillators must be phase-entrained daily by the SCN master clock to run in harmony with each other. Feeding–fasting cycles are the dominant Zeitgebers for cellular clocks in liver and many additional peripheral organs (1). It therefore appears that under normal conditions the SCN timekeeper synchronizes subsidiary oscillators in the periphery in an indirect fashion. By driving rest–activity rhythms it also imposes feeding–fasting rhythms, which in turn set the phase in most peripheral cells through molecular signaling pathways yet to be discovered.
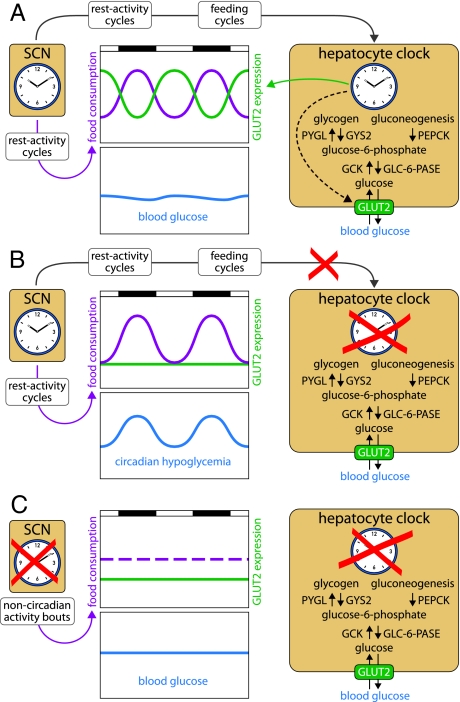
The synchronization of peripheral clocks by feeding time makes sense, because circadian metabolism is probably a major clock output in many if not most peripheral cell types. However, the work of Lamia et al. (6) revealed that feeding–fasting cycles also pose a challenge to the organism. Glucose plasma levels must be kept nearly constant throughout the day, to provide a constant source of fuel for brain cells and erythrocytes. During the absorptive phase carbohydrates are plentiful, and constant blood sugar levels can readily be adjusted through the insulin signaling pathway. High plasma glucose concentrations trigger the secretion of insulin by beta cells, which provokes the uptake of excessive glucose by liver and muscle and its polymerization into glycogen stores. During the postabsorptive phase, glucose is produced in the liver by glycogenolysis or gluconeogenesis and exported into the blood stream via the GLUT2 transporter. All these processes are under circadian control, but GLUT2-mediated export appears to be the rate-limiting step. In wild-type animals, the food-borne glucose supply largely meets the requirements during the absorptive phase, and a surge of GLUT2 expression associated with increased liver glucose export keeps blood sugar levels adequate during the postabsorptive phase (Fig. 1A). L-Bmal1−/− mice display normal rest–activity and feeding rhythms, because their SCN master clock is fully functional. However, these animals have disabled hepatocyte oscillators and express GLUT2 at constitutively low levels. Although gluconeogenesis and glycogenolysis still run at adequate rates in these animals, their liver cannot export sufficient amounts of glucose to keep the blood glucose level constant during the postabsorptive phase. The result is a temporally restricted hypoglycemia, despite acute mechanisms regulating glucose supply during the fasting phase, such as glucagon and glucocorticoid signaling (Fig. 1B). Bmal1−/− mice with disrupted Bmal1 alleles in all cells exhibit some phenotypes with regard to acute glucose regulation (ref. 15 and this paper). For example, they readjust normal blood glucose levels more sluggishly than wild-type mice after an i.v. glucose injection (glucose intolerance) and decrease blood glucose concentrations more dramatically after an i.v. insulin injection (insulin hypersensitivity). However, despite constitutively low hepatic GLUT2 expression, these animals have nearly normal resting blood glucose concentrations. Because Bmal1−/− mice are arrhythmic and ingest food in short temporal bouts throughout the day, the low hepatic levels of GLUT2 are supposedly sufficient for an adequate hepatic glucose delivery to the organism (Fig. 1C).
Fig. 1.
Hepatocyte oscillators drive the cyclic expression of many enzymes associated with glucose homeostasis, such as glycogen synthase (GYS2), glycogen phosphorylase (PYGL), phosphoenolpyruvate carboxykinase (PEPCK), glucokinase (GCK), glucose-6-phosphatase (GLC-6-Pase), and the glucose transporter GLUT2. (A) In wild-type animals, the SCN drives feeding rhythms and, thus, rhythms of glucose supply from nutrients. These cycles are counteracted by antiphasic rhythms of hepatic glucose export. (B) In L-Bmal1−/− mice, the floxed Bmal1 alleles have been disrupted only in hepatocytes. Because the SCN master clock is intact in these animals, their activity and feeding rhythms are normal. However, because the disabled liver clock can no longer compensate the lack of food-derived glucose by a burst of GLUT2 expression during the fasting phase, L-Bmal1−/− mice display a circadian hypoglycemia. (C) In Bmal1−/− mice, the circadian clocks are disabled in all tissues. These arrhythmic animals eat throughout the day, and the nutrient-derived glucose supply is nearly invariable over time. Although GLUT2 is expressed at low levels, it is sufficient to keep blood glucose at nearly normal (and invariable) concentrations.
Biological clocks have evolved to generate oscillations in physiology and behavior so as to anticipate and adapt to daily light–dark and temperature rhythms resulting from the rotation of the earth around its own axis. But for certain physiological parameters, such as blood glucose levels, large fluctuations are undesired. The study by Weitz and colleagues (6) provides compelling evidence for how the liver clock can counterbalance the oscillating sugar supply provoked by SCN-driven feeding–fasting cycles by governing antiphasic rhythms of hepatic glucose delivery. With their work, the authors have therefore established a convincing case for the importance of peripheral oscillators in modulating circadian physiology. A complication in assessing the function of autonomous peripheral clocks is that in one and the same tissue, cyclic gene expression can be driven by both local oscillators and/or rhythmic systemic timing cues (e.g., hormones, metabolites, body temperature rhythms, and inputs from the peripheral nervous system) (14). However, the dependence of oscillating Glut2 transcription on hepatocyte-derived BMAL1 strongly suggests that circadian glucose export is indeed driven by local liver clocks.
The further investigation of L-Bmal1−/− mice harboring Bmal1-null alleles specifically in hepatocytes is bound to unveil many additional physiological phenotypes. Indeed, rhythmically expressed liver genes revealed by genome-wide transcriptome profiling studies are associated with the metabolism of fatty acids, cholesterol, bile acids, amino acids, and xenobiotics (2–4, 16). It will be a challenging but enticing task to evaluate the contribution of liver clocks to the coordination of these various metabolic processes.
Footnotes
The authors declare no conflict of interest.
See companion article on page 15172.
References
- 1.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 2.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 4.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 5.Prasai MJ, George JT, Scott EM. Molecular clocks, type 2 diabetes and cardiovascular disease. Diab Vasc Dis Res. 2008;5:89–95. doi: 10.3132/dvdr.2008.015. [DOI] [PubMed] [Google Scholar]
- 6.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harb Symp Quant Biol. 2007;72:251–259. doi: 10.1101/sqb.2007.72.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Brewer JM, Lehman MN, Bittman EL. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: Effects of transplanting the pacemaker. J Neurosci. 2006;26:6406–6412. doi: 10.1523/JNEUROSCI.4676-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]