Abstract
Recognition sites for microRNAs (miRNAs) have been reported to be located in the 3′ untranslated regions of transcripts. In a computational screen for highly conserved motifs within coding regions, we found an excess of sequences conserved at the nucleotide level within coding regions in the human genome, the highest scoring of which are enriched for miRNA target sequences. To validate our results, we experimentally demonstrated that the let-7 miRNA directly targets the miRNA-processing enzyme Dicer within its coding sequence, thus establishing a mechanism for a miRNA/Dicer autoregulatory negative feedback loop. We also found computational evidence to suggest that miRNA target sites in coding regions and 3′ UTRs may differ in mechanism. This work demonstrates that miRNAs can directly target transcripts within their coding region in animals, and it suggests that a complete search for the regulatory targets of miRNAs should be expanded to include genes with recognition sites within their coding regions. As more genomes are sequenced, the methodological approach that we used for identifying motifs with high sequence conservation will be increasingly valuable for detecting functional sequence motifs within coding regions.
Keywords: computational biology, posttranscriptional regulation, comparative genomics, multiple-sequence alignment, evolutionary conservation
MicroRNAs (miRNAs) are endogenously encoded, single-stranded regulatory RNAs that bind to and inhibit the translation of transcripts with complementary sequence (1). Computational evidence suggests that miRNAs regulate at least 20% of human genes and have been implicated in the regulation of a wide range of biological systems (2). In plants, miRNA targets can be predicted with relatively high confidence because of the extensive base pairing between plant miRNAs and their target mRNAs (1). In animals, in contrast, miRNAs typically bind to their targets with significantly less complementarity, and the short target sequences are, therefore, difficult to identify on the basis of sequence alone. As a result, most computational approaches to predict miRNA–target interactions rely on conservation of target sites (3–6).
Although early studies reported some evidence for the targeting of miRNAs to sites within protein coding regions (4, 6), subsequent research has reported that there is minimal functionality for sites in ORFs or 5′ UTRs (7). A focus on miRNAs present within 3′ UTRs is supported by evidence suggesting that the G-cap/poly(A) tail interface (which connects the two ends of eukaryotic mRNAs during translation) is important for miRNA function (8) and that miRNAs tend to be more effective when localized at the end of the 3′ UTR rather than the middle (7, 9). Indeed, the protein translation machinery might be expected to displace an miRNA complex present within a gene's coding sequence. However, exogenously added siRNAs that target coding sequences, including siRNAs with imperfect base pairing, are effective at silencing (10). More recent reports have also shown that, contrary to other highly conserved coding region motifs, miRNA target sites are conserved in all three reading frames among Drosophila genomes (11), and that target sites introduced into the 5′ UTR of transcripts can repress translation (12). Further, the potential importance of sites embedded within the coding sequence of genes is supported by the nature of the genetic code itself, which has been shown to be nearly optimal for conveying information in parallel to the amino acid sequence (13). One recent report has shown downregulation via a coding region target directly (29).
We sought to investigate whether miRNA target sites within coding regions are functional, beginning with an unbiased screen for coding region sequence motifs that are more highly evolutionarily conserved than is required for amino acid conservation. We found that the motifs most highly conserved in coding regions are indeed miRNA target sites, and we experimentally confirmed that the endonuclease Dicer is targeted by the miRNA let-7 by means of sites within its coding region. We also found computational evidence to suggest that miRNA target sites in coding regions and 3′ UTRs may differ in mechanism.
Results and Discussion
We developed a computational algorithm to find conserved sequence motifs within coding regions (http://www.sitesifter.org). Our approach is based on the assumption that DNA sequences with a regulatory function should be evolutionarily conserved at the nucleotide sequence level over and above any conservation required to maintain the amino acid sequence of the encoded proteins. Most computational approaches for finding miRNA target sites put special emphasis on the miRNA target “seed” (corresponding to positions 2–7 of the miRNA) because complementarity at these base pairs has been shown to play an important role in target recognition (4, 14). In keeping with this approach, we chose to search for conserved motifs 8 bp in length, because we have found that this length increases specificity for miRNAs. Because coding sequences are under constraint by the canonical coding for amino acids, a larger number of genomes must be used when surveying conservation in coding regions compared to 3′ UTRs. We took advantage of a multiple-sequence alignment of 17 genomes (human, chimpanzee, macaque, mouse, rat, rabbit, dog, cow, armadillo, elephant, tenrec, opossum, chicken, frog, zebrafish, green spotted puffer, and fugu) obtained from the University of California Santa Cruz (UCSC) Genome Browser (15), used in conjunction with a list of coding region boundaries extracted from the UCSC Table Browser (16).
