Abstract
We outline a powerful method for the directed evolution of integral membrane proteins in the inner membrane of Escherichia coli. For a mammalian G protein-coupled receptor, we arrived at a sequence with an order-of-magnitude increase in functional expression that still retains the biochemical properties of wild type. This mutant also shows enhanced heterologous expression in eukaryotes (12-fold in Pichia pastoris and 3-fold in HEK293T cells) and greater stability when solubilized and purified, indicating that the biophysical properties of the protein had been under the pressure of selection. These improvements arise from multiple small contributions, which would be difficult to assemble by rational design. In a second screen, we rapidly pinpointed a single amino acid substitution in wild type that abolishes antagonist binding while retaining agonist-binding affinity. These approaches may alleviate existing bottlenecks in structural studies of these targets by providing sufficient quantities of stable variants in defined conformational states.
Keywords: integral membrane proteins, protein engineering, protein folding
G protein-coupled receptors (GPCRs) comprise ≈1% of the genes in mammalian genomes and constitute ≈60% of all drug targets (1). Given the critical importance of this class of integral membrane proteins, there is great interest in having detailed structures of these molecules. However, in the Protein Data Bank (2), which contains ≈18,000 nonredundant protein structures, the structures of only two GPCRs have been deposited, that of bovine rhodopsin (3), uniquely facilitated by its natural abundance in the retina (4), and very recently that of β2 adrenergic receptor (5), whose high resolution structure determination required the replacement of a loop by a whole protein (6).
Because most GPCRs are normally produced at very low levels, an overexpression system must be set up for each GPCR of interest to obtain quantities sufficient for biophysical analyses. Many attempts have been made to increase functional membrane protein expression levels by testing various combinations of variables defining the expression system, such as host organism, expression plasmid, fusion adducts, codon usage, and expression induction conditions (7). However, even if all of these parameters could be globally optimized, they still may not yield enough functional material because of inherent limitations in the protein sequence itself. Furthermore, the limited stability of solubilized GPCRs is another bottleneck for their biophysical investigation, and this cannot be influenced by the expression system. Thus, while it may be possible to solve a limited number of new GPCR structures whose sequences are sufficiently accommodating to support such a brute-force screening for expression and solubilization conditions, this method is unlikely to have general applicability in routinely generating large amounts of stable, solubilized protein for any given GPCR.
To directly address the importance of receptor sequence as an experimental variable in membrane protein expression and stability, we have developed a powerful approach, inspired by periplasmic expression with cytometric screening (8) and anchored periplasmic expression (9), in which we evolve the sequence of a GPCR, keeping all other variables constant, to yield more functionally expressed protein in a convenient heterologous host, Escherichia coli. We used as a model system the rat neurotensin receptor-1 (NTR1), which has been shown to give a detectable yield in E. coli (10, 11) but which still needs to be improved to allow more convenient preparation of milligram quantities of receptor.
Detailed characterization of the best variant from the selection reported here reveals that it exhibits an order-of-magnitude increase in expression level in both E. coli and Pichia pastoris, elevated expression in mammalian cells, and enhanced stability in detergent-solubilized form, yet it largely retains the biochemical properties of WT NTR1, including binding affinity, binding selectivity, and G protein-mediated signaling. We have further extended this approach to isolate mutants of NTR1 with altered ligand selectivity. This methodology should thus be of general utility in the directed evolution of stable variants of such proteins to high-level expression in multiple states of activity.
Results and Discussion
Setup and Optimization of Screening Methodology.
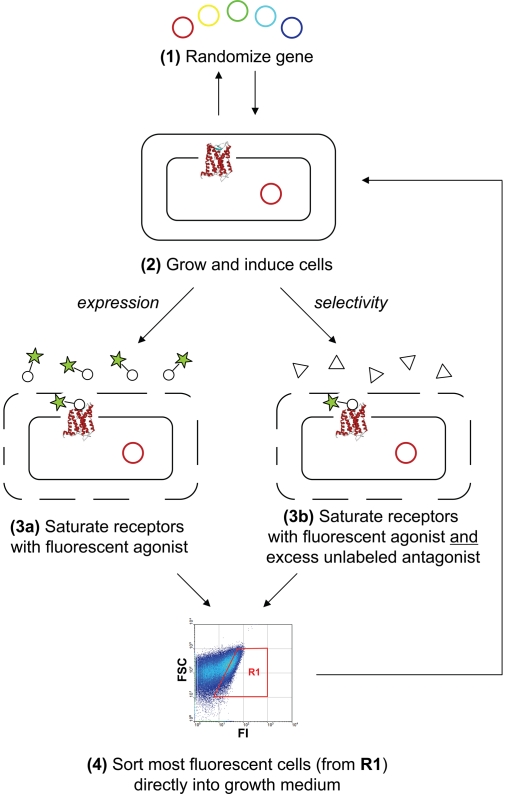
The general approach is given in Fig. 1. The expression vector containing the GPCR library of interest (e.g., from an error-prone PCR of the receptor gene) with two constant fusion partners (N-terminal maltose binding protein and C-terminal thioredoxin) is used to express the corresponding proteins in functional form in the inner membrane of E. coli DH5α (see Methods). After expression, cells are incubated at 4°C in an optimized buffer that renders the outer membrane permeable to small molecules to allow binding of fluorescent ligand to the receptors, and at the same time maximizes cell viability (see Methods). Chen and colleagues (8) have previously described conditions in which ligands as large as 10 kDa can enter the periplasmic space of E. coli without compromising cell viability. Here, the buffer was specifically optimized to allow for saturable, specific binding of fluorescently labeled agonist [BODIPY-NT(8–13)] to NTR1, while maximizing cell viability after fluorescence-activated cell sorting (FACS) [see supporting information (SI) Figs. S1–S4]. Although the expression conditions for WT NTR1 typically generate <1,000 functional receptors per cell (10, 11), optimization of the binding buffer and FACS gating conditions resulted in a specific signal in the gating window that was ≈900-fold above background (see Fig. S4).
