Abstract
The intrinsic antimicrobial resistance of the opportunistic human pathogen Pseudomonas aeruginosa is compounded in mutant strains that overexpress multidrug efflux pumps such as the prominent drug-proton antiporter, MexAB-OprM. The primary regulator of the mexAB-oprM operon is the MarR family repressor, MexR. An additional repressor, NalC, also regulates mexAB-oprM by controlling expression of ArmR, an antirepressor peptide that is hypothesized to prevent the binding of MexR to its cognate DNA operator via an allosteric protein–peptide interaction. To better understand how ArmR modulates MexR, we determined the MexR-binding region of ArmR as its C-terminal 25 residues and solved the crystal structure of MexR in a 2:1 complex with this ArmR fragment at 1.8 Å resolution. This structure reveals that the C-terminal residues of ArmR form a kinked α-helix, which occupies a pseudosymmetrical and largely hydrophobic binding cavity located at the centre of the MexR dimer. Although the ArmR-binding cavity partially overlaps with the small molecule effector-binding sites of other MarR family members, it possesses a larger and more complex binding surface to accommodate the greater size and specific physicochemical properties of a peptide effector. Comparison with the structure of apo-MexR reveals that ArmR stabilizes a dramatic conformational change that is incompatible with DNA-binding. Thus, this work defines the structural mechanism by which ArmR allosterically derepresses MexR-controlled gene expression in P. aeruginosa and reveals important insights into the regulation of multidrug resistance.
Keywords: gene regulation, MarR, mexAB-oprM, PA3719, protein peptide
Bacterial multidrug efflux pumps confer widespread antibiotic resistance by actively purging their cells of chemically diverse xenobiotics. The opportunistic human pathogen Pseudomonas aeruginosa possesses a number of these efflux systems, including at least 10 belonging to the resistance nodulation division (RND) superfamily (1, 2). The intrinsic resistance conferred by these pumps is compounded in P. aeruginosa because of a synergy of heightened drug efflux and low outer membrane permeability (3). Composed of three components (an inner membrane drug/proton antiporter, an outer membrane channel, and a periplasmic adapter), RND efflux systems appear to extrude noxious substrates by using a rotating peristaltic pump-like mechanism (4, 5). The best-characterized efflux system in P. aeruginosa is MexAB-OprM, which displays an expansive substrate profile that includes not only antibiotics and biocides, but organic solvents, dyes, detergents, and homoserine lactones involved in quorum sensing (3). Although pentachlorophenol exposure appears to up-regulate mexAB-oprM (6), expression of these efflux genes is generally considered constitutive in wild-type (WT) strains. However, mutations in any of three regulatory genes, mexR (7, 8), nalC (9, 10), or nalD (11), have been shown to cause hyperexpression of mexAB-oprM and increased resistance to various medically relevant antimicrobials (11, 12).
The mexAB-oprM operon is primarily regulated by the MarR family repressor, MexR, although the TetR family repressor, NalD, provides additional regulation at a weaker promoter (13). MexR forms a homodimer of 147 aa per subunit (total molecular mass of ≈34 kDa) and recognizes two operator sites that overlap promoters for both mexR and mexAB-oprM (14, 15). A second TetR family repressor, NalC, indirectly influences mexAB-oprM expression by repressing production of ArmR, a 53-aa antirepressor that appears to bind MexR at the exclusion of cognate DNA (9, 16). MexR has been shown to be modulated by a peptide effector, whereas all other MarR family effectors known to date are small hydrophobic (typically phenolic) compounds (17). The signal responsible for alleviating NalC repression in WT P. aeruginosa is currently unknown.
The MarR family of transcriptional regulators is widely distributed in bacteria and archaea and control various biological processes, including resistance to antimicrobials, sensing of aromatic xenobiotics, and virulence (18). Whereas MarR proteins are poorly conserved in amino acid sequence, they share a common fold that consists of a helical dimerization domain and two winged helix (or winged helix–turn–helix) DNA-binding domains (19). By conserving structure while varying amino acid sequence, the different members of this family have apparently diverged to recognize a large variety of signaling molecules and DNA targets. The crystal structure of MexR and other MarR family proteins have been determined (19–25), but detailed mechanistic information is currently limited by a paucity of structures of MarR family members in complex with their effectors.