Our algorithm first parses coding regions from the whole-genome multiple alignment. The fraction of coding region nucleotides for which the sequences of all 17 species are aligned (23.4%) is then searched for perfectly conserved sequences 8 bp in length. Conserved motifs are assigned a Sequence Level Conservation Score (SLCS) representing the degree to which the motif is conserved at the sequence level over and above the amino acid level. The SLCS is based on an empirical measure of the probability that a given codon is sequence-conserved, given that it is amino acid-conserved [see supporting information (SI) Table S1 for the full list of codons and their respective sequence level conservation probabilities]. For a given motif, the SLCS represents the logarithm of the probability of sequence conservation across the motif's constituent codons, taking into account codons that partially overlap the motif. If a motif is conserved multiple times throughout the genome, the score is summed over every conserved occurrence (see Materials and Methods).
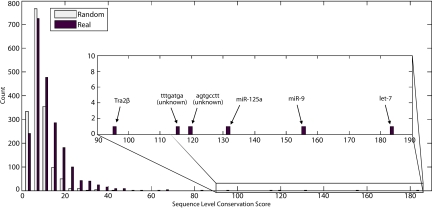
Results were obtained from the multiple alignment as well as a randomly permuted alignment in which the human genome sequence was left unchanged, but the choice of which codons were fully conserved at the sequence level in other species was assigned randomly. This randomization procedure alters the DNA sequence of genes in the multiple alignment while maintaining their amino acid sequences. As shown in Fig. S1, in the actual data, the overall distribution of spacing between fully conserved sites is similar to that in the permuted data, except that there is an overabundance of fully sequence-conserved codons that are adjacent to one another, which could reflect the presence of embedded regulatory motifs at the sequence level. Indeed, a comparison of the two result sets (Fig. 1) shows that there are significantly more high-scoring motifs from the real alignment compared with the permuted alignment (P < 10−49, Kolmogorov–Smirnov test), suggesting that these motifs have been functionally conserved within coding sequences even across such a large evolutionary distance (see Table S2 for a list of high-scoring motifs). Thirteen motifs from the real dataset had an SLCS >56.1, which was the maximum score obtained across 10 permuted datasets. There were also 41 miRNA target sites with an SLCS >0 in the real dataset, compared with a mean ± SD of 20.2 ± 2.2 in the permuted datasets (fewer than 5% of motifs have a non-zero SLCS in the real dataset). Four miRNA target sites, corresponding to those for let-7 (except for let-7d, which differs in its first base), miR-9, miR-125, and miR-153, scored above the maximum from the permuted datasets. The sixth-highest scoring motif matches the consensus binding site of the splicing factor TRA2β. We expected the TRA2β motif to be recognized by our algorithm, because it confers a regulatory signal by means of DNA sequence and was also identified by a previous report on coding region binding sites (17). For both datasets, discovered motifs were matched against known human miRNAs obtained from miRBase (18, 19). Among the top 15 scoring motifs in the real dataset are four miRNA target sites (for let-7, miR-9, miR-125a, and miR-153), which had SLCSs of 183.6, 153.2, 132.4, and 56.6, respectively, representing significant enrichment compared with the total set of scored motifs from the permuted dataset (P < 10−6, one-tailed hypergeometric test) (Fig. 1 Inset). Other motifs include highly conserved sequence elements of unknown function.
Fig. 1.
Motifs with a high SLCS are enriched for miRNA target sequences. The distribution of non-zero SLCSs (black bars) from a 17-genome alignment and the distribution that resulted from an analysis of a randomly permuted genome are plotted. The distribution demonstrates that there are significantly more high-scoring motifs within coding regions compared with the distribution of scores from a randomized alignment (white bars). The highest-scoring sequence motifs (Inset) are enriched for miRNA target sequences. Results are filtered so that conserved motifs are removed if they are within 3 bp of a higher-scoring motif.
