Abstract
Evolutionary change in gene regulation can result from changes in cis-regulatory elements, leading to differences in the temporal and spatial expression of genes or in the coding region of transcription factors leading to novel functions or both. Although there is a growing body of evidence supporting the importance of cis-regulatory evolution, examples of protein-mediated evolution of novel developmental pathways have not been demonstrated. Here, we investigate the evolution of prolactin (PRL) expression in endometrial cells, which is essential for placentation/pregnancy in eutherian mammals and is a direct regulatory target of the transcription factor HoxA-11. Here, we show that (i) endometrial PRL expression is a derived feature of placental mammals, (ii) the PRL regulatory gene HoxA-11 experienced a period of strong positive selection in the stem-lineage of eutherian mammals, and (iii) only HoxA-11 proteins from placental mammals, including the reconstructed ancestral eutherian gene, are able to up-regulate PRL from the promoter used in endometrial cells. In contrast, HoxA-11 from the reconstructed therian ancestor, opossum, platypus, and chicken are unable to up-regulate PRL expression. These results demonstrate that the evolution of novel gene expression domains is not only mediated by the evolution of cis-regulatory elements but can also require evolutionary changes of transcription factor proteins themselves.
Keywords: evolution of development, functional divergence, molecular evolution, trans-regulatory evolution
Developmental evolutionary studies have played an important role in elucidating the molecular mechanisms that give rise to phenotypic novelty and the evolution of gene regulation (1, 2). Mechanistically, embryonic development is governed by the precise temporal and spatial deployment of gene regulatory networks (1, 2). Thus, the evolution of development is fundamentally a question of the origin and evolution of gene regulation and regulatory networks. An emerging principle from evolutionary developmental studies is that adaptive mutations with phenotypic effects are much more likely to occur in cis-regulatory elements than in protein-coding genes (3–6). Indeed, there is ample evidence for the importance of cis-regulatory evolution (4–6), but it has also been shown that transcription factors do not remain functionally equivalent during evolution (7–9). This finding suggests that the evolution of transcription factors themselves may play an active role in the evolution of development and the origin of morphological innovations (10, 11). So far, however, no detailed evolutionary scenario has been elucidated that explains the role of transcription factor evolution. Here, we reconstruct some of the events that lead to the origin of pregnancy in eutherian mammals and demonstrate that adaptive changes in a transcription factor (HoxA-11) protein play an essential role.
One of the key evolutionary innovations of eutherian (placental) mammals is prolonged internal gestation and an invasive embryo (pregnancy), adaptations made possible by the evolution of uterus and placenta. Knockout and knockdown studies have identified many genes important for pregnancy, including transcription factors, growth factors, and cell signaling molecules, such as prolactin (PRL) (12). In humans, PRL is one of the most dramatically induced genes in decidualized (differentiated) endometrial stromal cells, and decidua-derived PRL is one of the most abundant secretory products in the amniotic fluid (13–17). Decidually expressed PRL has numerous functions, including regulation of uterine epithelial cell differentiation, trophoblast growth, angiogenesis, and modulating immune response (14, 16). A particularly important role for decidual PRL is the suppression of genes detrimental to pregnancy, such as interleukin 6 (IL-6) and 20α-hydroxysteroid dehydrogenase (20α-HSD), which promote inflammation and catabolize progesterone, respectively (13, 18, 19). Thus, the proper regulation of decidual PRL is essential for implantation and pregnancy.
Although the evolutionary mechanisms and timing of PRL recruitment into uterine expression are unclear, the molecular mechanisms leading to decidual PRL activation are well understood. Several genes have been identified that influence endometrial PRL expression, such as C/EBPβ, FOXO1A, ETS1 and the progesterone receptor (20–24). The Abd-B-related Hox gene HoxA-11 is required is to activate expression of PRL (V.J.L., B.G., and G.P.W., unpublished data). In addition to its role in PRL regulation, HoxA-11 is essential for the pre- and postnatal development of the female reproductive tract and is required for successful implantation of the mammalian blastocyst (25–28). Remarkably, a previous molecular evolutionary analysis of HoxA-11 suggested that an episode of positive Darwinian selection occurred in the stem-lineage of placental mammals, coincident with the origin of the uterus and pregnancy (29). This result suggests that adaptive evolution in HoxA-11 may have generated a novel PRL regulatory ability, possibly through the origin of new cofactor associations required for PRL expression in endometrium.
