Abstract
The active acquisition of epigenetic changes is a poorly understood but important process in development, differentiation, and disease. Our work has shown that repression of the p16/pRb pathway in human epithelial cells, a condition common to stem cells and many tumor cells, induces dynamic epigenetic remodeling resulting in the targeted methylation of a selected group of CpG islands. We hypothesized that cells in this epigenetically plastic state could be programmed by the microenvironment to acquire epigenetic changes associated with tumorigenesis. Here, we describe an in vitro model system where epigenetically plastic cells were placed in an environment that induced epithelial to mesenchymal transition (EMT) and led to a program of acquired de novo DNA methylation at targeted sites. In this model, we found that repression of E-cadherin transcription preceded the subsequent acquisition of methylated CpG sites. Furthermore, the induction of EMT was accompanied by de novo methylation of several other gene promoters, including those of the estrogen receptor and Twist. These data demonstrate that signals from the microenvironment can induce phenotypic and gene expression changes associated with targeted de novo epigenetic alterations important in tumor progression, and that these alterations occur through a deterministic, rather than stochastic, mechanism. Given the dynamic epigenetic reprogramming that occurs in these cells, DNA methylation profiles observed in human tumors may reflect the history of environmental exposures during the genesis of a tumor.
Keywords: epigenetic remodeling, human mammary epithelial cells, microenvironment, ras
The heritable regulation of gene expression changes that are critical to processes such as differentiation and disease can be controlled by epigenetic modifications of proteins and DNA sequences. We recently reported that the repression of p16INK4A in primary human mammary epithelial cells (HMEC) activates an E2F-mediated increase in proteins that remodel chromatin and causes targeted de novo DNA methylation at a non-random collection of loci (1). These studies show that cells can acquire epigenetic plasticity by altering the p16/pRb pathway, and that this program of acquired de novo methylation has a deterministic (predictable) rather than stochastic (random) pattern. Furthermore, the coordinated set of de novo DNA methylation events are preceded by, and dependent upon, the repression of gene expression. Thus, during cancer progression, one may envision that tumor cells can acquire epigenetic plasticity through repression of the p16/pRb pathway via mutations, deletions, or methyl a tion (2), which then provides the potential for programming epigenetic events. These observations are reminiscent of studies that show the acquisition of promoter hypermethylation upon modulation of estrogen or retinoic acid signaling (3, 4). In these cell population-based studies it is unclear whether the non-random hypermethylation events observed are due to induction or selection. To explore this question further, we chose a clinically relevant malignant phenotype and determined if repression of gene expression induced subsequent DNA methylation events or whether these occurred by selection.
Alterations in cellular phenotypes, such as epithelial to mesen chy mal transition (EMT), play a critical role in tumor progression (5), have been implicated in tumor recurrence (6), and are often associated with a poor prognosis in women with breast cancer (7). Consistent with this, there is now evidence demonstrating a link between EMT, basal-like tumors, the stem-cell phenotype, and the acquisition of tumorigenic and metastatic potential (8, 9). EMT is characterized by several molecular changes that include the loss of epithelial markers such as E-cadherin, and the induction of mesenchymal markers such as N-cadherin, fibronectin, and Snail (5). Though alterations in E-cadherin expression can occur through multiple mechanisms, including loss of heterozygosity and mutational inactivation, E-cadherin is frequently silenced through aberrant DNA hypermethylation of its promoter (10). Interestingly, when E-cadherin is silenced through promoter DNA hypermethylation, mammary cell lines often exhibit a mesenchymal morphology through the coordinated induction of a set of genes involved in EMT (11). In contrast, when E-cadherin is inactivated by mutation, the cells continue to exhibit an epithelial morphology, and these genes are not induced (11). This suggests that a program of molecular alterations leading to EMT, invasion, and metastasis can be modulated epigenetically.