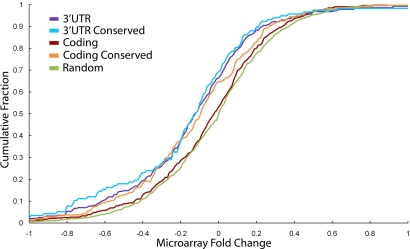
To investigate whether high-scoring sequence motifs matching miRNA target sites are indeed responsive to their associated miRNA, we performed a functional assay on the highest-scoring motif, which corresponds to the let-7 target site. The let-7 miRNA is highly conserved and was originally demonstrated to regulate developmental timing in the roundworm Caenorhabditis elegans (20). More recently, let-7 has been found to play a role in cell cycle regulation and cancer in humans (21). We transfected cultured human fibroblasts with a let-7 precursor or a control RNA and used microarrays to monitor changes in transcript levels after 24 h. All genes that contain the let-7 target site CTACCTCA within human coding regions were analyzed for down-regulation in response to let-7 addition. We discovered a significant correlation between the number of genomes in which the let-7 target site was conserved and the response to transfected let-7 (r = −0.196, P < 10−5, n = 521 for coding regions; r = −0.142, P = 0.017, n = 283 for 3′ UTRs), demonstrating that functional let-7 target sites within coding regions are more likely to be evolutionarily conserved. In addition, although previous reports have shown a statistically significant but small effect for miRNA target sites within coding regions (7), we discovered that the subset of target sites in coding regions that are highly evolutionarily conserved are functional as miRNA targets. As shown in Fig. 2, highly conserved target sites within coding regions resulted in down-regulation approaching that of the total set of 3′ UTR sites.
Fig. 2.
Highly conserved let-7 targets within coding regions are functional. Cultured human fibroblasts were transfected with pre-let-7b or a control RNA. Twenty-four hours after transfection, RNA was isolated, labeled, and hybridized to a whole-genome microarray. The fold change in expression between cells transfected with let-7b or the control was determined. The cumulative distribution of the fraction of genes down-regulated in the microarray data shows a significant difference between a random set of genes and genes with target sites in their 3′ UTR (P = 0.014, one-tailed Kolmogorov–Smirnov test), but not with genes with target sites in their coding region (P = 0.26). Genes with highly conserved target sites in their coding region, however, are significantly different from the set of random genes (P < 10−4), as are genes with highly conserved target sites in their 3′ UTRs (P < 10−3).
To determine whether the human genes containing a conserved let-7 target site are related in terms of biological function, we tested the 25% of genes with the most highly conserved let-7 target sites for enrichment in Gene Ontology categories (22). Genes were ranked by awarding one point for each genome in which the let-7 binding site is conserved. Scores were summed for genes with multiple let-7 target sites. We found significant enrichment among several Gene Ontology categories, including the processes cell cycle (multiple hypothesis-corrected P < 0.001), embryonic limb morphogenesis (corrected P < 0.002), chromatin modification (corrected P < 0.002), and posttranslational protein modification (corrected P < 0.01).
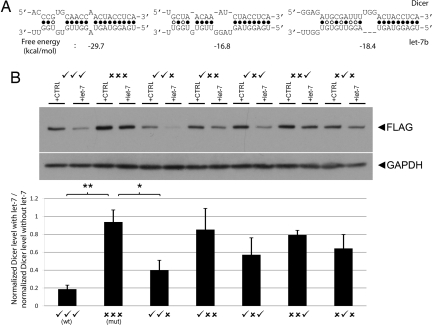
Next, we sought to verify directly that let-7 targets one of the genes with a conserved target site in its coding sequence. We selected the enzyme Dicer, which contains three highly conserved let-7 target sites (Fig. 3A) and performs the last processing step of miRNA maturation before the miRNA is loaded into the RNA-induced silencing complex (RISC). Dicer was recently found to be posttranscriptionally down-regulated by let-7, thus raising the possibility of a let-7-mediated negative-feedback loop (23, 30, 31).
Fig. 3.