Fig. 1.
General selection scheme for increasing expression level (steps 1, 2, 3a, 4, back to 2) and altering ligand selectivity (steps 1, 2, 3b, 4, back to 2).
After incubation with saturating concentrations of BODIPY- NT(8–13), bacteria expressing the largest number of functional receptors correspondingly exhibit the greatest fluorescence, and these cells were collected directly in growth medium and then expanded for a subsequent round. A single selection round, which consisted of library expansion, induced receptor expression, incubation with fluorescent ligand, and FACS to recover the most fluorescent bacterial cells, took approximately 1 day. The advantage of maintaining viable cells was that they could be immediately regrown after sorting, thus eliminating any preparative steps between selection rounds. Whenever additional diversity was desired after any FACS round, the sorted pool of cells was grown and harvested, the enriched plasmid collection was purified, the GPCR sequences (excluding the fusion partners) were further randomized, and fresh bacteria were transformed for the next selection. The flowchart for the selections on NTR1 is given in Fig. S5.
Selection of Variants with Increased Expression Level.
For increasing expression level, the initial randomized NTR1 library was subjected to four rounds of FACS. In each round, only the most fluorescent ≈0.1 to 1% of the cells were collected. Nonetheless, after these rounds, the evolved pool had a mean fluorescence intensity (MFI) no greater than that of the WT sequence. Error-prone PCR (epPCR) was used to overlay another set of random mutations on top of those that were enriched after the first four rounds of FACS, and this rerandomized library was again subjected to four rounds of sorting. In this second set of sorts, the MFI of the pool overtook that of WT NTR1. After a third randomization step followed by four more rounds of FACS, the evolved pool was split into two. One half was randomized by epPCR a fourth time and the other half was shuffled, using the staggered extension process (StEP) (12).
After these selections, the MFI was approximately five times that of WT NTR1. From the enriched pool, 96 single clones were sequenced and analyzed for receptor expression level (see Methods and Figs. S6 and S7). The clone with the best functional receptor expression level per cell, D03, exhibited approximately a 10-fold increase in specific signal, as assayed by [3H]-NT binding and flow cytometry (Table 1 and Fig. S8). D03 has 14 nucleotide substitutions scattered throughout its helices and loops (see Fig. S10 for a snake-like plot with the exact mutational positions). Five of these mutations are silent, suggesting that incorporation of nonsilent mutations was slow, approximately two amino acid substitutions per round of epPCR. This may be due to the seven-transmembrane helical topology of the protein, which limits the number and type of mutations that are possible, mandating a strategy with low mutational load.
Table 1.
Expression levels of NTR1, D03, and D03-L167R in multiple hosts
| NTR1 | D03 (fold of NTR1)* | D03-L167R (fold of NTR1)* | |
|---|---|---|---|
| E. coli,† no. per cell | 705 ± 101 | 6,155 ± 777‡ (8.7 ± 1.7) | 4,647 ± 879 (6.6 ± 1.6) |
| P. pastoris,§ pmol/mg | 18.3 ± 5.4 | 216.7 ± 36.9 (11.9 ± 4.1) | 51.1 ± 4.3 (2.8 ± 0.9) |
| HEK293T, no. per cell | 18,300 ± 1,300 | 58,800 ± 6,600 (3.2 ± 0.4) | 33,300 ± 5,400 (1.8 ± 0.3) |
*Parenthetical terms give the ratio of mutant expression to that of NTR1.
†Expression in 1 l cultures.
‡Expression in mutated vector pRGC7054G.
§Functional receptor (pmol) normalized to total membrane protein content (mg). NT binding cannot be measured with whole P. pastoris cells.
Evolved Receptor Retains Biochemical and Pharmacological Properties of WT.
