Abstract
The importance of core histones in the regulation of DNA function by chromatin is clear. However, little is known about the role of the linker histone. We investigated the role of H1 in Saccharomyces cerevisiae during extensive transcriptional reprogramming in stationary phase. Although the levels of linker histone Hho1p remained constant during growth to semiquiescence, there was a genome-wide increase in binding to chromatin. Hho1p was essential for compaction of chromatin in stationary phase, but not for general transcriptional repression. A clear, genome-wide anticorrelation was seen between the level of bound Hho1p and gene expression. Surprisingly, the rank order of gene activity was maintained even in the absence of Hho1p. Based on these findings, we suggest that linker histone Hho1p has a limited role in transcriptional regulation and that the dynamically exchanging linker histone may be evicted from chromatin by transcriptional activity.
Keywords: H1, nucleosome, yeast
The DNA in eukaryotes is packaged into chromatin, which is formed by arrays of a nucleoprotein complex, the nucleosome. The nucleosome is composed of ≈168 bp of DNA spooled as two negative superhelical coils onto a central octamer composed of two copies of each of the H2A–H2B and H3–H4 heterodimeric pairs. The repeating association of DNA with the histone octamer in chromatin renders the structure sensitive to the properties of the core histones, which are tailored by reversible chemical modifications, and by isotype permutations in octamer composition. A fifth histone, linker histone H1, binds to the outside of the nucleosome, contacting the nucleosomal DNA at positions close to the center and the ends of the superhelical gyres (1). Although this central binding of the linker histone to the nucleosome suggests an important role in the regulation of DNA function, many lower eukaryotes remain viable in the absence of a linker histone (2, 3). The continuous dynamic exchange of this histone has cast doubt on the canonical organization of the nucleosome, where the linker histone is accepted as a static structural feature (4).
The involvement of H1 in the condensation of the chromatin fiber is well established. H1 was shown to be required for the salt-dependent compaction of a nucleosome array into a regular 30-nm chromatin fiber in vitro (5). This compaction of chromatin, which appears to also involve the N-terminal tail of histone H4 (6), was proposed to be due to the steady-state level of the dynamically exchanging H1 molecule.
Given the connection between H1 binding and chromatin compaction, which limits unimpeded access to the DNA molecule, H1 was expected to influence global DNA function, and, in particular, gene expression, in a simple, direct manner. Surprisingly, absence of Hho1p in yeast did not result in an increase in basal transcription, as was expected for a global transcriptional repressor (3). Microarray analysis of transcription in a yeast strain in which the HHO1 gene was deleted, revealed that <1% of genes were affected by a factor of 2-fold or more (7). Similar results were obtained in mouse (8). H1 therefore appears to influence the expression of only a subset of genes.
Taken together, studies of H1 in a range of organisms suggest that linker histones are abundant and associated with the genome. Although a vast array of biochemical data show that linker histones facilitate chromatin condensation, the functional consequence of this activity in the cell is not entirely clear. The differentiation of cells in higher eukaryotes, where several closely related H1 isotypes exist, is associated with extensive chromatin remodeling (9) and a change in the expression profile of the H1 isotypes. We were therefore interested in investigating the relationship between linker histone binding and gene expression on a genome-wide scale in a cell undergoing widespread transcriptional reprogramming.
Saccaraomyces cerevisiae responds to nutrient starvation by exiting the cell cycle and entering stationary phase, exhibiting very low metabolic activity, low levels of gene expression, and low rates of protein synthesis (10). Here, we report on the role of the single, unique yeast linker histone in the extensive transcriptional changes that accompany entry and exit of the semiquiescent stationary phase in yeast.
Results
Hho1 Level Remains Constant into Stationary Phase.