The microRNA let-7 targets the coding region of Dicer. (A) Dicer contains three highly conserved let-7 target sites. Alignments between the let-7 miRNA and the target sites are shown. The first two sites are conserved in 16 of 17 genomes, and the last is conserved in all 17 genomes. The coding sequence of Dicer is shown above let-7b. (B) HEK 293 cells were cotransfected with Dicer and with let-7b pre-miRNA or a pre-miRNA control. Several variants of the Dicer construct were used, from which various combinations of the let-7 sites had been abrogated by site-directed mutagenesis. (Upper) An immunoblot of Dicer levels and GAPDH levels. Wild-type Dicer is in lanes 1 and 2, fully mutated Dicer is in lanes 3 and 4, and each subsequent pair of lanes is marked to indicate which sites were intact (✓) and which were abrogated (×). Cellular lysates were analyzed by immunoblotting with antibodies to the FLAG epitope (upper band) or GAPDH (lower band). (Lower) The fold reduction in the presence versus absence of exogenous let-7 for each Dicer variant. Means (n = 4) and standard errors are plotted. Wild-type Dicer is significantly down-regulated by let-7 compared with the Dicer variant with all three target sites abrogated (two-tailed t test, P < 0.01). The Dicer variant with the first two target sites intact is also significantly down-regulated (P = 0.02). The Dicer variant with the first and third target sites intact is down-regulated with the next-highest magnitude, but the effect did not reach significance. Dicer variants with a single site intact are down-regulated less strongly than variants with two sites.
To measure whether let-7 down-regulates Dicer via its coding region, we cotransfected human embryonic kidney 293 cells with an expression vector containing the coding sequence of Dicer (without its 3′ UTR and tagged with the FLAG epitope) and one of two pre-miRNAs (pre-let-7b or a control RNA). Additionally, to assess the relative effects of the three highly conserved let-7 target sites, we also created several variants of the Dicer construct by subjecting the let-7 target sites to site-directed mutagenesis. These mutations abrogated the let-7 binding sites while keeping the protein's amino acid sequence intact (see Materials and Methods). Cell lysates were collected after 48 h and analyzed by immunoblotting with an antibody to the FLAG epitope. Protein concentrations were normalized to GAPDH band intensity for quantification.
Our results show that wild-type Dicer is strongly down-regulated by the let-7 miRNA compared with transfection with a negative control (Fig. 3B, lanes 1 and 2), whereas let-7 has minimal effect on the version of Dicer in which the three coding region let-7 target sites are mutated (lanes 3 and 4) (wild type versus mutant compared by two-tailed t test, P < 0.01). The variant with the first two sites intact (lanes 5 and 6) is also significantly down-regulated compared with the fully mutated variant (two-tailed t test, P < 0.05), although to a lesser degree than wild-type Dicer. Although the variant with the first and third sites intact showed down-regulation of the next-greatest magnitude among the variants we tested (lanes 9 and 10), this effect did not achieve statistical significance. Each variant with a single intact site showed down-regulation of a lower magnitude compared with those with two intact target sites (lanes 7 and 8, 11 and 12, and 13 and 14). The increased importance of the first two let-7 target sites may be explained by their close proximity, because nearby miRNA target sites have been shown to act cooperatively (24).
Our findings also suggest that endogenous levels of let-7 are sufficient for down-regulating wild-type Dicer, because FLAG band intensity that results when wild-type Dicer is cotransfected with a control RNA is significantly lower than the band intensity that results when the fully mutated variant is cotransfected (wild type versus mutant compared by paired two-tailed t test, P = 0.019). To confirm these results, we transfected wild-type and fully mutated Dicer into two cell lines—the wild-type HCT116 human colorectal cancer cell line and a version of this cell line with targeted disruption of Dicer exon 5, which has the effect of lowering total intracellular mature miRNA concentrations (32). Down-regulation of wild-type versus mutated Dicer is larger in wild-type cells compared with cells with defective miRNA processing (Fig. S2). These results reinforce the importance of endogenous let-7 in regulation of Dicer levels.