Mutational analysis of MexR suggests that ArmR may act allosterically to competitively prevent DNA binding by interacting with MexR at a separate location from its DNA binding site (16). To better understand how interaction with ArmR alleviates MexR repression of mexAB-oprM, we determined the 1.8 Å crystal structure of the MexR double mutant Q106L/A110L (MexRLL) in a 2:1 complex with an ArmR fragment containing residues 29–53 (ArmRC). This structure reveals that a single ArmRC molecule occupies a pseudosymmetrical and largely hydrophobic binding cleft within the center of the MexRLL dimer. This effector binding site is separate from the MexR–DNA interface and partially overlaps with the small molecule effector-binding sites recently identified in other MarR family members. Comparison with the crystal structure of apo-MexR reveals that ArmR stabilizes a tertiary and quaternary conformational change that allosterically repositions the MexR DNA-binding lobes into an orientation that is incompatible with DNA binding. Thus, this structure defines the mechanism by which ArmR allosterically derepresses MexR-controlled gene expression in P. aeruginosa and provides important insights into the regulation of multidrug resistance.
Results and Discussion
Crystal Structure of the MexRLL–ArmRC Complex at 1.8-Å Resolution.
Initial attempts to crystallize the MexR–ArmR complex produced poor-quality crystals that were recalcitrant to standard optimization. Several strategies were pursued to improve diffraction quality, including truncating both MexR and ArmR, and engineering a hydrophobic crystallization epitope on the surface of MexR to generate additional crystal contacts (26). In our search for better diffracting crystals, we discovered that MexR preferentially protects ArmR residues 29–53 from proteolytic degradation. Analyses by NMR spectroscopy and isothermal titration calorimetry (ITC) revealed that this sequence is predominantly unstructured in isolation, albeit with helical propensity, and binds MexR with the same affinity as full-length ArmR [details provided in supporting information (SI) Text and Figs. S1 and S2]. Well ordered crystals were finally obtained after co-crystallization of this ArmR fragment (denoted ArmRC) with MexRLL (MexR truncated by five C-terminal residues and engineered with a Q106L/A110L crystallization epitope). The structure of the MexRLL–ArmRC complex was solved to 1.8-Å resolution by molecular replacement and refined to a Rwork/Rfree of 17.6%/22.9%.
Architecture of the MexRLL–ArmRC Complex.
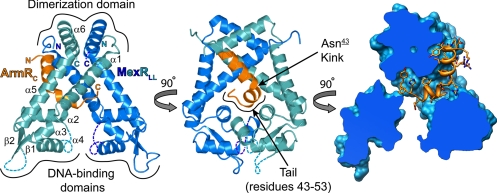
The structure of MexR closely resembles other members of the MarR family and consists of two winged helix DNA-binding domains, each linked to a helix-rich dimerization domain by a pair of long helices (25). As previously shown by ITC (16), ArmRC binds MexRLL in a 1:2 stoichiometry (Fig. 1). Consistent with NMR-based secondary structure propensities (see SI Text and Fig. S1), bound ArmRC forms an α-helix spanning residues 32–49 with a severe 75° kink at Asn-43 (underlined numbers indicate ArmRC residues). ArmRC associates with MexRLL between its DNA-binding and dimerization domains with residues C-terminal to the kink in the ArmRC helix (the C-terminal tail) buried between the two subunits of the MexRLL dimer. Of the two engineered leucines, only Leu-110 is involved in crystal contacts, forming a hydrophobic patch with His-107 and symmetry-related residues Val-126 and Ala-129. As expected from their native-like DNA and effector binding affinities (Fig. S3), these mutations are distant from both ArmRC and the DNA-binding domains.
Fig. 1.
The crystal structure of the MexRLL–ArmRC complex. Cartoon of the MexRLL dimer (blue) in complex with ArmRC (orange) and cross-section of MexRLL (blue surface) with ArmRC shown in ribbon and stick representation (C, N and O atoms in orange, blue and red, respectively). Disordered segments are indicated with dashed lines.
Interactions between ArmRC and MexRLL.