EMT has been shown to be induced in murine cells by oncogenic ras in cooperation with factors in serum (12). There is also evidence that exposing cells to serum induces a gene expression pattern that resembles that of a wounding response. This wound-response signature is strongly predictive of future invasive and metastatic behavior, both of which require EMT (13). To determine whether HMEC with repressed p16INK4A (vHMEC) could be programmed by the microenvironment to acquire epigenetic changes associated with tumorigenesis, immortalized vHMEC-expressing oncogenic ras (vHMEC-ras) were exposed to serum. When cultured in serum-rich media, vHMEC-ras cells underwent phenotypic changes indicative of EMT and became more motile. This morphological transition was accompanied by a program of directed de novo DNA methylation of genes such as E-cadherin. Thus, signals from the microenvironment can induce phenotypic changes that modify the epigenome in an active and deterministic manner. This valuable model system will allow further study of phenotypic and epigenetic modulation during malignant transformation, and provide targets for therapeutic epigenetic reprogramming.
Results
Immortalized HMEC Can Undergo EMT in a Serum-Rich Environment.
Because oncogenic ras has been shown to cooperate with factors in serum to induce EMT and promote tumorigenesis in murine cells (12), we exposed vHMEC-ras cells to either 0.5% (vHMEC-ras0.5) or 10% (vHMEC-ras10) serum. The cells grew equally well in both serum concentrations (Fig. 1A). In addition, we found that both cell populations were capable of continued proliferation after removal of serum [supporting information (SI) Fig. S1A, vHMEC-ras0.5 → 0 and vHMEC-ras10 → 0]. However, unlike the cells grown in 0.5% serum, those grown in 10% serum underwent a striking change in morphology that was associated with loss of E-cadherin expression, reorganization of the actin cytoskeleton, and upregulation of the mesenchymal markers N-cadherin and fibronectin (Fig. 1 B and C), demonstrating that the cells were undergoing EMT. Because EMT is associated with increased motility, we examined whether the cells that had undergone EMT were more motile than the vHMEC-vector, -ras, or -ras0.5 cells that maintained their epithelial morphology, and indeed, they were (Fig. 1D), indicating that the acquisition of this mesenchymal phenotype was biologically and functionally relevant. Moreover, the mesen chymal phenotype did not require constitutive extracellular serum stimulation, because it was maintained upon serum withdrawal (Fig. 1B, 10 → 0). The epithelial morphology of ras-expressing vHMEC grown in 0.5% serum and the mesenchymal morphology of the same cells grown in 10% serum was manifested both on plastic (2D) and in matrigel (3D). When cultured in matrigel, the epithelial-appearing vHMEC-ras0.5 cells organized into mammosphere-like structures, whereas the mesenchymal-appearing vHMEC-ras10 cells retained their spindle morpho logy (Fig. S1B).
Fig. 1.
Immortalized HMEC expressing oncogenic ras undergo EMT in a serum-rich environment. (A) Growth curves of vHMEC expressing Ha-rasV12 or control vector in the absence or presence of 0.5% or 10% serum. Arrows indicate time at which serum was added. (B) Phase contrast (10×) and immunofluorescence photomicrographs (63×) of vHMEC-ras cells grown in their original concentration of serum (0.5% and 10%, first and second columns, respectively) or after they were switched to no serum (10 → 0, third column). (C) Immunoblot analysis of fibronectin (Fn), N-cadherin (N-cad), and E-cadherin (E-cad) on cell lysates prepared from parental vHMEC (par), vHMEC-vector (vec), vHMEC-ras (ras), vHMEC-ras0.5 (0.5), and vHMEC-ras10 (10) cells. (D) Transwell motility assay depicting migration of cells toward MEGM + 10% FBS as a chemoattractant for 48 h.
TGFβ Can Phenocopy the Morphological Effects Induced by a Serum-Rich Environment.