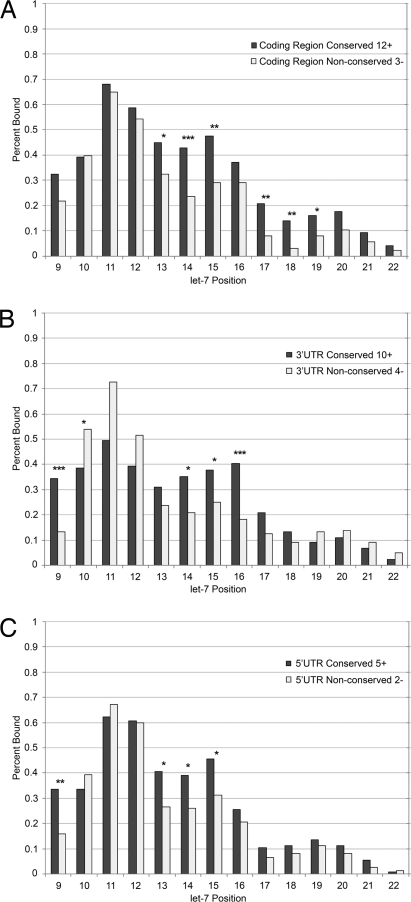
To assess whether conservation of target sites might give insight into the mechanism by which miRNAs silence transcripts at various target locations, we used the program RNAcofold from the ViennaRNA suite (25) to computationally fold let-7 to all of its seed matches in coding regions, 3′ UTRs, and 5′ UTRs. We compared base-pairing probabilities along the entire let-7 miRNA between conserved and nonconserved target seeds. Our results for 3′ UTRs (Fig. 4B) show a preference for looping at base pairs 10–12, followed by a preference for binding at positions 13–16, in conserved compared with nonconserved seed matches. These results closely recapitulate the binding preferences previously reported in papers by Kiriakidou et al. (26), which established a preference for a short bulge after the first nine bases of the miRNA, and Grimson et al. (7), which established the importance of base pairing at positions 13–16 of the miRNA. In sharp contrast, our data show a statistically significant preference for binding in positions 13–15 and 17–19 for conserved seed matches in coding regions and no preference for looping (Fig. 4A), suggesting a possible mechanistic difference between miRNA targeting in coding regions and in 3′ UTRs. Conserved coding region seeds, but not conserved 3′ UTR seeds, prefer a greater number of bound nucleotides in the remainder of the miRNA (P < 10−9, two-tailed Fisher exact test). We also tested binding preferences for seed matches in 5′ UTRs (Fig. 4C) and discovered a statistically significant preference for binding in positions 13–16. These data support the finding that miRNA target sites in 5′ UTRs may be functional.
Fig. 4.
Recognition sites at various positions within a transcript have different patterns of miRNA binding. Sites complementary to the let-7 seed were extracted from 3′ UTRs (A), coding regions (B), and 5′ UTRs (C), along with additional upstream sequence, and computationally folded to a consensus let-7 miRNA (see Materials and Methods). The fraction of target–seed interactions in which a particular base pair was bound was determined for each position of the miRNA outside of the seed. The fraction bound was compared for highly conserved versus poorly conserved target seeds. Numbers indicate the number of genomes in which a seed was conserved; 12+ indicates indicates conservation in 12 or more genomes, 3− indicates conservation in three or fewer genomes. Statistical comparisons were performed by using the Fisher exact test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Different cutoffs for the number of genomes in the “conserved” and “nonconserved” groups were used for each search region, and target seeds of 6 bp were used for 5′ UTRs to ensure an adequate number of sequences on which to perform statistical analysis (Fig. S3 shows results for various conservation levels).
Our findings demonstrate that there are regulatory sequence elements within coding regions that have been evolutionarily conserved through hundreds of millions of years of evolution (27) at a significantly higher level than would be expected by chance. These conserved sequence elements are enriched for miRNA-binding sites, thus supporting a potential role for miRNA targeting within coding regions in animals as well as plants. Further, we show that conserved let-7 target sites present within the coding regions of human genes are responsive to miRNAs, that the regulated genes are functionally related to one another, and that conserved miRNA target sites within coding regions are comparably effective at posttranscriptional repression relative to sites within 3′ UTRs in the microarray experiment described above. We demonstrate that the coding region of one gene in particular, Dicer, contains functional target sites for let-7 and loses its responsiveness to let-7 when target sites within its coding sequence are synonymously mutated. Regulation of the Dicer endonuclease by let-7 suggests a possible negative-feedback mechanism for miRNA processing. A similar mechanism has been discovered in Arabidopsis, in which an miRNA targets Dicer-like1 transcript degradation by means of a target sequence present in the middle of the coding sequence (28). We also provide computational evidence to suggest that miRNA function in coding regions and 3′ UTRs may differ mechanistically.
Our results provide evidence that roughly 700 genes in the human genome have conserved regulatory sites within coding regions. Because our investigation focused on conservation among 17 organisms with a large amount of evolutionary distance between them, this number almost certainly underestimates the number of human genes that contain conserved regulatory sites within coding regions. Also, the miRNAs that we identified with this approach are expected to be enriched for those that play a role in fundamental processes, such as early development or cell cycle regulation, a hypothesis supported by the fact that the highest-scoring miRNA in our computational screen was let-7, which is highly evolutionarily conserved and targets genes in coding sequences that are enriched for these biological functions. Our findings represent an important step toward a comprehensive understanding of regulatory DNA elements and the role of these elements in cellular networks and human health and disease.