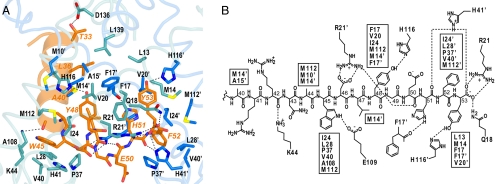
The high affinity of MexR for ArmR [Kd = 160–290 nM (16)] is reflected in the crystal structure of the MexRLL–ArmRC complex, which reveals an extensive set of interactions focused around the C-terminal tail of ArmRC (Fig. 2). These interactions are largely hydrophobic and include the aromatic ArmRC sidechains of Trp-45, Tyr-48, Phe-52, and Tyr-53. Each of these side chains fits into one of four corresponding hydrophobic pockets (respectively denoted I, II, I′, and II′), which are arranged in a 2-fold symmetrical fashion within the interior of the MexRLL dimer. The I and I′ hydrophobic pockets are comprised of a nearly identical set of residues from monomer chains A and B, respectively, including Ile-24, Leu-28, Pro-37, Val-40, and Met-112. The presence of Leu-28 and Met-112 in the ArmRC binding pocket explains why the mutation of either of these residues was previously shown to compromise ArmR binding in vivo (16). The II and II′ hydrophobic pockets are made up of residues from both chains of MexRLL, but likewise share a common set of residues, including Phe-17 from both subunits and Val-20 and Met-14. There are also a number of prominent polar interactions between ArmRC and MexRLL, including 4 hydrogen bonds to the backbone of ArmRC and 11 hydrogen bonds and salt bridges to ArmRC side chains. As with the hydrophobic interactions, identical polar residues from both chains of MexRLL are observed making similar interactions with residues in ArmRC. The side-chain imidazole of His-116 hydrogen bonds with the side-chain hydroxyl of Tyr-48 and the side-chain imidazole of His-116′ (where ′ denotes residues from MexR chain B) hydrogen-bonds with the side-chain hydroxyl of Tyr-53. Additionally, the guanidinium group of Arg-21 makes an electrostatic interaction with the side-chain carboxylate of Asp-46 and the guanidinium group of Arg-21′ makes an electrostatic interaction with the backbone carboxylate of the ArmRC C terminus.
Fig. 2.
Interactions between MexRLL and ArmRC. (A) Stick representation of ArmRC in its binding site (MexRLL C atoms in blue, ArmRC C atoms in orange; O, N and S atoms in red, dark blue and yellow, respectively). Hydrogen bonds are shown as dashed lines and ArmRC labels are italicized and colored orange to distinguish them from MexRLL (black labels; nonprimed, chain A; primed, chain B). (B) Schematic map of interactions between MexRLL and residues 40–53 of ArmRC, showing hydrogen bonds and salt bridges by dashed lines. MexRLL residues involved in hydrophobic contacts are listed in boxes.
N-terminal to the kink at Asn-43, the ArmRC helix is amphipathic and associates with the surface of MexRLL via a relatively weak hydrophobic interface. The most prominent interactions in this interface include van der Waal contacts between Leu-36 and MexR residues Leu-139 and Ala-15′ as well as Ala-40 and MexR residues Ala-15′ and Met-14′. In addition, the Thr-33 hydroxyl hydrogen-bonds with the Asp-136 carboxylate and the backbone carbonyl of Arg-42 hydrogen-bonds with Lys-44. The opposite face of the ArmRC helix is highly charged in this region because of the presence of six arginines, an aspartate, and a glutamate, all of which project their side chains into solvent. Deletion of ArmR residues 1–40 does not compromise MexR binding under ITC conditions (Table S1) and thus the majority of these interactions do not appear to contribute significantly to stabilizing the MexR–ArmR complex.
Physicochemical Symmetry Within ArmR.