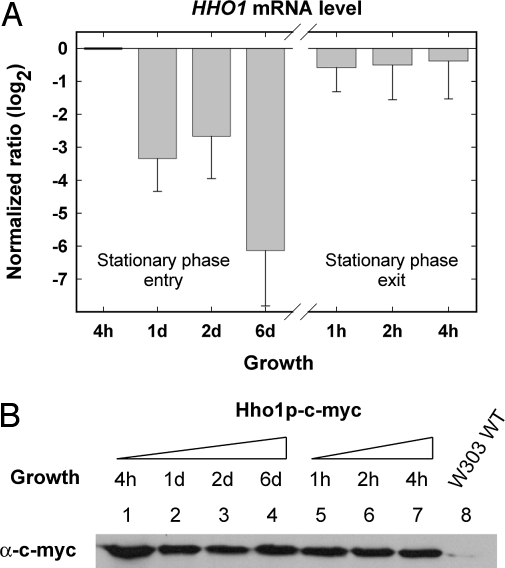
To elucidate the role of histone Hho1p in yeast cells during the extensive transcriptional reprogramming associated with growth to semiquiescent stationary phase, we incubated yeast cultures for 6 d, followed by reintroduction into rich growth media to allow reentry of the cell cycle. It was previously reported that S. cerevisiae entered semiquiescence only after 5 d of incubation (11). The steady-state level of HHO1 mRNA and total cellular Hho1p were determined at discrete time points during entry and exit of stationary phase (Fig. 1 A and B). Quantitative RT-PCR revealed that HHO1 mRNA decreased up to 250-fold at day six in stationary phase compared with the level during exponential growth (Fig. 1A). With reentry of the cell cycle, the HHO1 mRNA levels rapidly recovered to the level observed in logarithmically growing yeast cells. We next looked at the level of the Hho1 protein, making use of a strain expressing a single c-myc-tagged linker histone (Fig. 1B). The fused c-myc tag was previously shown not to affect the role of Hho1p in the suppression of recombination (12). Unlike the mRNA, the Hho1p protein remained at similar levels during 6 d of growth, from exponential phase to stationary phase, and during exit of the semiquiescent state. Although this finding demonstrates that the total linker histone-to-nucleosome ratio remains approximately constant, because the average nucleosome repeat length and, therefore, the genomic nucleosome density also remains constant during growth to stationary phase (13), it provides no information on possible changes in the binding of linker histone to chromatin during semiquiescence.
Fig. 1.
HHO1 mRNA and Hho1p levels during entry and exit of stationary phase. (A) HHO1 transcript levels were determined by quantitative RT-PCR. The determined amount of the HHO1 transcript is depicted as log2 ratio relative to the 4-h time point and represents the average of three independent experiments with the standard deviation indicated. (B) Total protein was extracted from a yeast strain expressing a unique, c-myc-tagged Hho1p from the native HHO1 promoter at the times indicated (lanes 1–7) and a Western blot performed using an anti-c-myc antibody. A representative result (n = 3) is shown. A protein sample from a native W303 strain expressing an untagged Hho1p was used as a negative control for antibody specificity (lane 8).
Increase in the Binding of Hho1p to Chromatin in Stationary Phase.
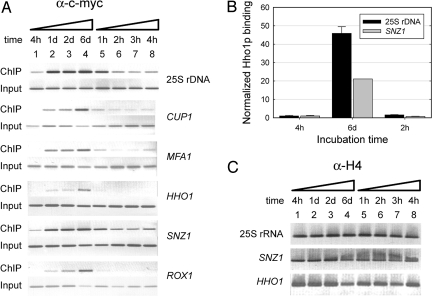
We performed ChIP of c-myc-tagged Hho1p at discrete times during the progression of a yeast culture to 6 d of stationary phase and also after transfer of the semiquiescent cells to new growth medium where cells exit stationary phase and reinitiate the cell cycle. ChIP gives information on both the fraction of DNA template associated with a protein and on the density of a protein at a specific genomic locus (14). We examined five different polymerase II-transcribed genes that are dispersed in the genome and of which two (ROX1 and HHO1) were down-regulated in stationary phase, two were up-regulated in stationary phase (SNZ1 and MFA1), and one remained repressed during growth from exponential to stationary phase (CUP1) (10). We also examined the polymerase I-transcribed 25S rRNA gene present in 100–150 copies on chromosome XII.
Looking first at the 25S rRNA locus, a significant increase in the density of Hho1p in this region is evident up to day six of stationary phase (Fig. 2A, lanes 1–4). Similarly, for each of the polymerase II-transcribed genes investigated, an increase in Hho1p binding is clearly evident during the same period (Fig. 2A, lanes 1–4). To obtain an accurate value for the fold change in Hho1p binding to chromatin in exponential compared with stationary phase, we quantitated the immunoprecipitated and input DNA for the 25S rRNA locus and SNZ1. As is evident in Fig. 2B, ≈20-fold and 45-fold changes in Hho1p binding signals were seen for the SNZ1 gene and 25S rRNA locus, respectively. It is not clear whether the higher signal measured for the 25S rRNA locus was due to a higher density of Hho1p at this locus or due to the multiple, tandem copies of this locus in the genome, which allowed linked, neighboring 25S rRNA genes to be precipitated and contributed to the measured amplification signal.
Fig. 2.