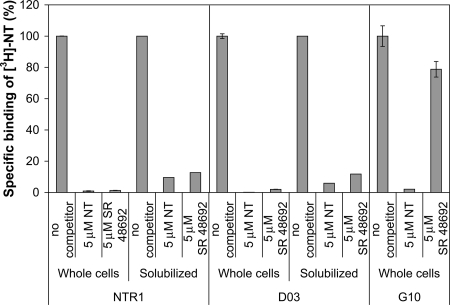
In the membrane of both E. coli and mammalian cells, D03 binds NT with affinities comparable to WT (Table 2), as determined by radioligand binding assays (see Methods). The radioactive signal is almost entirely competed away with a 50- fold excess of either unlabeled NT (agonist) or SR 48692 (antagonist) (13), suggesting that binding specificity is also faithfully retained (Fig. 2).
Table 2.
KD of full-length neurotensin binding to receptors on whole cells
| NTR1 | D03 | |
|---|---|---|
| E. coli | 0.14 ± 0.01 nM | 0.11 ± 0.01 nM |
| HEK293T | 3.7 ± 0.5 nM | 1.9 ± 0.2 nM |
Fig. 2.
Percentage radioligand binding to NTR1, D03 (evolved expression variant), and G10 (evolved selectivity variant) with unlabeled agonist (NT) and antagonist (SR 48692) competitors (used at 5 μM each). The radioligand was used at 10 nM. D03 retains the ligand binding properties of NTR1, whereas G10, which was evolved under selection pressure to abolish binding to SR 48692, no longer exhibits high affinity to antagonist.
We also compared the signaling properties of the evolved mutant to those of WT. NTR1 signals mainly via the Gq/11 subtype of G proteins (14), which triggers the mobilization of intracellular Ca2+ pools ([Ca2+]i) via phospholipase C (PLC)-generated IP3. The use of a fluorescent Ca2+ indicator, such as Fura 2, enables detection of variations in [Ca2+]i upon agonist binding to NTR1 (see Methods). Because of the different types of Ca2+ signaling patterns (oscillations, plateau, and transient; see Fig. S11), experiments with pooled cells cannot easily be evaluated, and we thus performed single-cell measurements of Ca2+ signaling in HEK293T cells transiently transfected with either WT or D03. It is well known that mutations in the highly conserved (D/E)R(W/Y) motif in helix III of Class A GPCRs can affect ligand binding and G protein coupling (15). D03 contains a substitution of Arg167 with Leu in this motif; this does not appear to influence the binding of NT to D03 (see Table 2), yet could potentially influence signaling. To check this, we also constructed a back mutant, D03-L167R, in which this important three amino acid motif was restored.
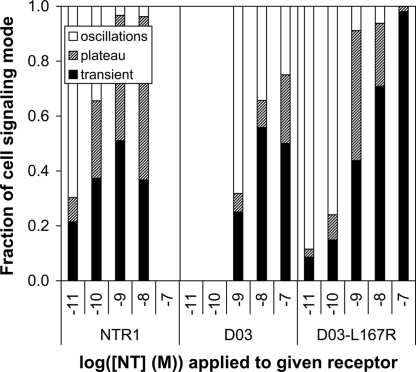
For WT NTR1, oscillations (the response at the lowest agonist concentration) were detected at 10−11 M NT, while for D03, these were only observed with a 100-fold higher concentration of agonist (Fig. 3), suggesting that the receptor can indeed still signal but that its coupling efficiency to Gq/11 is reduced. By contrast, the single amino acid revertant, D03-L167R, was capable of generating oscillatory behavior at the same minimal NT concentration (10−11 M) as WT. The concentration of NT (1 nM) needed to achieve strong transient and plateau responses (no oscillations) is the same for both WT and D03-L167R (see Fig. 3), while >100 nM agonist is needed for D03. For NTR1 and D03-L167R, the minimum concentration of agonist that leads to significant transient and plateau responses (> 40%) is similar to that at which the oscillatory response disappears, also approximately 1 nM, while it is again >100 nM for D03. These findings underscore the importance of the (D/E)R(W/Y) motif in G protein coupling (15). While these results have been obtained with constructs missing amino acids 1 to 42 of the GPCR, no major difference in signaling between the truncated and full-length forms was detected (see Fig. S12).
Fig. 3.
Ca2+ signaling in HEK293T cells expressing NTR1, D03, or D03-L167R. Single-cell measurements were performed and the resulting signal was classified as an oscillatory, plateau, or transient response (see Methods, SI Text, and Fig. S11).
That these evolved receptors can qualitatively mimic the signals generated by WT, including similar agonist concentration-dependent signaling of D03-L167R, is pleasantly surprising for two reasons: (i) the variants are significantly mutated (8–9 amino acid substitutions) and (ii) the ability to efficiently signal was never under the pressure of selection. More generally, this suggests that it is indeed possible to partially decouple the biophysical properties of a GPCR from its biochemical and pharmacological properties through mutagenesis, which thus enables the use of protein engineering approaches such as the present methodology to improve the robustness of these membrane proteins for structural studies.