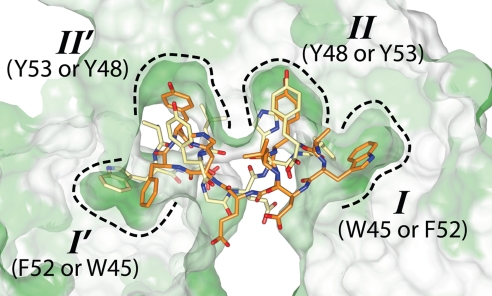
Because of the 2-fold symmetry of the MexR dimer, ArmR can potentially bind MexR in one of two mutually exclusive orientations. As only one complex was observed per asymmetric unit, only one orientation of ArmRC is observed (at full occupancy and with refined temperature factors similar to MexR). These two sites cannot be occupied simultaneously because they overlap in the interior region of MexR that binds the hydrophobic ArmR tail. This observation not only explains the stoichiometry of the native MexR–ArmR complex, but also reveals a surprising capacity for structural pseudosymmetry in the C-terminal tail of ArmR despite an absence of symmetry in its amino acid sequence. Approximating the two orientations of ArmRC by swapping the subunits of the MexRLL–ArmRC complex shows that ArmRC residues Trp-45 and Phe-52 occupy equivalent positions, as do residues Tyr-48 and Tyr-53 (Fig. 3). Consequently, the 2-fold symmetry of the MexR dimer is largely preserved in the resulting complex because of interactions with physicochemically similar aromatic residues in ArmRC. Indeed, the two MexRLL subunits superimposed with rmsd of 1.1 Å2 for all main-chain atoms.
Fig. 3.
ArmR can bind MexR in two mutually exclusive orientations. Regardless of the orientation, the four hydrophobic pockets of MexRLL (dashed outlines labeled I, II, I' and II') are filled by aromatic ArmRC residues, revealing physicochemical symmetry within the ArmRC C terminus. MexRLL is shown as a cross-sectioned hydrophobic surface where green represents relative hydrophobicity.
ArmR-Stabilized Conformational Change.
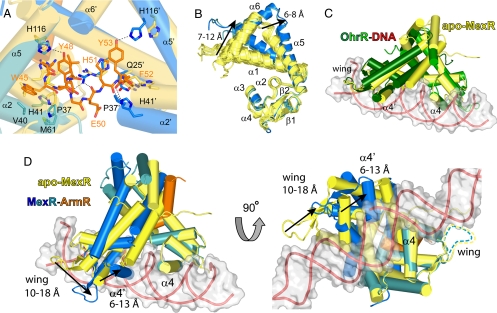
The intrinsic conformational flexibility of MexR and other members of the MarR family is well established (17). This flexibility is largely caused by the plasticity of loops within the dimerization domain and the hydrophobic nature of the dimerization interface. Moreover, whereas the winged helix DNA-binding domains are relatively rigid entities, the flexibility of the dimerization domain permits various spacing between these two DNA-binding lobes. The previously solved crystal structure of apo-MexR (25) provides an excellent example of this flexibility, displaying four separate conformations of the MexR dimer (denoted AB, CD, EF, and GH) within the asymmetric unit of the crystal. These apo conformations yield inter-DNA domain spacing ranging from 23 to 29 Å (Cα-Cα distance between Arg-73 and Arg-73′, i.e., the middle of the recognition helices α4 and α4′).
An examination of the structure of each apo-MexR dimer reveals that a conformational rearrangement is required to provide sufficient space for the bulky ArmR tail. This necessity is clearly shown by overlapping the dimerization domains of MexRLL-ArmRC and apo-MexR, which reveals that severe steric clashes would occur between ArmRC and the N-terminal residues of α2 and α2′ if positioned as in apo-MexR (Fig. 4A). To accommodate ArmRC-binding, each MexRLL subunit must undergo an α5 bend of ≈20° to move the C terminus of this helix (i.e., Ala-121) by 6–8 Å (depending on the particular apo-MexR dimer used for comparison). In addition, α1 pivots at Thr-22 to displace its N terminus (i.e., Asp-8) by 7–12 Å (Fig. 4B). These movements increase the distance between the dimerization and the DNA-binding domains of MexRLL and thereby open the I and I′ hydrophobic pockets. Concomitantly, the DNA-binding domains of ArmRC-bound MexRLL twist with respect to the dimerization domain to produce a sheared orientation that shrinks the distance between the N termini of the α4/α4′ recognition helices from 15–20 Å for apo-MexR to 9 Å for the MexRLL–ArmRC complex (Cα-Cα distance between Leu-67 and Leu-67′). These movements displace one domain relative to the other by 10–18 Å for the tip of the wing (i.e., Ser-88) and 6–13 Å for the midpoint of the recognition helix (i.e., Arg73; Fig. 4D).
Fig. 4.