Binding of Hho1p to chromatin increases in stationary phase. (A) Chromatin was prepared from a yeast strain expressing a unique, c-myc tagged Hho1p following growth for the times indicated (lanes 1–8), and ChIP performed using an anti-c-myc antibody. The recovered DNA and the corresponding control input DNA were amplified by PCR using oligonucleotide primer sets to the genes indicated and the PCR products resolved by agarose gel electrophoresis. A representative result is shown (n = 3). (B) ChIP samples were quantitatively amplified by using oligonucleotide primer sets to the SNZ1 and 25S rDNA genes. The determined levels were normalized to that of the corresponding input DNAs and the ratios expressed relative to that of the 4 h time points (average± SD, n = 3). (C) As a control for equal cross-linking efficiency throughout the growth period shown in A, ChIP was performed at the indicated times (lanes 1–8) by using an anti-H4 antibody. The recovered DNA samples were amplified and resolved as in A. A representative result is shown (n = 3).
The observed binding of Hho1p to chromatin in stationary phase was balanced by the expected dissociation of Hho1p from the chromatin of yeast cells that were exiting stationary phase and reentering the cell cycle. Approximately 2 h after leaving stationary phase, the density of Hho1p on chromatin was indistinguishable from that of exponentially growing cells [see Fig. 2 A (compare lanes 1 and 6) and B]. These data showed that there was a significant increase in Hho1p binding to chromatin during progression of the cell to semiquiescence, reaching a maximum increase of ≈20-fold for a polymerase II-transcribed gene after 6 d and that the linker histone Hho1p rapidly dissociated when the cell reentered the cell cycle and resumed exponential growth. The observed change in binding of Hho1p to chromatin during entry and exit of stationary phase was not due to growth state-dependent differences in cross-linking efficiency, because the measured density of histone H4 on the 25S rRNA locus and SNZ1 and HHO1 genes was constant during growth to stationary phase (Fig. 2C), in agreement with the reported invariant nucleosome repeat length (13).
Linker Histone Hho1p Is Evenly Distributed in the Genome.
To determine whether the increase in binding of Hho1p to chromatin in stationary phase was a general, genome-wide phenomenon, we determined the genome-wide distribution of Hho1p in S. cerevisiae as a function of growth phase. Six-day stationary-phase cultures and stationary-phase cultures that had been incubated in rich growth medium for 1 h, were cross-linked and c-myc-tagged, Hho1p-associated chromatin fragments were precipitated, and complementary probes were prepared and hybridized to microarray slides that contained ≈6,400 S. cerevisiae ORF sequences. To allow comparison between different slides, the data were normalized relative to the amount of DNF1, SNZ1, and ROX1 DNA in the precipitated samples, determined by quantitative PCR. Supporting information (SI) Fig. S1 shows the average level of Hho1p bound at every ORF in the yeast genome in stationary phase and 1 h after stationary-phase exit. It is evident that Hho1p is not simply reduced at the six loci determined above (Fig. 2A) but is present at a significantly lower level at every position in the genome during exponential growth. The variation in the relative level of Hho1p binding followed a normal distribution on all chromosomes, as shown by a quantile–quantile plot and Shapiro–Wilks test (see Fig. S2), demonstrating that there were neither single genes nor extended gene domains with levels of Hho1p significantly higher of lower than the genome average. The distribution was not due to the enrichment of sequence fragments from larger ORFs in the immunoprecipitated sample, because the relative level of Hho1p binding did not correlate with the ORF length (see Fig. S3). Although it was previously reported that H1 had a predisposition for binding to AT-rich sequences, we found no evidence for such differential binding (see Fig. S4).
Compaction of the Chromatin in Stationary Phase Is Mediated by Hho1p.
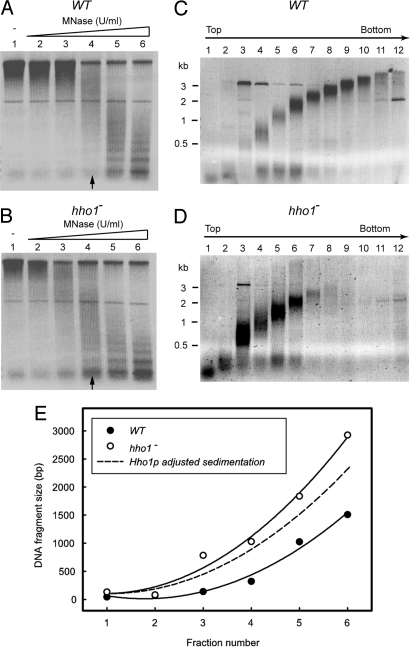
Because Hho1p binding to chromatin displayed a significant genome-wide increase in stationary phase, we investigated whether Hho1p was involved in chromatin condensation in stationary phase by determining the degree of chromatin compaction in WT and hho1− strains. Chromatin fragments were prepared from WT and hho1− nuclei and centrifuged through isokinetic sucrose gradients. The dependence of the sedimentation rate of a particle on its molar weight and buoyant density is a well established hydrodynamic principle. The more compact of two chromatin fragments of equal nucleosome density will sediment more rapidly (15).