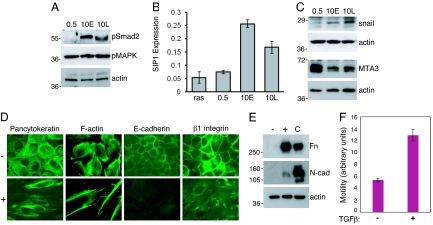
To gain insight into what factors in the serum-rich environment were responsible for the EMT observed in vHMEC-ras10 cells, we examined what signaling pathways were differentially activated in vHMEC-ras cells that exhibit a mesenchymal morphology relative to those that exhibit an epithelial morphology. Because TGFβ plays an important role in mediating EMT (14), we first examined the phosphorylation status of Smad2, one of the transcriptional mediators of TGFβ, by immunoblot analysis. As shown in Fig. 2A, basal Smad2 phosphorylation was increased in both early- (10E) and late- (10L) passage vHMEC-ras10 cells that exhibit a mesenchymal morphology, but was not detected in vHMEC-ras0.5 cells that exhibit an epithelial morphology. In contrast, phosphorylation of MAP kinase was elevated in all vHMEC-ras cells (independent of the serum concentration in the growth media), consistent with the activation of the ras signaling pathway in each of these cell populations. These data indicate that TGFβ signaling may be required for the phenotypic alterations induced by 10% serum. Consistent with this, in our analysis of several markers of EMT, including E47 and Slug, which remained unchanged (Fig. S2A), we observed an upregulation of smad-interacting protein 1 (SIP1), a protein that has been implicated in TGFβ-mediated EMT (15), in the vHMEC-ras10 cells that had undergone EMT (Fig. 2B). TGFβ has been shown to cooperate with oncogenic ras to induce Snail (16), a transcription factor that is a central mediator of EMT (17). Snail was also upregulated in the vHMEC-ras10 cells that had undergone EMT (Fig. 2C). In addition, the negative regulator of Snail, MTA3, a protein that forms part of chromatin complexes (18), was downregulated in vHMEC-ras10 cells (Fig. 2C). These data demonstrate that TGFβ signaling and several effectors of TGFβ-induced EMT are upregulated in vHMEC-ras cells grown in a serum-rich environment, and suggest that TGFβ may be involved in mediating serum-induced EMT.
Fig. 2.
TGFβ signaling is activated in vHMEC-ras cells with a mesenchymal morphology and can induce EMT in vHMEC-ras cells with an epithelial morphology. Immunoblot analysis of phospho-Smad2 and phospho-MAP kinase (A) or Snail and MTA3 (C) on cell lysates prepared from vHMEC-ras0.5 (0.5) and vHMEC-ras10 early- (10E) and late- (10L) passage cells. (B) Quantitative real-time PCR (qPCR) analysis of SIP1 expression. (D) Immunofluorescence analysis of vHMEC-ras0.5 untreated (−) or treated (+) with 2 ng/ml TGFβ for 72 h and immunostained as indicated. (E) Immunoblot analysis of fibronectin (Fn) and N-cadherin (N-cad) on cell lysates prepared from vHMEC-ras0.5 either untreated (−) or treated (+) as in (D). Cell lysates prepared from fibroblasts were used as a positive control (C). (F) Transwell motility assay depicting vHMEC-ras0.5 cell migration toward media not supplemented (−) or supplemented (+) with 10 ng/ml TGFβ as a chemoattractant for 48 h.
If TGFβ is one of the factors present in a serum-rich environment that cooperates with oncogenic ras to induce EMT, we reasoned that TGFβ could be used in lieu of 10% serum to induce EMT in vHMEC-ras0.5 cells that exhibit an epithelial morphology. To test this, we treated vHMEC-ras0.5 cells with TGFβ and assessed the expression of molecular markers associated with EMT by immunofluorescence and immunoblot analysis. Within 48 h of treatment with TGFβ, the cells showed signs of a morphological change, which were clearly manifested by 72 h. This morphological change was associated with a diminution in pancytokeratin expression, reorganization of the actin cytoskeleton, and disruption of both cell–cell and cell-matrix contacts, as evidenced by the loss of E-cadherin and β1-integrin expression, respectively (Fig. 2D). Coincident with the loss of epithelial marker expression was an upregulation in the expression of the mesenchymal markers N-cadherin and fibronectin (Fig. 2E). In addition, vHMEC-ras0.5 cells stimulated with TGFβ were more motile than their unstimulated counterparts in transwell migration assays, consistent with the expected increase in motility associated with EMT (Fig. 2F). These data suggest that TGFβ is one of the factors present in serum that cooperates with ras to induce EMT. However, unlike the serum-induced EMT, the TGFβ-induced EMT was rever sible upon withdrawal of TGFβ (Fig. S2B), as previously observed in other cell types (19).