Evolved Receptor Shows Increased Expression in Both Prokaryotic and Eukaryotic Hosts.
There are at least two conceivable mechanisms by which more functional D03 is obtained in E. coli as a result of selection: (i) the total amount of NTR1 and D03 per cell is comparable, but the fraction of properly folded and inserted D03 is significantly greater, or (ii) the total amount of D03 per cell is significantly greater than that of NTR1, but the fraction of functional receptor is similar. Whole-cell Western blots of E. coli proteins reveal that more D03 molecules are detected per cell (see Fig. S13). While this would be consistent with a greater rate of biosynthesis of D03, it more likely reflects the fact that noninserted and nonfunctional WT is degraded. Thus, the quantity of properly inserted, functional GPCR seems to correlate well with the total amount of receptor detected.
To determine whether D03 may have merely adapted to the biosynthetic pathway or the membrane of E. coli during selection or may have actually acquired traits of generally improved biophysical properties, we also expressed WT NTR1, D03, and D03-L167R in the methylotrophic yeast P. pastoris and compared functional and total expression yields. The functional expression level for D03, as assayed by specific radioligand binding to membrane preparations, was ≈12-fold higher than that for WT NTR1 (Table 1), while the improvement for the backmutated D03-L167R was more modest. This is in contrast to E. coli, where the effect of the back mutation was very small (Table 1). When total receptor protein levels in P. pastoris are compared by Western blot (Fig. 4), the increased expression of D03 is confirmed by the strong intensity of the band at ≈43 kDa (see Methods). In contrast, WT NTR1 shows a strong band that has not properly entered the gel, suggesting that this receptor is more aggregation-prone than the evolved D03 and D03-L167R. We found that the precursor form [unprocessed prepro- or pro-alpha factor fusion (16)] is detected at about equal intensity for WT and mutants (see Fig. S14), suggesting that protein biosynthesis is not changed, but that the expression level in P. pastoris is determined by a folding step after translocation and processing, further supporting the hypothesis that the biophysical properties have been improved.
Fig. 4.
Comparison of total GPCR yields in P. pastoris by immunoblot analysis. The C-terminal biotinylation was detected with a streptavidin-alkaline phosphatase conjugate. Three independent clones of each construct and a clone with an “empty” vector control (ctrl) integrated into the P. pastoris genome were analyzed (20 μg total membrane protein loaded per lane). Full-length, processed GPCR should run at the position of the arrow. The ≈60 kDa band present in all receptor samples represents the unprocessed precursor form (see Fig. S14). The ≈65 kDa band, which is more pronounced in the D03 samples, also reacts with both M1 and M2 anti-Flag antibodies (data not shown) and likely represents a compact dimeric form. The high molecular weight smear appears to be aggregated receptor, as it is diminished in D03 and D03-L167R. The band at ≈23 kDa, which also appears in the empty vector control, is a P. pastoris protein.
To test whether this improvement is also seen in mammalian cells, we transiently transfected HEK293T cells with NTR1 or D03. The evolved mutant was approximately threefold better expressed, as measured in whole-cell radioligand binding assays (see Table 1). Again, the effect of the back mutation L167R on expression is small (Table 1). Similar experiments in CHO cells and COS cells also revealed a two- to threefold increase in functional D03 expression relative to WT NTR1 (data not shown). In all of the experimentally tested hosts, neither the growth rate nor the final cell density was detectably different when expressing NTR1 and D03 in parallel cultures (data not shown). These results thus suggest that D03 is an intrinsically more robust protein in expressing and folding not only in E. coli, but also in multiple eukaryotic hosts. It should be noted that all receptors were expressed as fusion proteins with N-terminal maltose binding protein (MBP) and C-terminal thioredoxin (TrxA) fusions in E. coli, whereas these fusion partners were not present for expression in eukaryotic hosts (P. pastoris and HEK293T cells). Therefore, the similar trends in functional expression (Table 1) and biochemical affinity (Table 2) across different hosts suggest that the fusion partners used in bacteria are not involved in causing these evolutionary improvements.
Characterization of Solubilized and Purified Receptor.
To ascertain whether the observed enhancement in functional receptor expression in E. coli translates into a corresponding increase in pure, soluble material, we performed a side-by-side purification of the NTR1 and D03 fusion proteins (see Methods). Briefly, after detergent solubilization and purification by immobilized metal ion affinity chromatography (IMAC), the amount of functional GPCR was monitored at each step by a radiolabeled ligand-binding assay. As summarized in Table 3, the total amount of functional, purified D03 protein recovered from a 1 l expression is ≈6-fold that obtained for NTR1. A comparison of the gel filtration profiles for WT and D03 reveals almost identical elution behavior (see Figs. S17 and S18).
Table 3.