To clarify the evolutionary mechanisms that gave rise to uterine PRL expression, we have examined in detail the molecular and functional evolution of the PRL regulatory transcription factor HoxA-11. We show that endometrial PRL expression is a derived feature of placental mammals and that the episode of positive selection in HoxA-11 occurred coincident with the origin of its ability to interact functionally with another transcription factor protein (FOXO1A) and thereby up-regulate PRL from the promoter used in endometrial cells. In contrast, HoxA-11 from opossum, the reconstructed therian ancestor, platypus, and chicken is unable to up-regulate PRL expression. Remarkably, although PRL is not expressed in the uterus of the opossum (or chicken), genes normally silenced in the uterus during pregnancy (IL-6 and 20α-HSD) are expressed in the uterus of the pregnant opossum. These results identify some of the initial stages in the evolution of a novel gene regulatory network and demonstrate that gene recruitment is not only mediated by the evolution of cis-regulatory elements but also requires evolution of transcription factor proteins themselves.
Results and Discussion
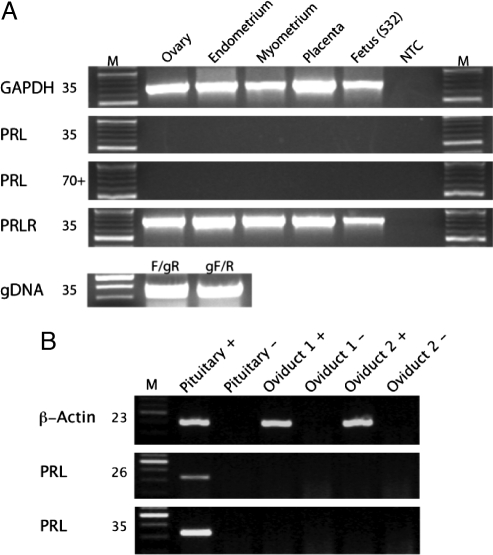
Although PRL and the prolactin receptor (PRLR) are expressed in the uterus and placenta of placental mammals (13–17), PRL has not been detected in the oviduct of fish, amphibians, and squamate reptiles (30–32). However, the PRLR is expressed in the oviducts of these species (30–32). Similarly, the PRLR is expressed in the uterus of marsupials (33), but the expression of PRL in the uterine tissues of marsupials has not been directly tested. We assayed PRL and PRLR expression by RT-PCR in the female reproductive tissues and placenta of a gestating marsupial, the gray short-tailed opossum (Monodelphis domestica), and found that PRL was not expressed in endometrium, myometrium, placenta, ovary, or whole-stage 32 fetus in this species [supporting information (SI) Fig. S1], although the RT-PCR primers efficiently amplify targets when paired with primers targeting introns and using genomic DNA as template (Fig. 1A). We also assayed PRL expression in chicken oviduct and pituitary and found that although PRL was expressed in pituitary it was not expressed in oviducts of egg-laying hens (Fig. 1B). To determine whether PRL is also expressed the female reproductive tract in basal mammalian lineages, we assayed PRL expression in the placental tissue of the African elephant (Loxodonta africana) by RACE and found that PRL expression is expressed in this tissue (Fig. S2). Thus, we conclude that PRL expression in the uterus is a derived feature of placental mammals.
Fig. 1.
PRL and PRLR expression in opossum and chicken female reproductive tracts. (A) Expression of PRL in tissues of the opossum female reproductive tract. Tissues are shown on top, genes to the left. Numbers next to gene names are the number of cycles used in PCRs. M, 100-bp ladder; NTC, no template control; gDNA, PCR of opossum genomic DNA using the RT-PCR primers (F and R) and intronic primers (gR and gF). The genomic amplifications show that the RT-PCR primers are functional. (B) Expression of PRL in the oviducts of two egg-laying hens. Note that PRL is only expressed in chicken pituitary.
An important role of PRL expression in the uterus of placental mammal is to silence the expression of genes that are detrimental to pregnancy, such as IL-6 and 20α-HSD, which promote inflammation and catabolize progesterone, respectively (13, 18, 19). The coincident origin in the stem-lineage of placental mammals of pregnancy and PRL expression in the female reproductive tract suggests that some of the functions of PRL during pregnancy may be absent outside of the Eutheria. To test this hypothesis we assayed IL-6 and 20α-HSD expression by RT-PCR in the female reproductive tissues of a gestating opossum (M. domestica) and found that both genes were expressed in the endometrium, ovary, and stage 32 fetus (Fig. S3). This result suggests that a major direct effect of recruitment of PRL into uterine expression may have been silencing the expression of IL-6 and 20α-HSD, allowing a more invasive embryo and prolonged gestation without eliciting a maternal immune response that was detrimental to the developing embryo.