Serum-Induced EMT Is Accompanied by Repression of E-cadherin Expression and Subsequent DNA Hypermethylation at the E-cadherin Promoter.
Because serum-induced EMT was irreversible and heritable (it was maintained upon the withdrawal of serum and transmitted to daughter cells), we examined whether epigenetic alterations were involved in permanently repressing E-cadherin expression in these cells. Epigenetic modifications in the promoter region of E-cadherin were assessed using methylation-specific PCR. Consistent with the heritable and morphological appearance of the cells, DNA methylation of the E-cadherin promoter was observed in the mesenchymal-appearing vHMEC-ras10 cells, but not in the epithelial-appearing vHMEC-ras0.5 cells (Fig. 3A, lanes 1–4). This methylation pattern was also maintained after the cells were switched to no (0.5 → 0) or low (10 → 0.5) serum growth conditions (Fig. 3A, lanes 5–8), consistent with the maintenance of their morphology under those conditions. In contrast, and as expected, we did not observe methylation of the E-cadherin promoter after TGFβ-induced EMT, which is reversible (Fig. S2C). These data suggest that TGFβ is sufficient to induce the phenotypic alterations observed in a serum-rich environment, but that additional factors (or time) are required for the epi genetic modifications that render EMT heritable to occur.
Fig. 3.
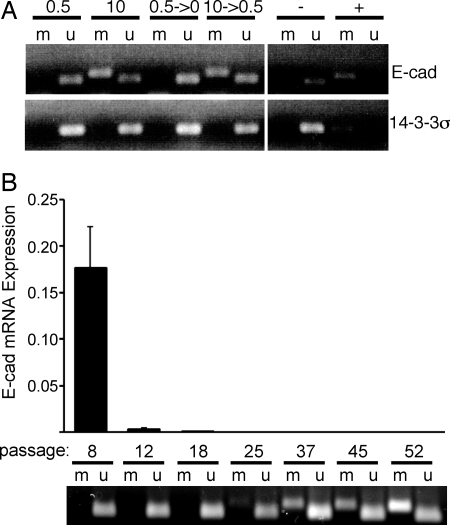
EMT is accompanied by repression of E-cadherin expression and epigenetic modifications at the E-cadherin locus. (A) Methylation-specific PCR (MSP) analysis of E-cadherin and 14–3-3σ on bisulfite-treated DNA isolated from vHMEC-ras cells grown in 0.5, 10, 0.5 → 0, or 10 → 0.5% serum using primer sets that specifically amplify either methylated (m) or unmethylated (u) DNA. Unmethylated (−) and methylated (+) control templates. (B) qPCR analysis of E-cadherin expression in vHMEC-ras10 cells at the indicated passages (Top); MSP analysis of the same cells (Bottom).
To rule out the unlikely possibility that the mesenchymal features in these cells were due to the selection and expansion of a rare contaminating population of fibroblasts, we analyzed the promoter gene region for the protein 14–3-3σ, which is a mammary epithelial-specific marker that is methylated in fibroblasts but unmethylated in epithelial cells (20). This locus remained unmethylated in all our cells, including those that acquired a mesenchymal morphology, confirming their epithelial origin (Fig. 3A). vHMEC-ras10 cells that exhibit a mesenchymal phenotype can also be distinguished from fibroblasts on the basis of their E-cadherin methylation as the E-cadherin gene promoter is repressed and unmethylated in human mammary fibroblasts (Fig. S2C, lane 2), but repressed and methylated in mesenchymal-appearing vHMEC-ras10 cells (Fig. 3). These data demonstrate that the acquisition of mesenchymal features by vHMEC-ras10 cells is the consequence of an active molecular process that occurs in epithelial cells.
To gain insight into the kinetics of E-cadherin silencing and methylation during serum-induced EMT, we examined E-cadherin mRNA expression and methylation at multiple passages after exposure to serum. This analysis indicated that E-cadherin gene expression was dramatically reduced after eight passages in serum (≈50-fold less than ras0.5), and was even further repressed before methylation, which was detected at passage 25 and only became clearly evident in later passages (Fig. 3B). This demonstrates that the repression of E-cadherin and emergence of mesenchymal characteristics precede methylation of the E-cadherin promoter.