Materials and Methods
Extracting Coding Regions.
The 17-genome multiple alignment used in this study was obtained from the UCSC Genome Browser (multiple alignment files are available directly from http://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz17way/). Chromosomes M and Y were excluded from the analysis. Coding regions were identified by using the annotated gene list available from http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/refGene.txt.gz (column names and definitions are available from http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/refGene.sql). Genes were excluded if the cdsStartStat or cdsEndStat was not complete (“cmpl”) to filter out genes with imprecise sequences. When coding regions overlapped, the longer coding region was used in the analysis. Multiple alignments for coding regions were directly parsed from the multiple alignment files.
Scoring Model.
Our scoring method is designed to detect excess sequence conservation over and above that required for amino acid conservation. The probabilities for each of the codons in a motif are then used to assign the motif a SLCS, as described below. For a given conserved codon in a motif, its codon sequence level conservation (CSLC) probability is calculated as follows:
where consseq denotes that the codon is sequence-conserved and consaa denotes that the codon is amino acid-conserved. When a codon is amino acid-conserved but not sequence-conserved, the human sequence is used to determine the codon assignment.
We also measured these probabilities for partial codons, because conserved motifs will span both full and partial codons in the multiple alignment. If x bases of the codon are within the conserved motif, the probability is measured as follows:
where consx,pos,seq denotes that the first or last (denoted by pos) x bases of the codon are sequence-conserved. The portion of a codon that is part of a conserved motif must be fully sequence-conserved to be scored, but the remainder of the codon need not necessarily be conserved. As a result, we must also measure partial codon probabilities in the case where x bases within the codon are fully sequence-conserved, but the full codon is not amino acid-conserved:
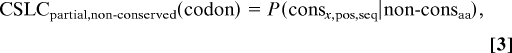 |
where nonconsaa denotes that the codon is not amino acid-conserved. Our resulting table (Table S1) contains the probability that a given full or partial codon is sequence-conserved, further broken down by whether or not the codon is amino acid-conserved.
Motif Scoring.
To score a conserved motif, we looked up the appropriate probability in Table S1 for each partial or full codon. The logs of the reciprocal of the probabilities are summed across the motif:
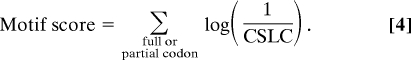 |
When a motif is scored more than once across the genome, this motif score is further summed across occurrences:
Permutation Testing.
For each amino acid that was perfectly conserved at the amino acid level in all 17 genomes, we replaced it with perfect sequence conservation or with imperfect sequence conservation at the empirically determined rate. The regenerated genome was analyzed, and sequence level conservation scores were determined as for the actual human genome.
let-7 Conservation.
To find genes containing the let-7 target 8-mer, the human sequence from the multiple alignment was searched for the motif CTACCTCA in such a way that gaps in the human sequence of the alignment were ignored (i.e., the human sequence CTACC---TCA in the alignment was included in the analysis). For each occurrence of this motif, a conservation score was calculated by counting the number of genomes in which this motif was conserved in the same frame as the human sequence. If the motif appeared in a gene more than once, the conservation score was summed across occurrences.
Binding Profiles Downstream of the let-7 Seed.
All instances of the 8-mer let-7 target seed were retrieved from each search region (coding regions, 3′ UTRs, and 5′ UTRs) with 27 bp of upstream sequence. Sequences were discarded if less than 22 bp of upstream sequence was available (e.g., if the transcription start site was encountered for target sites in the 5′ UTR). The seed and upstream sequence were computationally folded to let-7 using the RNAcofold program of the Vienna RNA package (25) (using the −noGU option to disallow G/U base pairing, and the −C option to enforce seed binding). Because there are several members of the let-7 miRNA family, we constructed a consensus let-7 sequence, which is identical to let-7a.
Genome Coverage.
To estimate the number of genes that contain high-scoring motifs conserved at the DNA sequence level, we subtracted the total number of genes containing a conserved motif in the permuted dataset from the number of genes containing a conserved motif in the nonpermuted dataset. Only motifs with an SLCS >15 were considered.