Our finding that PRL expression in the uterus is an innovation of placental mammals is consistent with the recent finding that the enhancer driving decidual PRL (dPRL) expression in human endometrial stromal cells is a eutherian-specific transposable element (MER20) (34). We have identified this element from all major mammalian lineages by searching mammalian genome databases. Using a Bayesian relaxed-clock method, we dated the insertion of the MER20 element upstream of PRL to ≈166–155 MYA, shortly after the placental–marsupial divergence at ≈173–175 MYA (35, 36) and coincident with a reported period of rapid evolution in HoxA-11 (29), suggesting that the earliest time that PRL could have been expressed in the uterus of placental mammals was shortly after their origin.
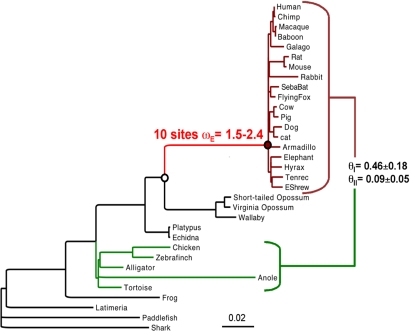
To test directly for an episode of positive Darwinian selection in HoxA-11 in the stem lineage of placental mammals, we used likelihood models of dN/dS ratio variation among lineages and sites (37–39) on a large HoxA-11 dataset. Under the simplest model of rate variation, which constrains all lineages in the phylogeny to have the same dN/dS ratio, the dN/dS ratio was 0.146 (Table S1). To test for an episode of accelerated evolution in the stem-lineage of placental mammals we used a two-ratios model that estimated the rate of the eutherian stem-lineage (ωE) separately from all other lineages (ω0). Under this model, the dN/dS ratio in eutherian stem-lineage was significantly higher than the background rate (P = 0.011) (Table S1). A three-ratio model that estimates a separate dN/dS ratio for the eutherian stem-lineage, crown group eutherians (ωCE) and all other animals found that although the eutherian stem-lineage evolved significantly faster than nonplacentals, the intensity of purifying selection has increased moderately within the crown group eutherians (P = 0.027) (Table S1). We also tested for amino acid sites that were positively selected in the eutherian stem-lineage by comparing branch-site model A, which allows for a class of codons to have dN/dS >1, with the null model (M1a), which does not allow for positively selected sites. Branch-site model A, which is a significantly better fit to the data than the model that does not allow for selected sites (P = 0.005) (Table S1), identified 10 sites under positive selection (ω = 1.51–2.41) in the stem-lineage of placental mammals (Fig. 2). Although these sites are under relatively weak negative selection in nonplacental mammals (ω = 0.44), they are under extremely strong selective constraints within the extant placental mammals (ω = 0.08) (Fig. 3). Together, these findings suggest that the HoxA-11 protein has acquired an additional functional constraint during the origin of placental mammals.
Fig. 2.
Positive selection on HoxA-11 exon 1 in the stem-lineage of placental mammals. Eutherians are shown in dark red, with the stem-lineage of eutherians in red. Ten sites were identified under positive selection with ωE = 1.5–2.4 depending on how codon frequencies were modeled (equal or empirical). Type I (θI) and type II (θII) functional divergence was estimated between eutherians and sauropsids (shown in green). θI, P < 0.01; θII, P < 0.05. Branch lengths are proportional to the number of nonsynonymous substitutions per codon. The red filled and white filled circles indicate the location of the ancestral eutherian and therian sequences recreated for functional tests.
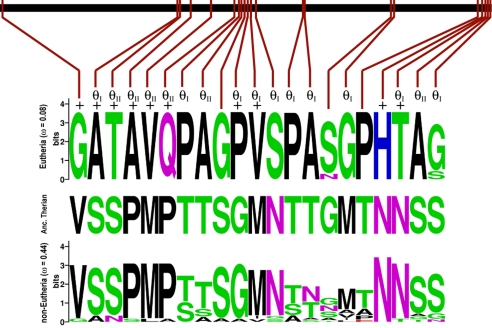
Fig. 3.
Location of positively selected and functionally divergent sites in exon 1. The black bar represents exon 1 of HoxA-11, red marks indicate the relative locations of sites that were substituted in the stem-lineage of eutherians. The two sequence logos show site variation within eutherians (Top) and the noneutherians (Bottom). The inferred amino acids at these sites is shown for the reconstructed therian ancestor (Middle). Amino acids are color coded by physicochemical property. +, site identified under positive selection; θI, type I site; θII, type II site. The rate of evolution of these sites is shown for eutherians and noneutherians.