To confirm that DNA methylation events were causal (and not simply correlative), for the long-term repression of E-cadherin expression, late-passage vHMEC-ras10 cells exhibiting hyper methylated E-cadherin promoter sequences were exposed to the DNA methyltransferase (DNMT) inhibitor 5-aza-2′deoxycytidine (AZA). As shown in Fig. S3, inhibition of DNA methylation restored E-cadherin expression, but only modestly. Because Snail expression was upregulated in vHMEC-ras10 cells that exhibit a mesenchymal morphology, and Snail can mediate E-cadherin repression by the recruitment of histone deacetylases (HDAC) (21), we treated the cells with the HDAC inhibitor trichostatin A (TSA) to determine whether deacetylation was also involved in E-cadherin repression. Inhibition of HDACs resulted in an increase in E-cadherin expression, which was even more robust when the cells were treated with both inhibitors. This suggests that deacetylation and DNA hypermethylation at the E-cadherin promoter both contribute to the mechanistic basis of long-term silencing of E-cadherin expression.
To determine whether the mechanism by which E-cadherin is initially repressed before permanent silencing through DNA methylation also involved deacetylation, early-passage vHMEC-ras10 cells were exposed TSA and E-cadherin expression was examined. The inhibition of HDACs increased expression of E-cadherin (Fig. S3). These data suggest that deacetylation is implicated in repressing E-cadherin expression before long-term silencing by DNA methylation.
Methylation of the E-cadherin Promoter Occurs de Novo.
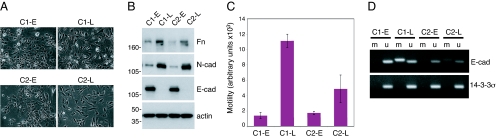
To determine if methylation of the E-cadherin promoter was a de novo event in vHMEC-ras10 cells, or the result of the selection of a pre-existing population of epithelial cells with a methylated E-cadherin promoter region, we isolated two independent clones of vHMEC-ras0.5 cells that exhibit an epithelial morphology and exposed them to media containing 10% serum. Early-passage clones (C1-E and C2-E) retained their epithelial morphology after short-term exposure to 10% serum (Fig. 4A) and expressed E-cadherin (Fig. 4B). Upon continued exposure to media containing 10% serum, both clones gradually acquired a mesen chymal morphology (C1-L and C2-L), E-cadherin expression was lost, the mesenchymal markers N-cadherin and fibronectin were upregulated (Fig. 4B), and the cells became more motile (Fig. 4C). Subsequent to the induction of EMT, their E-cadherin promoter became hypermethylated (Fig. 4D). These data demonstrate that the changes in cellular morphology and DNA methylation observed when vHMEC-ras cells are exposed to 10% serum occur de novo.
Fig. 4.
EMT and epigenetic modifications at the E-cadherin locus occur de novo. (A) Photomicrographs (10×) of two early- (C1-E and C2-E) and late- (C1-L and C2-L) passage clones. (B) Immunoblot analysis of fibronectin (Fn), N-cadherin (N-cad), and E-cadherin (E-cad) on cell lysates prepared from each clone. (C) Transwell motility assay depicting migration of clones toward MEGM + 10% FBS as a chemoattractant for 48 h. (D) MSP analysis of E-cadherin and 14–3-3σ.
Methylation of the E-cadherin Promoter Is Part of a Nonrandom Program of Targeted Epigenetic Changes.