The micrococcal nuclease (MNase) digestion of chromatin under identical conditions in nuclei purified from a WT and a hho1− strain in stationary phase consistently resulted in a size distribution depleted for larger chromatin fragments in the hho1− strain (compare lanes 9–12 of Fig. 3 C and D). This finding suggested that stationary phase chromatin was more accessible to MNase in the absence of Hho1p, indicative of a more open conformation. This difference was not observed for exponential phase WT and hho1− cells (data not shown). The average size of the DNA isolated from chromatin fragments that had migrated equal distances through the sucrose gradient was larger in the case of the hho1− strain compared with the WT strain (Fig. 3E). The observed increase in the sedimentation rate of equal-sized DNA molecules in the presence of Hho1p can be due to the increase in the molar weight of the fragment due to Hho1p binding, a decrease in the frictional coefficient of the fragment due to Hho1p-induced compaction, or a combination of the two effects. The additional weight of Hho1p will increase the sedimentation rate by a factor equal to the ratio of the buoyant mass of equal-sized chromatin fragments in the WT to the hho1− strain. When we adjusted the sedimentation rate of the chromatin lacking Hho1p by a factor of 1.1 (see Materials and Methods), the predicted sedimentation rate due to the added weight of the Hho1p molecule was still significantly less than that observed for the WT chromatin (Fig. 3E). This finding shows that the increase in sedimentation rate of equal-sized chromatin fragments from the hho1− and WT strains contained a contribution from a decrease in the frictional coefficient of the chromatin. This is consistent with an increase in chromatin compaction and shows that Hho1p is essential to compact yeast chromatin in stationary phase.
Fig. 3.
Hho1p is required for chromatin compaction in stationary phase. (A and B) Chromatin was digested with MNase for different times (lanes 1–6) in the nuclei of WT (A) and hho1− (B) strains in stationary phase. Chromatin was subsequently prepared under conditions that gave the most even size distribution of fragments (indicated by the arrows) and centrifuged on isokinetic sucrose gradients. (C and D) The DNA purified from the fractionated sucrose gradients of the chromatin samples isolated from WT (C) and hho1− (D) stationary-phase yeast nuclei were resolved on an agarose gel and the average DNA size in each fraction determined from the size standard. (E) The average size of the DNA in each fraction. Fractions 7–12, depleted of chromatin in the hho1− sample, were omitted from the analysis. A second-order polynomial was fitted to the data points (r2 = 0.99 and 0.98 for the hho1− and WT plots, respectively). The predicted sedimentation rate of the hho1− chromatin after correcting for the increase in molar weight due to Hho1p binding is shown by the dashed line. A representative result (n = 3) is shown.
Histone Hho1p Is Not a General Transcriptional Repressor in Stationary Phase.