To test whether HoxA-11 genes in the Eutheria evolve under different selective constraints than in other vertebrate lineages, we estimated the coefficient of functional divergence (θ), which measures the difference in evolutionary rate at amino acid sites between two clades. Rejection of the null hypothesis (θ = 0) is evidence for altered functional constraints between clades (40, 41). We found significant evidence of type I functional divergence between eutherians and sauropsids (θI = 0.46 ± 0.18, P < 0.01; Fig. 1) and significant but weak evidence of type II functional divergence (θII = 0.09 ± 0.05, P < 0.05; Fig. 1). Type I sites (40) are amino acids that are conserved within the Eutheria but variable in the sauropsids, indicating they evolved new or stronger structural and functional constraints in Eutheria, whereas type II sites (41) are amino acids that are fixed in eutherians and sauropsids for different amino acids, indicating a functional switch. The greater degree of type I (θI = 0.46) than type II (θII = 0.09) functional divergence suggests that selection recruited weakly constrained amino acids into a novel function rather than amino acids that were under strong preexisting functional constraints. These data suggest that selection acted within existing structural and functional constraints on proteins to generate novel functions (Fig. 3).
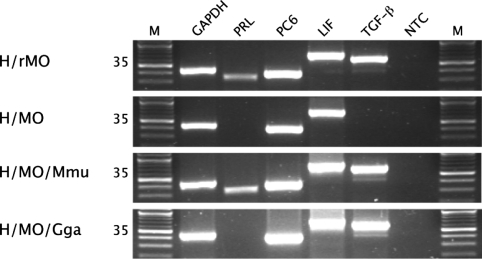
The episode of adaptive evolution in HoxA-11 in the stem-lineage of eutherians and the absence of PRL expression in the uterus and oviduct of nonplacental mammals suggests that the ability to regulate PRL expression in the decidua might be a novel function of HoxA-11 in eutherians. We initially tested for functional differences between human and chicken HoxA-11 genes by using an in vivo knockdown/xenogene rescue system in cultured human endometrial stromal cells (hESCs). Briefly, hESCs were induced to decidualize with physiological levels of 17β-estradiol and progestin while intrinsic HoxA-11 expression was knocked down with a morpholino, followed by rescue with either mouse or chicken HoxA-11. Eight days after differentiation, genes known to be expressed in decidualized stroma, but not expressed in undifferentiated cells, were highly expressed in cells treated with both random-sequence control morpholino (Fig. 3) and untreated cells (data not shown). However, decidualized cells treated with the HoxA-11-specific morpholino failed to express PRL and TGF-β. To test whether HoxA-11 knockdown was reversible, we rescued the knockdown with mouse HoxA-11, which recovered both PRL and TGF-β expression (Fig. 3). We next rescued the knockdown with chicken HoxA-11, which does not have the derived amino acid replacements of placental mammals, and we recovered TGF-β expression but failed to rescue PRL expression (Fig. 4). These results suggest that the HoxA-11 protein underwent amino acid substitutions necessary for the derived ability to regulate PRL but that the ability to regulate TGF-β is ancestral for the amniote HoxA-11 protein.
Fig. 4.
Partial functional nonequivalence of human, mouse, and chicken HoxA-11 genes in endometrial cells. The expression of GAPDH (endogenous control) and marker genes of decidualized stromal cells (PRL; PC6, proprotein convertase 6; LIF, leukemia inhibitory factor; TGF-β; NTC, no template control) were assayed by RT-PCR after hormone-induced differentiation and morpholino treatment. Treatments are shown on the left. H/rMO, hormone and random morpholino. H/MO, hormone and HoxA-11-specific morpholino. H/MO/Mmu, hormone and HoxA-11-specific morpholino rescued with mouse HoxA-11. H/MO/Gga, hormone and HoxA-11-specific morpholino rescued with chicken HoxA-11. M, 100-bp ladder. The number of PCR cycles used in the PCR is shown next to the treatments.