The binding of ArmRC stabilizes a MexRLL conformation that is incompatible with binding of DNA. (A) Steric clashes prevent binding of ArmRC to apo-MexR, demonstrated by superimposing the dimerization domains of apo-MexR dimer CD (yellow cylinders, PDB ID 1LNW) and MexRLL-ArmRC [blue cylinders with ArmRC shown as sticks coloured by atom type (C, orange; N, blue; oxygen, red; sulfur, yellow)]. (B) Superimposition of the eight chains from the crystal structure of apo-MexR (yellow ribbons) and the two ArmRC-bound MexRLL subunits (blue) reveals the conformational change due to ArmRC binding. (C) Superimposition of the DNA-binding domains of apo-MexR dimer CD (PDB ID 1LNW; yellow cylinders) and DNA-bound OhrR from Bacillus subtillus (PDB ID 1Z9C; green cylinders with red ribbons) indicates a highly similar DNA-binding conformation for MexR. (D) Superimposing apo-MexR dimer CD (yellow cylinders), as a model for DNA-bound MexR, with the DNA-binding domain of MexRLL-ArmRC (blue and orange cylinders) reveals that the latter cannot bind DNA due to severe steric clashes.
Mechanism of Antirepression.
Of the four conformations available for the apo-MexR dimer, the widest spacing between the DNA-binding lobes was observed in dimer CD (Cα-Cα distance of 29 Å between Arg-73 and Arg-73′). Considering the spacing between major grooves in regular B-DNA is similar (34 Å), it was previously proposed that apo-MexR dimer CD resembles the DNA-bound conformation of MexR (25). Additionally, the shortest spacing was observed in dimer AB (Cα-Cα distance of 23 Å between Arg-73 and Arg-73′), which was found to bind the C-terminal residues of another MexR molecule in the asymmetric unit. Based on these observations, it was speculated that a protein or peptide effector could modify the spacing of the DNA-binding domains and thereby regulate mexAB-oprM expression by preventing binding of MexR to its cognate DNA (25). It is now clear that the effector binding site predicted from the AB dimer of apo-MexR partially overlaps with the experimentally observed ArmR binding cleft. The side chains of Asp-146 and Ile-147 from the apo-MexR C terminus appear to reasonably mimic the backbone carboxylate and side chain of the ArmRC C terminus (i.e., Phe-53). Despite these common interactions, however, the DNA-binding domains of apo-MexR dimer AB did not adopt the sheared orientation that we observe in the MexRLL–ArmRC complex.
Currently, the only structure of a MarR family member in complex with its cognate DNA is OhrR from Bacillus subtilis [BsOhrR (23)]. The binding of BsOhrR to its pseudopalindromic DNA operator was shown to induce a global bend of 10° and a slight under-twisting of the otherwise regular B-form DNA. These conformational changes shorten the spacing requirement between the DNA-binding lobes of BsOhrR from 34 Å (to fit regular B-DNA) to 31 Å (Cα-Cα distance between Lys-76 and Lys-76′). In fact, similar protein-induced DNA conformational changes may be typical of the MarR family, as suggested by studies using atomic force microscopy with ExpG (27) and circular dichroism spectroscopy with HucR (28). These studies support the proposal that apo-MexR dimer CD may closely resemble the DNA-bound conformation. Indeed, a comparison of the structures for apo-MexR dimer CD and DNA-bound BsOhrR (Protein Data Bank ID code 1Z9C) reveals that the orientation of their DNA-binding lobes is highly similar (rmsd of 2.2 Å2 for MexR Cα atoms corresponding to DNA-binding residues 37–99; Fig. 4C). This conclusion is consistent with the structures of reduced apo-OhrR from Xanthomonas campestris [XcOhrR (20)], HucR (21), and ST1710 (22), all of which have been observed in conformations that are preconfigured for DNA binding.