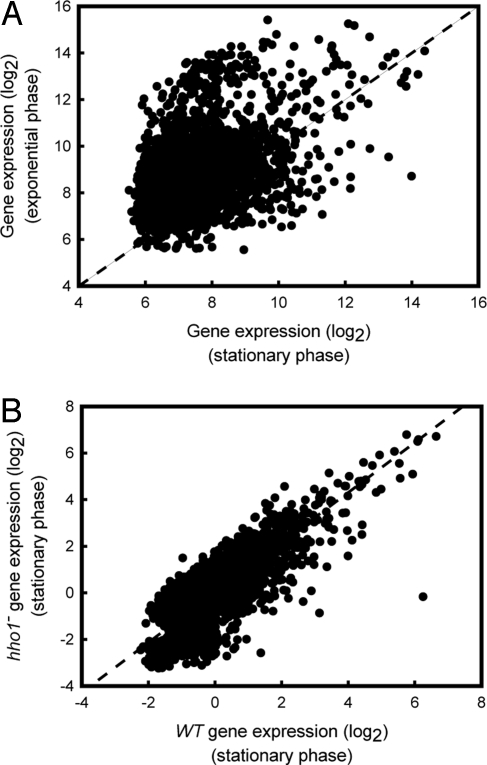
It was previously reported that Hho1p was not a general transcriptional repressor in exponentially cycling yeast cells (7). However, the reduced binding of Hho1p to chromatin during active growth reported here may have limited the regulatory effect of the linker histone in exponential phase. To investigate the role of Hho1p on the regulation of gene expression during increased binding and extensive transcriptional reprogramming in the semiquiescent state, microarray analysis was performed by using cyanine-labeled cDNA prepared from a WT strain grown to a 6-d stationary-phase culture and a matched stationary-phase culture that was incubated in rich growth medium for 1 h before RNA isolation. The expression data were normalized to the average expression levels of the DNF1, SNZ1, and ROX1 internal control genes, determined by quantitative PCR. Similar to previous studies (7, 16), we also observe that the expression level of <1% of genes change by >2-fold in the absence of Hho1p in yeast in exponential phase. Fig. 4A shows the relationship between the genome-wide level of gene expression in stationary- and exponential-phase cultures. The offset in the data distribution relative to the diagonal demonstrates a general reduction in the level of gene expression in the stationary-phase culture compared with the exponential phase culture (Fig. 4A). To determine whether this reduction, which was previously reported (17), was due to the elevated binding of Hho1p to chromatin and concomitant transcriptional repression, we also performed a microarray analysis of gene expression in a stationary-phase WT and a strain that lacked Hho1p. Fig. 4B shows that the level of gene expression remained constant between the WT and the hho1− strains in stationary phase. This result shows that, even though Hho1p bound at higher levels to stationary-phase chromatin, the absence of Hho1p in the hho1− strain did not cause a general transcriptional derepression in stationary phase.
Fig. 4.
Hho1p is not required for repression of transcription in stationary phase. (A) The relative transcript level for each of the ≈6,400 yeast genes were normalized to the population median and is shown plotted on a log2 scale for stationary phase versus exponential phase (n = 6). (B) The level of gene expression, normalized to the population average, is shown on a log2 scale in stationary phase in the WT strain versus stationary phase in the hho1− strain (n = 4).
Hho1p May Be Displaced from Chromatin by Transcriptional Activity.
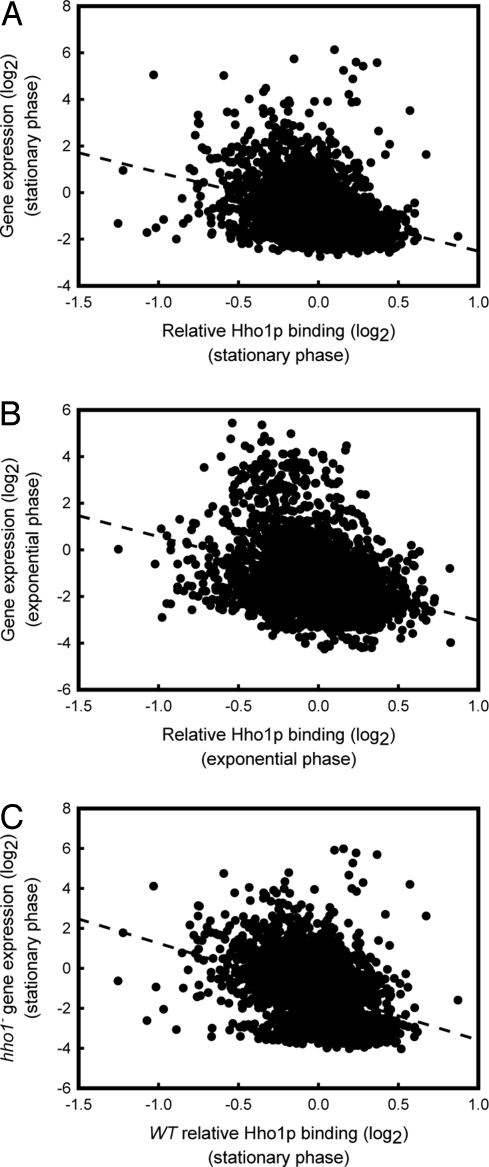
To gain further insight into the transcriptional regulatory role of Hho1p, we analyzed the level of gene expression as a function of relative Hho1p binding on a genome-wide scale. Fig. 5A shows the connection between Hho1p binding and gene expression in stationary phase. A statistically significant (Spearman rank correlation, ρ = −0.4, P < 0.001) relation is visible, showing that a decreased level of Hho1p binding to chromatin is associated with increased transcriptional activity. In exponential phase, where a significant proportion of Hho1p dissociated from chromatin (see Fig. 2 and Fig. S1), a larger spread of transcriptional activities (Fig. 5B), and a weaker relationship (Spearman rank correlation ρ = −0.3) between gene expression and Hho1p chromatin binding was visible. Nonetheless, the correlation between Hho1p binding and transcriptional activity was still statistically highly significant (P < 0.001). To test whether the high levels of Hho1p binding related to a decreased transcriptional activity, we also analyzed the genome-wide level of gene expression in the hho1− strain in stationary phase as a function of Hho1p binding to each gene in the WT strain. This analysis (Fig. 5C) clearly showed (Spearman rank correlation ρ = −0.4; P < 0.001) that genes that were associated with higher levels of Hho1p in stationary phase retained a low level of transcriptional activity, even in the absence of Hho1p. This finding is a strong indication that the cause-and-effect relationship here may be a decrease in Hho1p binding due to higher transcriptional activities, as opposed to higher transcriptional activities resulting from a decrease in Hho1p binding, because a general increase in expression of genes associated with high levels of Hho1p in WT stationary phase cells was not observed in the hho1− strain.