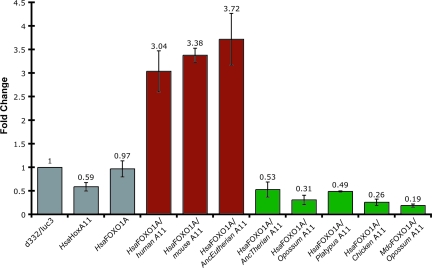
The knockdown/rescue approach described above is a powerful technique to test for functional divergence, particularly in nonmodel organisms, but it can be complicated by complex auto- and cross-regulatory interactions that can interfere with the knockdown, the rescue, or both. Therefore, it is desirable to test functional divergence in more controlled assays, particularly outside of the cell type of interest, once functional differences are identified. To this end, we used a luciferase reporter assay in HeLa cells to test specifically the ability of HoxA-11 genes from the major amniote lineages to activate transcription from the decidual PRL enhancer. Transfection with HoxA-11 alone repressed reporter gene expression, consistent with previous studies that found that the HoxA-11 protein acts as an intrinsic repressor (42). Cotransfection of mouse or human HoxA-11 with FOXO1A led to dramatic activation from the PRL enhancer (Fig. 5), consistent with the rescue results in hESCs. Next, we tested whether the opossum, platypus, and chicken HoxA-11, which lack the derived placental-specific adaptive amino acid substitutions, activated luciferase expression from the decidual PRL enhancer. Remarkably, unlike mouse and human HoxA-11, the orthologs from opossum, platypus, and chicken, species that lack PRL expression in uterus and oviducts, repressed luciferase expression when cotransfected with FOXO1A (Fig. 4). Finally, we tested whether opossum HoxA-11 could activate expression from the dPRL enhancer when paired with the opossum FOXO1A gene. Similarly to the opossum HoxA-11 paired with human FOXO1A, opossum HoxA-11/opossum FOXO1A failed to activate luciferase expression (Fig. 5).
Fig. 5.
Functional divergence in amniote HoxA-11 genes ability to up-regulate expression from the decidual PRL enhancer. The ability of HoxA-11 genes to activate expression from the decidual PRL enhancer was determined in a luciferase reporter construct using dPRL-332/luc3 (d332/luc3). Transfection with human HoxA-11 alone (HsaHoxA11) repressed reporter gene expression. Transfection with human FOXO1A alone (HsaFOXO1A) had no effect on reporter gene expression. Cotransfection of human FOXO1A with human HoxA-11 (human A11) or mouse HoxA-11 (mouse A11) activated reporter gene expression. Cotransfection of human FOXO1A with opossum HoxA-11 (opossum A11), platypus HoxA-11 (platypus A11) or chicken HoxA-11 (chicken A11) repressed reporter gene expression. Cotransfection of the reconstructed ancestral eutherian HoxA-11 gene (AncEutherian A11) with the human FOXO1A gene activated reporter gene expression, conversely cotransfection of the reconstructed ancestral therian HoxA-11 gene (AncTherian A11) with the human FOXO1A did not lead to activation of reporter gene expression. Expression levels are shown as fold changes relative to luciferase expression in cells transfected with the reporter gene (d332/luc3) and empty vector (pcDNA3.1). n = 6, mean ± SEM.
To test specifically whether the amino acids identified from the statistical analysis as under positive selection and/or functional divergence were sufficient to confer PRL regulatory ability on a HoxA-11 gene that lacks PRL regulatory functions, we reconstructed the ancestral therian (i.e., the protein from the last common ancestor of humans and opossum) HoxA-11 gene by maximum likelihood and tested its ability to activate luciferase expression from the decidual PRL enhancer. As expected, the ancestral therian HoxA-11 gene, which lacks the selected and functionally divergent amino acid sites, was unable to cooperate functionally with FOXO1A to up-regulate PRL expression (Fig. 5). However, the ancestral eutherian HoxA-11 gene, which has the selected and functionally divergent amino acid sites and was reconstructed without ambiguity (Bayesian posterior probability of 1.0), did have PRL activation ability (Fig. 5). These results show that at least some of the amino acid substitutions in the stem-lineage of placental mammals generated a novel functional interaction with FOXO1A that was used to recruit PRL expression into the uterus of placental mammals.
Although protein-mediated evolution of developmental pathways is rarely excluded as a means of gene regulatory evolution, the contribution of protein change to the origin of novel gene regulatory networks is generally not considered to play a major role in evolution. The primary argument against protein-mediated evolution of gene regulatory networks is the negative pleiotropic effects of mutations that are ascribed to changes in protein-coding genes (3, 4). For example, given the multiple functions of HoxA-11 in blood cell differentiation (43) and development of the body axis (44), limbs (25, 45), kidney (46, 47), and male (27, 28) and female reproductive systems (25, 27, 28), it seems unlikely that a novel function could emerge in endometrial cells without simultaneously having deleterious effects in these other contexts. However, our data indicate that selection acted to maintain ancestral functions during the emergence of a novel function by recruiting amino acid sites that were previously under weak functional constraints and thus free to acquire novel functions. The identification of an episode of adaptive evolution in HoxA-11 coincident with the origin of a novel function demonstrates a clear link between adaptive protein evolution and the emergence of a novel function. This and other examples of functional divergence among transcription factors (7–9) indicate that the evolution of proteins themselves actively contributes to the evolution of development.