Using the structures of MexRLL–ArmRC and apo-MexR dimer CD as models for the effector- and DNA-bound states of MexR, respectively, an allosteric mechanism for the antirepression of MexR can now be proposed. The C-terminal tail of ArmR binds into a cavity between MexR subunits and stabilizes a sheared orientation of the DNA-binding lobes. Superimposition of one DNA-binding domain from apo-MexR dimer CD with the MexRLL–ArmRC complex reveals a displacement of the α4′ recognition helix by 13 Å (measured from the Cα of Arg-73′), which would result in severe steric clashes with the DNA backbone (Fig. 4 and Movie S1). Moreover, the DNA-binding wing is displaced by as much as 18 Å (measured from the Cα of Ser-88′) from its likely position in the DNA minor groove. The wings of BsOhrR were observed making numerous minor groove interactions in the crystal structure of DNA-bound BsOhrR (23), and mutational analysis has established the importance of MexR wing residues Arg-83 and Arg-91 to DNA binding (29). Finally, Glu-50 of bound ArmRC projects into the gap between the two MexR DNA-binding domains, which places a negatively charged carboxylate group in close proximity to the helix–helix motif, a third DNA-binding element identified in the BsOhrR-DNA structure that primarily uses positively charged residues to bind the negatively charged DNA backbone. As such, we predict Glu-50 is specifically positioned to provide electrostatic repulsion of the DNA backbone in this region, further weakening the affinity of the MexR repressor for its cognate DNA. Taken together, these features define an allosteric mechanism of MexR regulation in which ArmR and DNA binding is mutually exclusive, in accordance with the absence of a ternary complex after combination of MexR, ArmR, and cognate DNA (16).
Comparison with Other MarR Family Members.
To date, MexR is the only MarR family member known to bind a peptide effector. The ligands for the remaining MarR family members are lipophilic (typically phenolic) compounds such as salicylate or uric acid (17). In several cases, these lipophenolic effectors are organic hydroperoxides that induce structural changes through cysteine oxidation. For example, the crystal structures of reduced and oxidized XcOhrR recently revealed that a nearly perpendicular arrangement of the DNA-binding lobes results from the formation of two intersubunit disulfide bonds (20). This oxidized conformation bears no resemblance to the sheared orientation of the MexRLL–ArmRC complex despite the fact that the oxidizable XcOhrR cysteines occupy locations that are proximal to several elements of the ArmR-binding site in MexR. A number of other MarR family members, such as BsOhrR and MgrA, contain a single oxidizable cysteine and are unlikely to form disulfide bonds (23, 30). The oxidized conformation of these MarR family members remains to be determined, but without added stabilization from disulfides, the perpendicular orientation of XcOhrR is highly unlikely (20). Instead, overoxidation of the reactive cysteine or the formation of either cyclic sulfenamides or protein-effector mixed disulfides (31) is expected to stabilize rearrangements of the DNA-binding lobes, potentially via an ArmR-like mechanism involving steric clashes with residues residing between the dimerization and DNA-binding domains.
The majority of MarR family members, such as MexR, MarR, and HucR, do not contain oxidizable cysteines and are instead regulated by the noncovalent binding of low molecular weight ligands. In the first structure of a MarR family member, salicylate (cocrystallized with MexR at a high concentration of 250 mM) was observed bound to Escherichia coli MarR in two sites (SAL-A and SAL-B) positioned on either side of the α4 recognition helix (19). This structure suggested a mode of regulation in which DNA-binding is prevented by steric occlusion of the DNA–protein interface. However, the physiological significance of either effector binding site could not be confirmed because the SAL-A salicylate was involved in crystal contacts and the SAL-B salicylate was highly solvent-exposed. Interestingly, the conformation of MarR in the MarR–salicylate crystal structure closely matches that of MexRLL–ArmRC, superimposing to 2.8 Å2 rmsd (for 225 common Cα residues). This similarity includes not only the relative positions of the recognition helices, but also much of the cavity corresponding to the ArmR binding site. Despite a lack of conserved residues in this region, the presence of a cavity indicates a potential effector binding site. Indeed, recent structures of a MarR family member from Methanobacterium thermoautotrophicum, MTH313, revealed two unique salicylate binding sites, one of which corresponds to the oxidizable cysteine (i.e., Cys-15) in BsOhrR (24). These binding sites appear to be mutually exclusive between the two subunits of the MTH313 dimer, producing an asymmetrical conformational change that enlarged the spacing of the DNA-binding lobes relative to apo-MTH313. From these data, it was postulated that the apo form of MTH313 (with narrow spacing) represents the “active” conformation (capable of DNA binding) and the salicylate-bound structure (with wide spacing) represents the “inactive” conformation (incapable of DNA binding). These conclusions are puzzling as they contradict the wide spacing observed in the structure of DNA-bound BsOhrR (23). In fact, the salicylate-bound MTH313 structure appears well configured for DNA binding, superimposing with the structure of DNA-bound BsOhrR to 2.3 Å2 rmsd (for 238 common Cα residues). Moreover, the salicylate-bound E. coli MarR structure is clearly ill suited for binding a DNA double helix as its DNA-binding lobes share an orientation that is similar to that of the MexRLL–ArmRC complex and its DNA recognition helices are even linked by a pair of salt bridges (19). Regardless, one of the MTH313 salicylate-binding sites appears to partially overlap with the ArmRC-binding site of MexRLL, occupying a small pocket between the α1 and α2′ helices. This position situates salicylate directly between the dimerization and DNA-binding domains of MTH313, in a location that is proximal to the I/I′ hydrophobic pockets of MexRLL and the oxidizable cysteines of OhrR and MgrA. In contrast to the MTH313 salicylate-binding sites, MexRLL possesses a larger and more complex binding surface to accommodate the greater size and specific physicochemical properties of a peptide effector. Despite these differences, the various members of the MarR family appear to share a similarly positioned effector-binding cavity, suggesting that the MarR family may possess significant mechanistic similarities as well.