Fig. 5.
Relationship between the level of Hho1p binding and gene expression. (A) Plot of the relative level of Hho1p binding versus the transcriptional activity of each gene in a 6-d stationary-phase culture. Both the binding and expression datasets were normalized to the population average and are shown on a log2 scale. (B) Plot of the relative level of Hho1p binding versus the transcriptional activity of each gene in an exponential phase culture. Normalization and scaling is identical to A. (C) Plot of the relative level of Hho1p binding of a WT culture versus the transcriptional activity of each gene in an hho1− culture in 6-d stationary phase. Normalization and scaling is identical to A. The average of several independent replicates (n = 4) is shown in each image.
Discussion
The packaging of DNA into specialized chromatin structures at transcriptionally silenced regions is well established. It is also generally assumed that binding of H1 and associated condensation of chromatin facilitates repression of gene expression. Several studies have shown a direct connection between H1 and transcriptional inactivity, both in vitro (18) and in vivo (19), including the diminished ability of SWI/SNF to remodel nucleosome arrays assembled in vitro in the presence of H1 (20). However, the groups of Turcotte (7) and of Skoultchi (8) have shown that the absence of Hho1p in S. cerevisiae and of histone isotypes H1c, H1d, and H1e in mouse, did not cause a general, genome-wide transcriptional derepression. This suggests that the generally accepted transcriptional regulatory function of H1 may be more subtle in vivo. However, the absence of H1 binding data in these previous studies (7, 8) did not allow the investigators to directly relate the association of H1 to gene activity at the level of individual genes.
Here, we investigated both the distribution of Hho1p and transcriptional activity on a genome-wide scale in S. cerevisiae during extensive transcriptional reprogramming with entry and exit of stationary phase. We found a strong inverse relationship between the level of Hho1p binding and the transcriptional activity of a gene. A central question is whether this anticorrelation is caused by the requisite removal of H1 to facilitate transcriptional activity, or whether transcriptional activity, associated with the modification of the core histones and the passage of the elongating polymerase complex, actively expel the linker histone. A genome-wide study of nucleosome positions in S. cerevisiae showed that transcribed genes generally contained a well defined nucleosomal array downstream of the transcription start site (21). This suggests that Hho1p depletion was not due to a drop in nucleosome density on transcribed genes. It is likely that the passage of the polymerase complex transiently displaces or disrupts the nucleosome, which will abolish the structure recognized by Hho1p. Mistelli and colleagues (22) have shown that H1 continuously repartitioned in the genome. Thus, an elongating polymerase may never be challenged by the lengthy presence of an H1 on nucleosomal template DNA, or the nucleosome disruptive/displacement activity associated with a transcribing polymerase may cause eviction of H1.
We have shown that the rank order of the expression level of genes was maintained in a strain that lacked histone Hho1p. If the inverse relationship between Hho1p binding density and gene expression level simply reflected a repressive effect of Hho1p on transcription then we did not expect the expression rank order of the genes to be conserved in the absence of Hho1p. The anticorrelation and the preservation of expression order in the absence of Hho1p are consistent with the displacement of Hho1p from transcriptionally active genes. This idea is also supported by the finding that expression of a dominant-negative ISWI mutant in Drosophila larvae, which caused extensive transcriptional derepression, was associated with depleted levels of chromatin-associated H1 and with chromatin decondensation (23).