Materials and Methods
Gene Expression Surveys in Elephant, Opossum, and Chicken.
Expression of PRL, PRLR, FOXO1A, IL-6, and 20α-HSD in the pregnant (stage 32) opossum (M. domestica) endometrium, myometrium, placenta, ovary, and fetus was assayed by RT-PCR. Similarly, the expression of PRL in sexually mature (egg-laying) female chickens (Gallus gallus) and in the placental tissue of an African elephant (L. africana) was assayed by RT-PCR. Details are provided in the SI Text.
Molecular Evolutionary Analysis of HoxA-11.
HoxA-11 genes were identified from BLAST searches of whole-genome databases at National Center for Biotechnology Information, BLAT University of California at Santa Cruz, and BLAST ENSEMBL; see ST Text for a list of included species. We used codon-based maximum-likelihood models of coding sequence evolution implemented in CODEML in the PAML 4 package of programs (49) to test for lineages and amino acid sites under positive selection. Sites were classified as being under positive selection if they were identified from the Bayes Empirical Bayes (BEB) method with a posterior probability of >0.90 under model A. Initial branch lengths for the input trees were determined under the one-ratio model (M0) and used in further analyses. To ensure convergence of the maximum-likelihood estimates generated under the branch-specific and branch-site models we altered the starting values of the dN/dS ratio, the transition:transversion ratio, and the equilibrium codon frequencies (f =, f61) and reanalyzed the data. Difference in selective pressure between putatively positively selected sites in extant mammals and other species was obtained by averaging the sitewise dN/dS values from those sites with the site-specific model M3 with three rate categories. Ancestral sequences were inferred with CODEML, which uses maximum likelihood and an empirical Bayes approach to estimate ancestral character states, and the phylogeny shown in Fig. 2. The Bayesian posterior probability of the reconstructed therian ancestral sequence (AncTherian) was 0.96, and the probability of the eutherian (AncEutherian) ancestral sequence was 1.0. Functional divergence was tested with DIVERGE 2 (50). A limitation of functional divergence analyses is the requirement of at least four taxa per clade for estimation of the coefficient of functional divergence (θ). Given this limitation, we only tested for functional divergence between placental mammals and birds/reptiles.
HoxA-11 Expression Vector Construction.
We amplified axon 1 of HoxA-11 by PCR from the genomic DNA of human (Homo sapiens), mouse (Mus musculus), opossum (Didelphis virginiana), platypus (Ornithorynchus anatinus), and chicken (G. gallus) by using previously described degenerate primers (29). Exon 2 is identical in amino acid sequence for these species, therefore we fused the human exon 2 sequence to exon 1 of the above species by using primer overlap PCR mutagenesis; C-terminal His6 tags were added by PCR mutagenesis. His-tagged PCR products were cloned into the mammalian expression vector pcDNA3.1(+) (Invitrogen) and verified by sequencing. Proper expression of tagged HoxA-11 genes was checked in HeLa cells with immunocytochemistry using an anti-His antibody. The ancestral therian (AncTherian) and ancestral eutherian (AncEutherian) genes were synthesized by GeneScript Corp. with human optimized codon usage and ligated into pcDNA3.1.
In Vivo HoxA-11 Knockdown/Xenogene Rescue.
Tests of functional nonequivalence were performed in a telomerase-immortalized human endometrial stromal cell line (CRL-4003; American Type Culture Collection) treated either with an antisense morpholino (GeneTools) generated against the human HoxA-11 5′-UTR or a random sequence morpholino. Morpholinos were delivered by using EndoPorter (GeneTools) according to the manufacturer's protocols. Twelve hours after morpholino treatment, cells were differentiated with 10−9 M 17β-estradiol and 10−8 M medroxyprogesterone acetate, and total RNA was collected after 8 days. HoxA-11 knockdown in endometrial cells was rescued with either a mouse HoxA-11 or chicken HoxA-11 expression vector. Efficacy of morpholino knockdown and rescue was determined by assaying the expression of genes known to be induced during differentiation, including PRL, leukemia inhibitory factor, proprotein convertase 6, and TGF-β, and the expression of the housekeeping gene GAPDH by RT-PCR after RNA extraction (RNeasy; Qiagen) and cDNA preparation (High-Capacity Reverse Transcriptase kit, Applied Biosystems). RT-PCR primers were designed to span at least one intron.