The de novo DNA hypermethylation observed at the E-cadherin locus could be due to a generalized increase in DNMT function that results in a generalized hypermethylation of CpG islands, or a program of more directed events. Exposing vHMEC-ras cells to 10% serum led to an increase in DNMT3a expression (Fig. S4); however, only selected CpG islands subsequently acquired DNA hypermethylation. Previous studies have shown that the E-cadherin and estrogen receptor (ER) promoter sequences are often hypermethylated in the same samples (22, 23). Additionally, a number of genes have been reported to be more frequently hypermethylated in invasive breast carcinomas (24, 25) and in basal-like breast cancers that have a poor prognosis (26). Because EMT is associated with invasive behavior, and has been correlated with the basal-like phenotype (8), we investigated whether genes that have been reported to be methylated in invasive and basal-like breast cancers also acquired hyper methyl ated promoter sequences in vHMEC-ras10 cells that exhibit a mesenchymal morphology. Several genes, including those that encode ER and Twist, which have been reported to be methylated in basal-like and/or invasive breast cancers (25, 26), were hypermethylated in vHMEC-ras10 cells that exhibit a mesenchymal morphology, but not in vHMEC-ras or vHMEC-ras0.5 cells that exhibit an epithelial morphology (Table 1 and Fig. S4B). The promoter region for CST6, another locus that is hypermethylated in basal-like tumors, was also hypermethylated in cells that had acquired a mesenchymal phenotype, but its methylation occurred before methylation of the E-cadherin promoter. In contrast, other loci, known to be frequently hyper methyl ated in breast tumors, such as BRCA1, GATA3, TIMP3, and DKK3 did not exhibit DNA promoter hypermethylation in these cells, thus confirming a nonrandom rather than generalized process (Table 1 and Fig. S4B). These data suggest that there is a sequential and progressive increase in promoter gene methylation along the progression to malignancy that reflects the effect of signals from the surrounding environment.
Table 1.
MSP analysis of genes hypermethylated in breast cancer
| vec | ras | ras0.5 | ras10E | ras10L | |
|---|---|---|---|---|---|
| ECAD | U | U | U | U | M |
| ER | U | U | U | U | M |
| TWIST | U | U | U | U | M |
| CST6 | U | U | M | M | M |
| BRCA1 | U | U | U | U | U |
| GATA3 | U | U | U | U | U |
| TIMP3 | U | U | U | U | U |
| DKK3 | U | U | U | U | U |
U, unmethylated; M, methylated.
Discussion
The accumulation of aberrant DNA methylation events that arise during tumorigenesis could occur through random epi genetic changes within the tumor cell population that are then selected for expansion because of a selective advantage or, alternatively, they could occur through more deterministic events and be programmed by specific repression of targeted genes. The data in this study provide a mechanism by which premalignant cells can acquire de novo DNA methylation at biologically relevant sites early in the carcinogenic process in a deterministic manner. Individual clones with an epithelial phenotype were reprogrammed by exogenous signals to acquire a mesenchymal phenotype associated with increased motility and predictable de novo DNA methylation events. Conceptually, our data suggest that the profile of methylation events in tumors is the sum of environmental exposures during their inception. Different environmental conditions can be expected to elicit different programs of epigenetic changes that create a memory of processes that drive carcinogenesis in each particular tumor. Thus, these cells represent a valuable model in which to study phenotypic and epigenetic plasticity, and the acquisition of directed de novo DNA methylation events.
The association between E-cadherin and ER promoter methyl ation we report in our in vitro system has been observed previously in human breast tumors and correlates with clinical parameters. Examination of several methylated gene loci for associations with clinicopathological features revealed that methylation at the E-cadherin and ER gene loci showed a strong positive correlation with each other and an additional association with a younger patient age at diagnosis (27). Parrella et al. (23) examined the frequency of methylation at these loci in benign and malignant tissue, and reported an association between E-cadherin and ER methylation that increased with progression. Likewise, Nass et al. (22) reported that E-cadherin and ER promoter methylation as individual events were detectable in early lesions such as DCIS, were more frequent in invasive lesions, and that the coincident methylation of both promoters occurred in both types of lesions. Our in vitro studies provide a rationale for the association between E-cadherin and ER promoter methylation. Under specific conditions, both genes are targeted for hypermethylation as part of an epigenetic program that is induced by EMT. These genes, as well as other genes, are targeted for epigenetic remodeling via an initial repression of gene expression and the subsequent recruitment of DNA methyl trans ferases.