Purification of NTR1 and D03
| NTR1, pmol | D03, pmol | D03/NTR1* | |
|---|---|---|---|
| Whole cell material | 2,300 | 15,800 | 6.9 |
| Solubilized material | 1,700 ± 80 | 11,500 ± 1250 | 6.8 |
| IMAC eluate | 950 ± 5 | 6,020 ± 175 | 6.3 |
*Ratio of purification yields.
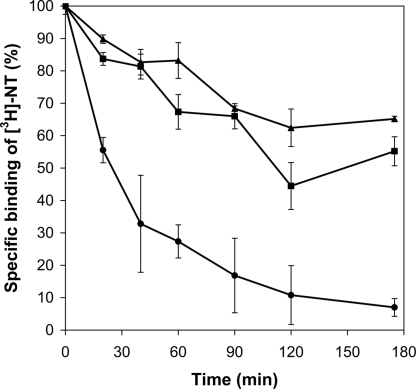
We then investigated whether the evolved receptors were also more stable in solubilized form. Solubilized and purified NTR1, D03, and D03-L167R were incubated at 45°C and the remaining activity was measured, after cooling, at various time points (Fig. 5). D03 and D03-L167R (in which the highly conserved DRY motif is restored) were both found to be significantly more thermostable than NTR1, suggesting that evolution with selection for increased functional expression favored proteins with improved biophysical properties. This implicit correlation between stability and functional expression level has been noted by others as well (17); thus, the present platform may serve to generate more stable variants of a given receptor.
Fig. 5.
Kinetics of thermal denaturation of purified NTR1 (●), D03 (▲), and D03-L167R (■) as fusion proteins with maltose binding protein and thioredoxin. Equal amounts of each receptor were incubated at 45°C and specific activities were measured at various time points by saturation binding assays (see Methods). While NTR1 retains only ≈7% of its specific binding activity after 175 min, the evolved receptors D03 and D03-L167R retain ≈65% and ≈55% of their respective activities after the same interval of time. Results are duplicate measurements from a representative experiment.
Tracing the Effects of Single Substitutions.
Why is D03 better expressed and more stable? To understand the contribution of each of the mutations in D03 in enhancing expression level, a systematic, two-pronged site-directed mutagenesis strategy was used: (i) each of the substitutions in D03 was individually introduced into the WT sequence, and (ii) each of the existing substitutions in D03 was individually reverted back to the corresponding WT nucleotide. The first approach should elucidate which individual mutations can substantially increase the expression level on their own, whereas the second approach should reveal any additional mutations that also may be beneficial but whose effect may be masked by unfavorable WT amino acids. For the 28 variants that were generated in this manner, the receptor expression level was determined by saturation radioligand binding (see Fig. S19) and the impact of each single mutation—whether added to WT or subtracted from D03—is relatively small. We refer to this phenomenon as a “staircase effect,” because the improved phenotype of D03 is the sum of many incremental enhancements that leads to an increase in expression level.
In addition to the R167L mutation discussed previously, other unusual substitutions in NTR1 and one unintended mutation in the expression vector arose during the evolution of D03. Five of the nucleotide substitutions in NTR1 were silent, but two of these surprisingly introduced rare leucine codons (CTA) into the sequence (see Fig. S10). While it was recently shown that a synonymous single-nucleotide polymorphism in the MDR1 gene can change the conformation of the corresponding protein because of altered kinetics of cotranslational folding and insertion (18), in our case none of the rare codons appears to alter protein expression (see Fig. S19) (although we cannot rigorously exclude that they might subtly influence protein conformation by altering biosynthesis kinetics). This result suggests that codon optimization or the use of rare tRNA-overexpressing strains may not be fruitful if other, inherently larger bottlenecks exist in heterologous protein expression.
Lastly, an unintentional mutation arose in the origin of replication in the plasmid harboring D03. This mutation (C7054G, for numbering see Fig. S15), close to the origin of replication and close to a previously identified mutation influencing the copy number (19), increases the copy number of the plasmid by 100%, as determined by measuring the total amount of DNA from plasmid isolation experiments from standard expressions at 20°C (see Fig. S16). However, when D03 is expressed in the mutated pRG vector (pRGC7054G), its expression level increases by only ≈25% as compared to the original vector (pRGWT), and when WT NTR1 is expressed in the mutated vector, there is even no noticeable increase in its expression level (see Fig. S16). We hypothesize that the major bottlenecks in functional protein expression for WT NTR1 are cotranslational folding and insertion and, therefore, any other improvements along the expression pathway remain masked.
Selection of Variants with Altered Ligand Selectivity.