Transient Transfection and Luciferase Reporter System.
HeLa cells were grown in 24-well culture plates in DMEM supplemented with 10% FBS. At 80% confluence, cells were transiently transfected with trans-IT-LT1 (Mirus) according to the manufacturer's instructions by using 100 ng of the dPRL luciferase reporter construct dPRL-332/Luc3, which contains the wild-type dPRL promoter sequence −332 to +65 relative to the human dPRL transcriptional start site (51). Depending on treatment, cells were cotransfected with either 200 ng of empty pcDNA3.1(+) (vehicle control) or 100 ng of the FOXO1A expression vector pcDNA3.1(+)-FOXO1A (Addgene plasmid 13507) and 100 ng of one of the HoxA-11 expression vectors described above. In addition, the effect of both FOXO1A alone and HoxA-11 alone on reporter gene expression was assayed by transfection with the reporter plasmid and 100 ng of pcDNA3.1(+)-FOXO1A/100 ng of empty pcDNA3.1(+), or 100 ng of pcDNA3.1(+)-hHoxA-11/100 ng of empty vector. Forty-eight hours after transfections, total mRNA was harvested, and cDNA was prepared by using a High-Capacity Reverse Transcriptase kit (Applied Biosystems). Luciferase expression was assayed with quantitative real-time PCR (qRT-PCR) by using a luciferase-specific and GAPDH TaqMan primer/probe pair (Applied Biosystems). Each experiment was repeated twice, with three replicates per experiment. qRT-PCRs were also performed in triplicate.
Supplementary Material
Acknowledgments.
We thank Dr. J. J. Roth for assistance in making expression constructs and tissue culture and Dr. T. Williams for help with immunocytochemistry. We also thank Dr. K. Smith (Duke University, Durham, NC) for providing pregnant opossum uterus samples and Dr. D. Wildman (Wayne State University, Detroit, MI) for providing RACE ready cDNA from elephant placenta. This work was supported by a grant from the John Templeton Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802355105/DCSupplemental.
References
- 1.Carroll S. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Malden, MA: Blackwell; 2005. [Google Scholar]
- 2.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. 1st Ed. Burlington, MA: Academic; 2006. [Google Scholar]
- 3.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prud'homme B, Gompel N, Carroll SB. Colloquium papers: Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 6.Stern DL. Perspective: Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 7.Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 8.Grenier JK, Carroll SB. Functional evolution of the Ultrabithorax protein. Proc Natl Acad Sci. 2000;97:704–709. doi: 10.1073/pnas.97.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 10.Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00440.x. in press. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi A, Behringer RR. Developmental genetics of the female reproductive tract in mammals. Nat Rev Genet. 2003;4:969–980. doi: 10.1038/nrg1225. [DOI] [PubMed] [Google Scholar]
- 13.Bao L, et al. Decidual prolactin silences the expression of genes detrimental to pregnancy. Endocrinology. 2007;148:2326–2334. doi: 10.1210/en.2006-1643. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Jonathan N, Mershon JL, Allen DL, Steinmetz RW. Extrapituitary prolactin: Distribution, regulation, functions, and clinical aspects. Endocr Rev. 1996;17:639–669. doi: 10.1210/edrv-17-6-639. [DOI] [PubMed] [Google Scholar]
- 15.Gellersen B, Brosens IA, Brosens JJ. Decidualization of the human endometrium: Mechanisms, functions, and clinical perspectives. Semin Reprod Med. 2007;25:445–453. doi: 10.1055/s-2007-991042. [DOI] [PubMed] [Google Scholar]
- 16.Jabbour HN, Critchley HO. Potential roles of decidual prolactin in early pregnancy. Reproduction. 2001;121:197–205. doi: 10.1530/rep.0.1210197. [DOI] [PubMed] [Google Scholar]
- 17.Labied S, et al. Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol Endocrinol. 2006;20:35–44. doi: 10.1210/me.2005-0275. [DOI] [PubMed] [Google Scholar]
- 18.Deb S, et al. The expression of interleukin-6 (IL-6), IL-6 receptor, and gp130-kilodalton glycoprotein in the rat decidua and a decidual cell line: Regulation by 17β-estradiol and prolactin. Endocrinology. 1999;140:4442–4450. doi: 10.1210/endo.140.10.7063. [DOI] [PubMed] [Google Scholar]