The clinical ramifications of these methylation events are critical. Many human breast cancers and breast cancer cell lines display features of EMT (11, 28), and recent studies have shown that this phenotype is prominently featured in human basal-like breast tumors (8). Consistent with this, studies have reported that loss of E-cadherin expression is positively correlated with advanced histological grade, metastasis, and decreased survival (29, 30), characteristics that are typically associated with the basal-like subtype. Thus, methylation of the E-cadherin promoter leading to loss of E-cadherin expression and increased motility in vHMEC-ras10 cells may be an important event in the transformation of these cells. Coleman and coworkers (26) reported a methylation signature that exists in basal-like breast tumors. This panel of six genes is frequently hypermethylated in basal-like tumors and distinguishes them from other breast tumor subtypes. Of these six genes, the three that we examined—E-cadherin, ER, and CST6—were hypermethylated in the cells that underwent EMT.
Our data also suggest that there are consequences to the silencing of gene loci by DNA hypermethylation when compared with transcriptional repression via deacetylase activity. In this report we show that the repression of E-cadherin in early-passage vHMEC-ras10 cells (that have undergone EMT, but do not exhibit methylation of the E-cadherin promoter) is dependent on deacetylation, as inhibition of HDACs by TSA resulted in an increase in E-cadherin expression in these cells. In contrast, late-passage vHMEC-ras10 cells (that have undergone EMT as well as E-cadherin promoter methylation) appear to repress E-cadherin through the dual action of histone deacetylation and DNA promoter hypermethylation. The consequences of differentially repressing E-cadherin are twofold. First, early-passage (P8) vHMEC-ras10 cells exhibit a higher level of E-cadherin expression than late-passage vHMEC-ras10 cells, and though they are more motile than vHMEC-ras cells that exhibit an epithelial morphology, they are less motile than vHMEC-ras10 cells that exhibit both a mesenchymal morphology and E-cadherin promoter methylation (Fig. 3 and data not shown, respectively). Second, and most importantly, the heritable nature of DNA methylation events ensures that subsequent generations of cell progeny will also exhibit the invasive phenotype associated with sustained induction of EMT even in the absence of inducing signals.
Finally, this in vitro model system provides unique insights into the timing and induction of methylation events that may occur during tumor progression. In this system, there is an ordering to the potential epigenetic reprogramming that occurs. Exposure of vHMEC to 10% serum or TGFβ in the absence of ras overexpression evokes a pronounced cell cycle arrest (unpublished data) rather than the EMT that is observed in the vHMEC-ras0.5 cells used in this study. This suggests that EMT is a process that occurs later in the transformation process and that an increase in genomic instability as well as additional molecular alterations are required before invasive phenotypes such as EMT can occur. This is of particular significance because though EMT has been observed in a few rodent cell lines in culture, a survey of 14 human epithelial cell lines and two primary human cells revealed that none of the human cells could undergo EMT in response to 48 h of TGFβ treatment (31). Thus, our data provide previously undescribed evidence that extracellular cues (leading to immortalization) can cooperate with TGFβ signaling and oncogenic stress during transformation to induce EMT in human cells. These data suggest that genetic and epigenetic events, provoked by ras overexpression, set the stage for the program of gene repression and DNA hypermethylation that can subsequently occur. Translating these observations to a biological setting, we postulate that damaging agents, such as ionizing radiation, which activate TGFβ, would typically induce a proliferative arrest in the bulk of breast epithelial tissue, but activate targeted epi genetic reprogramming in foci of cells that had already acquired preparatory molecular events such as ras overexpression and immortalization. Understanding these processes may provide avenues to effective intervention and prevention.
Materials and Methods
Cell Culture.
Immortalized vHMEC expressing pLXSP3-Ha-rasV12 or the control pLXSP3 vector (N.D., et al., unpublished data) were grown in MEGM (Lonza) supplemented with either 0.5% or 10% FBS and 2 μg/ml puromycin, as previously described (1). vHMEC-ras0.5 clones were isolated using standard ring cloning procedures. All experiments performed with early passage clones were conducted between 5 and 15 passages following exposure to serum; experiments with late passage clones were conducted 40 or more passages following exposure to serum. For 3D cultures, 5 × 103 cells were resuspended in media containing 2% Matrigel (BD Biosciences) and seeded into eight-well glass chamber slides on top of a thin layer of Matrigel.
Immunofluorescence Analysis.