The increase in the association of Hhop1p with chromatin in stationary phase may therefore not be due to an active regulatory process, such as an activation of a hypothetical linker histone chaperone, or dephosphorylation of Hho1p. The increased binding may simply reflect the reduction in displacement activity that is due to a decrease in the expression of the majority of genes in stationary phase. This implies that condensation of chromatin confers a selective advantage to organisms under conditions of reduced activity. Intriguingly, Hho1p was shown to inhibit homologous recombination in S. cerevisiae (12). Together, these findings are strongly suggestive of a protective role for H1 in lower eukaryotes, where transcriptional down-regulation allows increased binding of H1, facilitating compaction of chromatin to maintain the integrity of the genome during extended periods of semiquiescence.
Materials and Methods
Plasmid Construction, Yeast Strains, and Cultures.
All routine molecular biology procedures were performed by established procedures (24). A S. cerevisiae W303 strain, in which the single, unique linker histone HHO1 gene was disrupted, was constructed as previously described (3) and denoted GSY1. Yeast cultures were routinely grown in yeast–peptone–dextrose (YPD) medium to an OD600 of ≈0.6 for exponential phase, or for 6 d (stationary phase) at 30°C with rotary shaking.
ChIP.
A culture of JDY43 (12), expressing a unique, c-myc-tagged Hho1p, was incubated in YPD medium and an aliquot (109 cells) taken after 6 d, or the culture was reinoculated into prewarmed YPD medium and 109 cells taken after 1 h. Chromatin samples were prepared as described (16), where chromatin fragments with an average size of 300–500 bp were recovered after sonication. To minimize nonspecific binding, the chromatin extracts were precleared with protein A Sepharose beads (Amersham Biosciences). Precleared chromatin extracts (1 mg of protein) were adjusted to a final volume of 200 μl and precipitated with 2 μl of anti-c-myc antibody (Roche). Immunocomplexes were collected and washed as described, resuspended in TE buffer [10 mM TRIS-HCl (pH 7.5), 1 mM EDTA], 1% SDS, and 250 mM NaCl and the cross-link reversed as described (16). Immunoprecipitated DNA was purified and analyzed by PCR using 1 μl of either the precipitated DNA or the corresponding input control sample. The nucleotide sequences of the primers used are given in Table S1. PCR products were electrophoresed on an agarose gel and quantitated with Quantity One software (Bio-Rad). Controls for equal cross-linking efficiency in exponential and stationary phases were performed with an anti-histone H4-antibody (Abcam), and the anti-c-myc antibody specificity was verified with a W303 strain that lacked the c-myc-tagged Hho1p. For genome-wide ChIP, DNA was purified as described and labeled with Cy-3 or Cy-5 dye (25). Hybridization and visualization of the S. cerevisiae microarray slides (University Health Network, Canada) were as described below. The genomic distribution of Hho1p was analyzed with Pyxis software (26).
RT and quantitative PCR.
Total yeast RNA was isolated with Trizol (Invitrogen) and reverse transcribed by using a reverse-transcription system kit (Promega) as specified by the manufacturer. RT products as well as ChIP samples were analyzed by quantitative PCR using the FastStart DNA Master SYBR green I kit (Roche) according to the manufacturer's instructions. The RT products and ChIP samples were amplified by using primers specific for the HHO1, SNZ1, or 25S rDNA genes (Table S1). Data were acquired at 1-sec intervals at 80°C and quantitative levels determined from a standard curve using known quantities of purified PCR products of HHO1, SNZ, or 25S rDNA, respectively.
Microarray Analysis.
Total RNA was isolated with a RNeasy Midi kit (Qiagen) from 6-d stationary-phase cultures of strains W303 and YGS1 or from stationary-phase cultures that were transferred into 200 ml of prewarmed YPD medium and incubated for 1 h. Aminoallyl-UTP (Sigma) was incorporated during RT of the RNA by using a Reverse Transcription System (Promega) and Cy-5 or Cy-3 ester (Amersham Pharmacia) attached according to the manufacturer's instructions. Pooled pairs of Cy-5- and Cy-3-labeled cDNA were added to 60 μl of DIG Easy Hyb solution (Roche) containing 80 μg/ml yeast tRNA (Invitrogen) and 10 μg/ml human cot1-DNA (Invitrogen), incubated at 65°C for 2 min and then allowed to cool to room temperature. The hybridization mixture was then applied to a microarray slide containing ≈6,400 yeast ORFs (University Health Network, Canada). Hybridization and washing was performed according to the slide manufacturer's instructions. Scanning was performed with a GenePix 4000B (Molecular Dynamics) system and analyzed by using GenePix Pro Version 5 and with scripts using the R programming language (www.r-project.org).