For selections of ligand selectivity on NTR1 (see Fig. 1), two ligands were used, agonist BODIPY-NT(8–13) and antagonist SR 48692. In principle, selectivity selections could be performed independently of selections for expression level. However, in the case of NTR1, the FACS signal of the WT receptor was so weak with BODIPY- NT(8–13) alone, that the addition of excess SR 48692 would drop the signal of any positive clones into the background. Thus, the selectivity screen was only used after an initial selection for expression level to ensure that the MFI of the pool was significantly above background (after the fourth epPCR; see Fig. S5). Sequencing of the library before the fourth epPCR revealed expression-specific mutations; thus, any mutations that were enriched after the fourth epPCR in this screen could be readily identified as selectivity mutations and not expression mutations.
After performing selections for NTR1 sequences that still bound BODIPY-NT(8–13) in the presence of 100-fold excess SR 48692, 96 single clones were sequenced and analyzed for receptor expression level. The best expressing clone, G10, exhibited approximately a fivefold higher expression level than the WT, could be fully competed by NT, but could not be fully competed by 100-fold excess SR 48692 (see Fig. 2 and Fig. S9).
In stark contrast to the staircase effect observed with D03 and other expression variants, there was only one consensus mutation observed in G10 and other selectivity variants. This “elevator effect” in changing selectivity arose from a single mutation, F358S (see Fig. S10). The effects of mutation at Phe-358 have been studied by others (20), and reveal that a substitution to alanine at this position results not only in decreased antagonist affinity, but also in spontaneous basal inositol phosphate production in a receptor-dependent manner (21). Based upon a sequence alignment with bovine rhodopsin, Phe-358 may play an important role in maintaining the interaction between transmembrane helices 6 and 7, which in turn keeps NTR1 in an inactive conformation (21). Disruption of this residue leads to the observed constitutive activity. Thus, more generally, the present methodology has the ability to rapidly isolate mutations that may trap receptors in the active or inactive state, an approach that is complementary to previous work in engineering GPCR selectivity and activity (22, 23).
Conclusion
There have been recent advances in membrane protein engineering, including technologies for the screening of high-expressing members in a diverse pool of eukaryotic membrane proteins (24), identification of functionally critical amino acids in a GPCR (25–27), manual blot screening of randomly mutated membrane proteins for increased expression (28), and introduction of thermostabilizing mutations, individually identified by trial and error, in a GPCR (29). Complementary to such approaches, we present here a powerful, high-throughput platform for the directed evolution of a GPCR to enhance both expression level and stability while retaining function and to tailor ligand selectivity. This methodology should be applicable to other integral membrane proteins as well, for which specific binding ligands are available, and may help to facilitate biophysical studies by allowing milligram-level production of these proteins in multiple states of activity. Importantly, no basal heterologous expression of a WT GPCR sequence is necessary for the approach to work, as long as there exist expressing mutants that can be recovered by FACS. In the present study, a highly expressing, stable NTR1 variant displays WT biochemical properties—as assayed by binding affinity, binding selectivity, and G protein mediated signaling—and therefore provides a biologically meaningful template for structural studies. Such evolved membrane proteins may facilitate X-ray crystallography trials not only because they can be produced more abundantly but also because they remain functional in detergent micelles for significantly longer periods of time. If the proteins are more stable because of enhanced rigidity, they may also be more likely to generate properly diffracting crystals for structural determinations. The ability to obtain such high-resolution structures may help to elucidate the molecular basis for activation, inactivation, or pathology associated with that receptor, and may also provide templates for drug design.
Methods
Library Design and Selection.
The rat neurotensin receptor-1 gene (NTR1; amino acids 43–424) has previously been expressed in E. coli using a vector that generates an N-terminal fusion of the receptor to MBP and C-terminal fusion to TrxA to enhance expression. The fusion protein contains tobacco etch virus (TEV) protease cleavage sites on both ends of the receptor and a C-terminal His10 tag. This derivative of the expression vector pRG/III-hs-MBP containing the NTR1 gene with these fusions was a kind gift from R. Grisshammer (National Institutes of Health). Expression of NTR1 in E. coli DH5α was essentially as described in ref. 30. Details of the preparation of fluorescently labeled neurotensin [BODIPY-NT(8–13)] and construction of NTR1 libraries are given in the SI Text. To allow binding of BODIPY-NT(8–13) to NTR1 in the inner membrane of E. coli while maximizing cell viability, an optimized binding buffer (50 mM Tris·HCl, pH 7.4, and 150 mM KCl) was identified (see SI Text). During each round of FACS, only the most fluorescent ≈0.1 to 1% of the cells in the population (≈107–108 cells) were recovered during sorting for regrowing and further selection. Individual cells from the final selections were sorted directly into 96-well plates during FACS and regrown to perform single clone analysis of expression levels by radioligand binding assays.
Radioligand Binding Assays.
Details of the experimental protocols are given in the SI Text. Briefly, quantitative measurements of receptor number in E. coli, P. pastoris, HEK293T cells, and detergent solution were performed using a saturating concentration of radioactive agonist [3H]-NT (10 nM) (Perkin-Elmer). For determining equilibrium binding affinities, a dilution series of radioligand was used (0.04–20 nM). Nonspecific binding was determined in the presence of 5 μM unlabeled NT. For competition experiments, antagonist SR 48692 (Sanofi Aventis) was used at a concentration of 5 μM.