The structure of MexRLL in complex with ArmRC reveals the allosteric mechanism responsible for derepressing production of MexAB–OprM, the major multidrug efflux pump in P. aeruginosa. This structure also adds to our understanding of an important family of bacterial transcriptional regulators, many members of which are involved in antimicrobial resistance and virulence. Such knowledge may prove vital in controlling the rise in drug-resistant bacterial infections.
Materials and Methods
Protein Production.
The MexRLL–ArmRC complex was produced by coexpression of MexRLL [vector pET41a (Novagen), MexR residues 1–142 with point mutations Q106L and A110L] and ArmRC [vector pTYB12 (New England Biolabs), ArmR residues 29–53 (i.e., ARRDYTEQLRRAARRNAWDLYGEHFY)] in BL21 Star (DE3) E. coli (Invitrogen). The intact complex was purified via three chromatographic steps: (i) chitin affinity/intein tag cleavage (New England Biolabs), (ii) Mono-Q anion exchange (Amersham Biosciences), and (iii) superdex-75 gel filtration (GE Healthcare). Using an Amicon Ultra-15 5K concentrator (Millipore), the pure complex (in 20 mM Tris·HCl, 150 mM NaCl, 5 mM TCEP-HCl, pH 7.5) was concentrated to 9.4 mg/ml as estimated with a predicted ε280 = 12,950 M−1·cm−1 (33). More details are provided in SI Text.
Crystallization and Structure Determination.
Crystals of the MexRLL–ArmRC complex were obtained by screening 480 microbatch conditions at 8°C and 18°C by using the Oryx 6 Crystallization Robot (Douglas Instruments). The highest quality crystals grew as clusters from a mixture of 0.5 μl of protein at 5 mg/ml and 0.5 μl of 20% (wt/vol) PEG 3000, 0.1 M sodium citrate (pH 5.5) after 2 days under paraffin oil at 18°C. Single crystals were separated, cryoprotected in 20% (wt/vol) PEG 3000, 10% (wt/vol) PEG 1000, and 0.1 M sodium citrate (pH 5.6) and flash-frozen in N2(l).
Data were collected to 1.8 Å at beamline 8.2.2 of the Advance Light Source (Berkeley, CA) and processed with the program HKL2000 (34). Despite the dramatic conformational differences between apo-MexR and the MexRLL–ArmRC complex, the structure was successfully phased by molecular replacement. To accomplish this, it was necessary to use only the dimerization domain of apo-MexR (residues 3–31 and 105–139) as an initial search model in the program Phaser (35). The position of the dimerization domain was then fixed and the DNA-binding domain (residues 35–98) was used as a search model in the program Molrep (36). The structure was built by using the program Coot (37) and refined by using the programs Phenix (38) and Refmac (39) with a final R/Rfree of 17.6%/22.9%, respectively. Data collection and refinement statistics are given in Table S2. Graphics were prepared with Pymol (40) and Chimera (41).
Supplementary Material
Acknowledgments.