Chromatin Preparation, Sucrose Gradient Sedimentation, and Analysis.
Nuclei were prepared from 6-d stationary-phase WT and hho1− cultures (27) and digested at 37°C for 15 min with 10 units/ml of MNase (Sigma). The nuclei were resuspended in TEEP20 [10 mM Tris·HCl (pH 7.6), 0.1 mM EDTA, 0.1 mM EGTA, 0.25 mM PMSF, 20 mM NaCl] containing 300 μg/ml α-lysolecithin (Sigma) and lysed at 4°C overnight. Nuclear debris was removed by centrifugation (12,000 × g for 5 min at 4°C). Equal amounts of soluble chromatin were fractionated on 5–40% (wt/vol) isokinetic sucrose gradients containing TEEP80 [10 mM Tris·HCl (pH 7.6), 0.1 mM EDTA, 0.1 mM EGTA, 0.25 mM PMSF, 80 mM NaCl] by centrifugation at 4°C for 3 h at 36,000 rpm in a SW40Ti rotor (Beckman). DNA was purified from the individual gradient fractions and resolved on a 1% (wt/vol) agarose gel. The average size of the DNA fragments in each fraction was calculated relative to size standards by using Quantity One software (Bio-Rad). The contribution of the mass of Hho1p to the sedimentation rate of chromatin was calculated from the relation SWT/SKO = MbWT/MbKO, where S and Mb represent the sedimentation coefficient and buoyant mass of chromatin in the presence and absence of Hho1p, denoted by the WT and KO subscripts, respectively. The buoyant mass was calculated from the Svedberg equation with partial specific volumes of 0.75 ml/g and 0.55 ml/g for protein and DNA, respectively, and molecular masses of 106.4 kDa for a 160-bp DNA fragment, 107.7 kDa for the histone octamer, and 27.8 kDa for Hho1p, assuming a solvent density of 1g/ml.
Supplementary Material
Acknowledgments.
We thank J. Downs (University of Sussex, Sussex, U.K.) and S. Buratowski and K. Struhl (Harvard University, Cambridge, MA) for yeast strains and R. Girlano (National Institutes of Health, Bethesda, MD) for helpful discussions. This work was supported in part by grants from Wellcome Trust and the National Research Foundation (to H.-G.P.). G.S. was supported by a German Academic Exchange Service (DAAD) fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806337105/DCSupplemental.
References
- 1.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 2.Ramon A, Muro-Pastor MI, Scazzocchio C, Gonzalez R. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol Microbiol. 2000;35:223–233. doi: 10.1046/j.1365-2958.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 3.Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J Biol Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 4.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 5.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 7.Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 9.Rasmussen TP. Embryonic stem cell differentiation: A chromatin perspective. Reprod Biol Endocrinol. 2003;1:100–107. doi: 10.1186/1477-7827-1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez MJ, et al. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: Gene expression and identification of novel essential genes. Mol Biol Cell. 2004;15:5295–5305. doi: 10.1091/mbc.E03-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuge EK, Braun EL, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 13.Lohr D, Ide G. Comparison on the structure and transcriptional capability of growing phase and stationary yeast chromatin: A model for reversible gene activation. Nucleic Acids Res. 1979;6:1909–1927. doi: 10.1093/nar/6.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert N, et al. Chromatin architecture of the human genome; gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homolog Hho1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- 18.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 19.Kamakaka RT, Thomas JO. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990;9:3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn PJ, et al. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol. 2002;9:263–267. doi: 10.1038/nsb776. [DOI] [PubMed] [Google Scholar]
- 21.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 22.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 23.Corona DF, et al. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel FM, et al. Current Protocols in Molecular Biology. New York: Wiley; 2007. [Google Scholar]
- 25.Bohlander SK, Espinosa R, III, Le Beau MM, Rowley JD, Diaz MO. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 1992;13:1322–1324. doi: 10.1016/0888-7543(92)90057-y. [DOI] [PubMed] [Google Scholar]
- 26.Chang CF, Wai KM, Patterton HG. Calculating the statistical significance of physical clusters of co-regulated genes in the genome: The role of chromatin in domain-wide gene regulation. Nucleic Acids Res. 2004;32:1798–1807. doi: 10.1093/nar/gkh507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterton HG, Simpson RT. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol Cell Biol. 1994;14:4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.