Mammalian Cell Culture and Transfection.
For expression in mammalian cells, receptors were cloned into the vector pcDNA3.1 (Invitrogen) encoding C-terminal Myc and His6 tags. The HEK293T cell line (31) (a clonal line of HEK293 cells stably expressing SV40 large T antigen) was grown as described in ref. 32. Cells were routinely seeded into 6-well culture plates or 10 mm Petri dishes and grown for 24 h, reaching 70 to 80% confluence before transfection. Cells for binding assays were transiently transfected with DNA using calcium phosphate precipitation as described in ref. 32.
Single-Cell Monitoring of Variations in Intracellular Free Calcium, [Ca2+]i.
Changes in [Ca2+]i in response to NT were measured in individual HEK293T cells using the indicator Fura2-acetoxymethyl ester, as described in ref. 33. Different types of Ca2+ responses were observed and were classified as transient, oscillatory, or plateau (see SI Text).
Expression in Pichia pastoris.
The P. pastoris strain SMD1163 (Invitrogen) was used for all experiments. A modified version of the yeast shuttle vector pPICZαC (Invitrogen) was previously designed in which the Saccharomyces cerevisiae alpha-factor prepro sequence (34), under the control of the AOX promoter, was followed by a Flag-M2 tag, a His10 tag, a TEV cleavage site, the GPCR, the linker sequence EFELGTRGS, and a biotin acceptor (BioAcc) domain (SwissProt P02904, amino acids 50–123). As a negative control in all experiments, the plasmid without GPCR insert was used. Each expression vector was integrated in the P. pastoris genome under the control of the AOX1 promoter, and three independent clones of each GPCR construct and a negative vector-only control were analyzed. Yeast cultures were incubated at 22°C at 250 rpm for 15 h after induction, harvested by centrifuging the cultures at 1,500 × g at 4°C for 10 min, and the cells were resuspended in 5 ml TBS containing 1% protease inhibitor mixture (Sigma-Aldrich) and stored at −80°C. Details of membrane preparations are given in the SI Text.
Immunoblot Analysis.
For immunoblot analysis, yeast membranes (≈20 μg of total membrane protein) were diluted with TBS to a concentration of 4 μg/μl and an equivalent volume of 2× SDS loading buffer was added. The samples were incubated at 42°C for 30 min in the presence of 10 mM DTT before separating the proteins by SDS/PAGE (4–12% Bis-Tris gels; Invitrogen). Proteins were transferred to Immobilon-P transfer membranes (Millipore), and membranes were blocked in TBST (TBS with 0.5% Tween-20) with 5% milk powder for 1 h at room temperature. The C-terminal biotinylation was detected with a streptavidin-alkaline phosphatase conjugate (Roche Diagnostic GmbH). While the expected size of the GPCR is ≈55 kDa, results from N-terminal sequencing and from in-gel digestions followed by mass spectrometry of the P. pastoris construct showed that the band at ≈43 kDa indeed corresponds to the full-length GPCR, correctly processed at the N terminus (data not shown).
Receptor Solubilization and Purification.
This was performed essentially as published by Grisshammer and Tucker (30). The detailed protocol is provided in the SI Text.
Thermal Stability.
Receptors were expressed as fusion proteins (MBP-GPCR-TrxA-His10) in E. coli and purified by IMAC and size exclusion chromatography (Superdex 200). Thermal stability was assayed in buffer STAB30 [50 mM Tris•HCl, pH 7.4, 30% glycerol, 200 mM NaCl, 1 mM EDTA, 0.05% dodecyl-β-d-maltopyranoside (DDM), 0.5% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS), 0.1% (wt/vol); cholesteryl hemisuccinate (CHS)]. Samples were incubated at 45°C for the indicated period (up to 175 min) and were then put on ice. Radioligand binding assays were performed as described in the SI Text.
Supplementary Material
Acknowledgments.
The authors are grateful to Eva Niederer for invaluable assistance with flow cytometry, to Dr. Reinhard Grisshammer (National Institutes of Health) for helpful discussions and sharing vectors, and to Dr. Randy Schekman (University of California Berkeley) for antiprepro alpha factor antibodies. This work was supported by a National Institutes of Health postdoctoral fellowship (to C.A.S.), a predoctoral fellowship from the For schungskredit of the University of Zurich (to I.D.), and by grants from the Transregio Program and from the Swiss National Science Foundation (NCCR in Structural Biology) (to A.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 14747.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803103105/DCSupplemental.