We thank Dr. Raz Zarivach for assistance with x-ray data collection and the U.S. Department of Energy and the Advanced Light Source (Berkeley, CA) for access to beamline 8.2.2. The work was funded by the Howard Hughes Medical Institute, Canadian Institutes of Health Research, Michael Smith Foundation for Health Research, Canadian Foundation for Innovation, and the University of British Columbia Blusson Fund. M.S.W. was supported by the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research. M.H. was supported by the Alexander von Humboldt Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ECH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805489105/DCSupplemental.
References
- 1.Mima T, Sekiya H, Mizushima T, Kuroda T, Tsuchiya T. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol Immunol. 2005;49:999–1002. doi: 10.1111/j.1348-0421.2005.tb03696.x. [DOI] [PubMed] [Google Scholar]
- 2.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J Bacteriol. 2007;189:7600–7609. doi: 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poole K, Srikumar R. Multidrug efflux in. Pseudomonas aeruginosa: Components, mechanisms, and clinical significance. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 4.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 5.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 6.Muller JF, Stevens AM, Craig J, Love NG. Transcriptome analysis reveals that multidrug efflux genes are up-regulated to protect Pseudomonas aeruginosa from pentachlorophenol stress. Appl Environ Microbiol. 2007;73:4550–4558. doi: 10.1128/AEM.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito K, Yoneyama H, Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol Lett. 1999;179:67–72. doi: 10.1111/j.1574-6968.1999.tb08709.x. [DOI] [PubMed] [Google Scholar]
- 8.Srikumar R, Paul CJ, Poole K. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-oprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao L, Srikumar R, Poole K. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: Identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol Microbiol. 2004;53:1423–1436. doi: 10.1111/j.1365-2958.2004.04210.x. [DOI] [PubMed] [Google Scholar]
- 10.Llanes C, et al. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother. 2004;48:1797–1802. doi: 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobel ML, Hocquet D, Cao L, Plesiat P, Poole K. Mutations in. PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutoille D, et al. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing β-lactam resistance. FEMS Microbiol Lett. 2004;230:143–146. doi: 10.1016/S0378-1097(03)00882-6. [DOI] [PubMed] [Google Scholar]
- 13.Morita Y, Cao L, Gould VC, Avison MB, Poole K. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol. 2006;188:8649–8654. doi: 10.1128/JB.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans K, Adewoye L, Poole K. MexR repressor of the. mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: Identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K, Eda S, Maseda H, Nakae T. Molecular mechanism of MexR-mediated regulation of MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2001;195:23–28. doi: 10.1111/j.1574-6968.2001.tb10492.x. [DOI] [PubMed] [Google Scholar]
- 16.Daigle DM, et al. A protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol. 2007;189:5441–5451. doi: 10.1128/JB.00543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 18.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3-A resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 20.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J Mol Biol. 2006;360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Kumarevel T, et al. Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7. J Struct Biol. 2008;161:9–17. doi: 10.1016/j.jsb.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: High-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum. J Mol Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the. mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 26.Yamada H, et al. “Crystal lattice engineering,” an approach to engineer protein crystal contacts by creating intermolecular symmetry: Crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines. Protein Sci. 2007;16:1389–1397. doi: 10.1110/ps.072851407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumgarth B, Bartels FW, Anselmetti D, Becker A, Ros R. Detailed studies of the binding mechanism of the Sinorhizobium meliloti transcriptional activator ExpG to DNA. Microbiology. 2005;151:259–268. doi: 10.1099/mic.0.27442-0. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson SP, Grove A. Negative cooperativity of uric acid binding to the transcriptional regulator HucR from Deinococcus radiodurans. J Mol Biol. 2005;350:617–630. doi: 10.1016/j.jmb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 29.Saito K, Akama H, Yoshihara E, Nakae T. Mutations affecting DNA-binding activity of the MexR repressor of mexR-mexA-mexB-oprM operon expression. J Bacteriol. 2003;185:6195–6198. doi: 10.1128/JB.185.20.6195-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen PR, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 31.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soonsanga S, Lee JW, Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol Microbiol. 2008;20:131–141. doi: 10.1111/j.1365-2958.2008.06200.x. [DOI] [PubMed] [Google Scholar]
- 33.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J App Crystallgr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagin AA, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallgr. 1997;30:1022–1025. [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project No. 4. The CCP4 suite programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.