Monitoring Dicer Levels.
The CS2+ expression vector containing the coding sequence of Dicer with His and FLAG tags was transfected into 293 cells along with control pre-miRNA, pre-let-7b, control anti-miRNA, or anti-let-7b. Levels of Dicer or the loading control GAPDH were determined by immunoblotting of protein lysates, collected at 48 h, with antibodies to FLAG or GAPDH. The three let-7-binding sites were mutated with the Stratagene multisite-directed mutagenesis kit, and the experiment was repeated.
Microarray Analysis.
To monitor changes in transcript levels associated with let-7 overexpression, dermal fibroblasts were transfected with pre-let-7b or a control RNA. Samples were collected after 24 h. Total RNA was isolated by using the mirVana miRNA Isolation kit (Ambion) according to the manufacturer's protocol. High-quality RNA was confirmed by using a Bioanalyzer 2100 (Agilent Technologies), and the amount was determined with a NanoDrop spectrophotometer (Thermo Scientific). A total of 325 ng of total RNA was amplified by using the Low RNA Input Fluorescent Labeling Kit (Agilent Technologies) according to the manufacturer's protocol. Cyanine 3-CTP (Cy-3) or cyanine 5-CTP (Cy-5) (Perkin–Elmer) was directly incorporated into the cRNA during in vitro transcription. Cy-3-labeled cRNA was used as reference RNA to compare with corresponding Cy-5-labeled let-7b-overexpressing sample cRNA. The mixed labeled cRNA was hybridized to whole Human Genome Oligo Microarray slides (Agilent Technologies) at 60°C for 17 h and subsequently washed according to the Agilent Oligo Microarray Hybridization Kit. Slides were scanned with a dual laser scanner (Agilent Technologies). The Agilent feature extraction software, in conjunction with the Princeton University Microarray database, was used to compute the log ratio of the difference between the two samples for each gene after background subtraction and dye normalization. Of the ≈44,000 probes on the microarray, 25,972 probes generated signal in four of five arrays. Data for these probes were mapped to genes based on UniGene Clusters. If multiple probes mapped to a single gene, the values were averaged, resulting in 14,590 genes.
Site-Directed Mutagenesis.
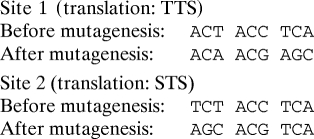
Primers (synthesized by Integrated DNA Technologies) were designed to introduce mutations into all three let-7 target sites in the Dicer coding region such that the let-7 target site would be abrogated without changing the amino acid translation of the Dicer gene. The sites were mutated as follows:
 |
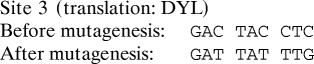 |
Mutagenesis was performed with the Stratagene multisite-directed mutagenesis kit according to the manufacturer's instructions. The msDicer construct was verified to contain all three mutated sites and no other mutations by primer extension sequencing performed by GENEWIZ.
Supplementary Material
Acknowledgments.
We thank Jason Myers, David Hendrickson, and Jim Ferrell for providing the Dicer expression vector; Burt Vogelstein for providing the HCT116 Dicerex5 cells; and Curtis Huttenhower, Peter Wei, Amy Caudy, Leonid Kruglyak, and the entire Coller and Kruglyak labs for helpful discussions. H.A.C. is the Milton E. Cassel scholar of the Rita Allen Foundation. This work was supported in part by grants from the PhRMA Foundation, National Institutes of Health/National Institute of General Medical Sciences Grant 1R01 GM081686 to H.A.C, National Institutes of Health/National Institute of General Medical Sciences Grant P50 GM071508 to H.A.C. and A.L-M., and National Cancer Institute Training Grant 5T32 CA009528 to J.J.F.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803230105/DCSupplemental.
References
- 1.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 11.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itzkovitz S, Alon U. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res. 2007;17:405–412. doi: 10.1101/gr.5987307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karolchik D, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Down T, Leong B, Hubbard TJ. A machine learning strategy to identify candidate binding sites in human protein-coding sequence. BMC Bioinformatics. 2006;7:419. doi: 10.1186/1471-2105-7-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Boyle EI, et al. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 24.Saetrom P, et al. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhart SH, et al. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 28.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 29.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 31.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 32.Cummings J, et al. The colorectal microRNAome. PNAS. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.