References
- 1.Lundström K. Structural genomics of GPCRs. Trends Biotechnol. 2005;23:103–108. doi: 10.1016/j.tibtech.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Okada T, et al. X-ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J Struct Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarramegna V, Talmont F, Demange P, Milon A. Heterologous expression of G-protein-coupled receptors: Comparison of expression systems from the standpoint of large-scale production and purification. Cell Mol Life Sci. 2003;60:1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, et al. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS) Nat Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- 9.Harvey BR, et al. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc Natl Acad Sci USA. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grisshammer R, Duckworth R, Henderson R. Expression of a rat neurotensin receptor in Escherichia coli. Biochem J. 1993;295:571–576. doi: 10.1042/bj2950571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tucker J, Grisshammer R. Purification of a rat neurotensin receptor expressed in Escherichia coli. Biochem J. 1996;317:891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 13.Gully D, et al. Biochemical and pharmacological profile of a potent and selective nonpeptide antagonist of the neurotensin receptor. Proc Natl Acad Sci USA. 1993;90:65–69. doi: 10.1073/pnas.90.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisshammer R, Hermans E. Functional coupling with Gαq and Gαi1 protein subunits promotes high-affinity agonist binding to the neurotensin receptor NTS-1 expressed in Escherichia coli. FEBS Lett. 2001;493:101–105. doi: 10.1016/s0014-5793(01)02281-5. [DOI] [PubMed] [Google Scholar]
- 15.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: Beyond the ground state. Mol Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 16.Julius D, Schekman R, Thorner J. Glycosylation and processing of prepro-α-factor through the yeast secretory pathway. Cell. 1984;36:309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- 17.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 18.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 19.Lin-Chao S, Chen WT, Wong TT. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol. 1992;6:3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 20.Labbe-Jullie C, et al. Mutagenesis and modeling of the neurotensin receptor NTR1: Identification of residues that are critical for binding SR 48692, a nonpeptide neurotensin antagonist. J Biol Chem. 1998;273:16351–16357. doi: 10.1074/jbc.273.26.16351. [DOI] [PubMed] [Google Scholar]
- 21.Barroso S, Richard F, Nicolas-Etheve D, Kitabgi P, Labbe-Jullie C. Constitutive activation of the neurotensin receptor 1 by mutation of Phe358 in helix seven. Br J Pharmacol. 2002;135:997–1002. doi: 10.1038/sj.bjp.0704546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ault AD, Broach JR. Creation of GPCR-based chemical sensors by directed evolution in yeast. Protein Eng Des Sel. 2006;19:1–8. doi: 10.1093/protein/gzi069. [DOI] [PubMed] [Google Scholar]
- 23.Sommers CM, et al. A limited spectrum of mutations causes constitutive activation of the yeast α-factor receptor. Biochemistry. 2000;39:6898–6909. doi: 10.1021/bi992616a. [DOI] [PubMed] [Google Scholar]
- 24.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, et al. Rapid identification of functionally critical amino acids in a G protein-coupled receptor. Nat Methods. 2007;4:169–174. doi: 10.1038/nmeth990. [DOI] [PubMed] [Google Scholar]
- 26.Scarselli M, Li B, Kim SK, Wess J. Multiple residues in the second extracellular loop are critical for M3 muscarinic acetylcholine receptor activation. J Biol Chem. 2007;282:7385–7396. doi: 10.1074/jbc.M610394200. [DOI] [PubMed] [Google Scholar]
- 27.Sommers CM, Dumont ME. Genetic interactions among the transmembrane segments of the G protein coupled receptor encoded by the yeast STE2 gene. J Mol Biol. 1997;266:559–575. doi: 10.1006/jmbi.1996.0816. [DOI] [PubMed] [Google Scholar]
- 28.Molina DM, et al. Engineering membrane protein overproduction in Escherichia coli. Protein Sci. 2008;17:673–680. doi: 10.1110/ps.073242508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grisshammer R, Tucker J. Quantitative evaluation of neurotensin receptor purification by immobilized metal affinity chromatography. Protein Expr Purif. 1997;11:53–60. doi: 10.1006/prep.1997.0766. [DOI] [PubMed] [Google Scholar]
- 31.DuBridge RB, et al. Analysis of mutation in human cells by using an Epstein–Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott D, Frischknecht R, Plückthun A. Construction and characterization of a kappa opioid receptor devoid of all free cysteines. Protein Eng Des Sel. 2004;17:37–48. doi: 10.1093/protein/gzh004. [DOI] [PubMed] [Google Scholar]
- 33.Vermeiren C, et al. Loss of metabotropic glutamate receptor-mediated regulation of glutamate transport in chemically activated astrocytes in a rat model of amyotrophic lateral sclerosis. J Neurochem. 2006;96:719–731. doi: 10.1111/j.1471-4159.2005.03577.x. [DOI] [PubMed] [Google Scholar]
- 34.Talmont F, Sidobre S, Demange P, Milon A, Emorine LJ. Expression and pharmacological characterization of the human μ-opioid receptor in the methylotrophic yeast Pichia pastoris. FEBS Lett. 1996;394:268–272. doi: 10.1016/0014-5793(96)00971-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.