Cells were fixed with 100% methanol for 7 min at −20°C (for E-cadherin, β1-integrin, and cytokeratin staining) or with 4% paraformaldehyde/PBS for 20 min at room temperature (RT) (for F-actin staining). Paraformaldehyde-fixed cells were permeabilized with 0.1% Triton X-100/PBS for 10 min at RT and washed with PBS. Nonspecific binding sites were blocked and the cells were then incubated with primary antibodies overnight at 4°C, followed by a FITC-conjugated secondary antibody for 1 h at RT, or incubated with Alexa Fluor 488-Phalloidin (Molecular Probes, A12379) for 1 h at RT and appropriately washed. Images were captured using a Nikon epifluorescence inverted microscope and a cooled CCD digital camera (Optronics).
Motility Assay.
Motility assays were conducted using Chemicon's QCM Fluorometric Chemotaxis Assay Kit (8 μm) according to the manufacturer's protocol.
Methylation-Specific PCR.
Genomic DNA was isolated from cells using the Wizard Genomic DNA Isolation Kit (Promega). Approximately 750 ng of DNA was bisulfite treated with the EZ DNA Methylation-Gold Kit according to the manufacturer's protocol (Zymo Research). Methylation-specific PCR (MSP) was performed on bisulfite-modified DNA using previously described primer pairs and PCR cycle conditions (see SI Text for additional details on this and other experimental procedures).
Supplementary Material
Acknowledgments.
We thank Dr. Beverly Emerson for the generous gift of TGFβ and Dr. Stephen Baylin for GATA3 MSP primers. This research was supported by the Susan G. Komen Foundation Postdoctoral Award PDF124906 (to N.D.), the National Cancer Institute Institutional Training Grant T32 CA009043 (to M.B.W.), the Department of Defense Breast Cancer Research Program Concept Award BC023982 (to T.D.T.), and National Institutes of Health National Cancer Institute Grants CA097214–01A1 and CA122024–01 (to T.D.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807146105/DCSupplemental.
References
- 1.Reynolds PA, et al. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 3.Leu YW, et al. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–8192. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- 4.Ren M, et al. Impaired retinoic acid (RA) signal leads to RARbeta2 epigenetic silencing and RA resistance. Mol Cell Biol. 2005;25:10591–10603. doi: 10.1128/MCB.25.23.10591-10603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs IB, et al. The prognostic significance of epithelial-mesenchymal transition in breast cancer. Anticancer Res. 2002;22:3415–3419. [PubMed] [Google Scholar]
- 8.Sarrio D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 9.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Droufakou S, et al. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. Int J Cancer. 2001;92:404–408. doi: 10.1002/ijc.1208. [DOI] [PubMed] [Google Scholar]
- 11.Lombaerts M, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oft M, et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 13.Chang HY, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 15.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: Mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 17.Davidson NE, Sukumar S. Of Snail, mice, and women. Cancer Cell. 2005;8:173–174. doi: 10.1016/j.ccr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21:2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 20.Moreira JM, Ohlsson G, Rank FE, Celis JE. Down-regulation of the tumor suppressor protein 14–3-3sigma is a sporadic event in cancer of the breast. Mol Cell Proteomics. 2005;4:555–569. doi: 10.1074/mcp.M400205-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nass SJ, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–4348. [PubMed] [Google Scholar]
- 23.Parrella P, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann U, et al. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol. 2002;160:605–612. doi: 10.1016/S0002-9440(10)64880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fackler MJ, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–975. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 26.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006;237:272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Bankfalvi A, et al. Immunophenotypic and prognostic analysis of E-cadherin and beta-catenin expression during breast carcinogenesis and tumour progression: A comparative study with CD44. Histopathology. 1999;34:25–34. doi: 10.1046/j.1365-2559.1999.00540.x. [DOI] [PubMed] [Google Scholar]
- 30.Park D, et al. Expression pattern of adhesion molecules (E-cadherin, alpha-, beta-, gamma-catenin and claudin-7), their influence on survival in primary breast carcinoma, and their corresponding axillary lymph node metastasis. Apmis. 2007;115:52–65. doi: 10.1111/j.1600-0463.2007.apm_524.x. [DOI] [PubMed] [Google Scholar]
- 31.Brown KA, et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6:R215–231. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.