- 19.Zhong L, Parmer TG, Robertson MC, Gibori G. Prolactin-mediated inhibition of 20α-hydroxysteroid dehydrogenase gene expression and the tyrosine kinase system. Biochem Biophys Res Commun. 1997;235:587–592. doi: 10.1006/bbrc.1997.6833. [DOI] [PubMed] [Google Scholar]
- 20.Buzzio OL, Lu Z, Miller CD, Unterman TG, Kim JJ. FOXO1A differentially regulates genes of decidualization. Endocrinology. 2006;147:3870–3876. doi: 10.1210/en.2006-0167. [DOI] [PubMed] [Google Scholar]
- 21.Grinius L, Kessler C, Schroeder J, Handwerger S. Forkhead transcription factor FOXO1A is critical for induction of human decidualization. J Endocrinol. 2006;189:179–187. doi: 10.1677/joe.1.06451. [DOI] [PubMed] [Google Scholar]
- 22.Brar AK, Kessler CA, Handwerger S. An Ets motif in the proximal decidual prolactin promoter is essential for basal gene expression. J Mol Endocrinol. 2002;29:99–112. doi: 10.1677/jme.0.0290099. [DOI] [PubMed] [Google Scholar]
- 23.Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology. 1999;140:4809–4820. doi: 10.1210/endo.140.10.7070. [DOI] [PubMed] [Google Scholar]
- 24.Christian M, et al. Cyclic AMP-induced forkhead transcription factor, FKHR, cooperates with CCAAT/enhancer-binding protein β in differentiating human endometrial stromal cells. J Biol Chem. 2002;277:20825–20832. doi: 10.1074/jbc.M201018200. [DOI] [PubMed] [Google Scholar]
- 25.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 26.Gendron RL, et al. Abnormal uterine stromal and glandular function associated with maternal reproductive defects in Hoxa-11-null mice. Biol Reprod. 1997;56:1097–1105. doi: 10.1095/biolreprod56.5.1097. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh-Li HM, et al. HoxA-11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- 28.Wong KHH, Wintch HD, Capecchi MR. HoxA-11 regulates stromal cell death and proliferation during neonatal uterine development. Mol Endocrinol. 2004;18:184–193. doi: 10.1210/me.2003-0222. [DOI] [PubMed] [Google Scholar]
- 29.Lynch VJ, et al. Adaptive evolution of HoxA-11 and HoxA-13 at the origin of the uterus in mammals. Proc R Soc London Ser B. 2004;271:2201–2207. doi: 10.1098/rspb.2004.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imaoka T, Matsuda M, Mori T. Expression of prolactin messengers ribonucleic acid in the mouse gonads during sexual maturation. Life Sci. 1998;63:2251–2258. doi: 10.1016/s0024-3205(98)00510-4. [DOI] [PubMed] [Google Scholar]
- 31.Kato K, Ikemoto T, Park MK. Identification of the reptilian prolactin and its receptor cDNAs in the leopard gecko, Eublepharis macularius. Gene. 2005;346:267–276. doi: 10.1016/j.gene.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, et al. cDNA sequence and spatio-temporal expression of prolactin in the orange-spotted grouper, Epinephelus coioides. Gen Comp Endocrinol. 2004;136:134–142. doi: 10.1016/j.ygcen.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Eckery DC, et al. The corpus luteum and interstitial tissue in a marsupial, the brushtail possum (Trichosurus vulpecula) Mol Cell Endocrinol. 2002;191:81–87. doi: 10.1016/s0303-7207(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 34.Gerlo S, Julian RE, Davis Dixie L, Kooijman RM. Prolactin in man: A tale of two promoters. BioEssays. 2006;28:1051–1055. doi: 10.1002/bies.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedges SB, Kumar S. Genomic clocks and evolutionary time scales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 36.Hugall AF, Foster R, Lee MSY. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- 37.Messier W, Stewart C-B. Episodic adaptive evolution of primate lysozymes. Nature. 1997;385:151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 40.Gu X. Functional divergence in protein (family) sequence evolution. Genetica. 2003;118:133–141. [PubMed] [Google Scholar]
- 41.Gu X. A simple statistical method for estimating type II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 42.Roth JJ, Breitenbach M, Wagner GP. Repressor domain and nuclear localization signal of the murine Hoxa-11 protein are located in the homeodomain: No evidence for role of polyalanine stretches in transcriptional repression. J Exp Zool. 2005;304:B468–B475. doi: 10.1002/jez.b.21061. [DOI] [PubMed] [Google Scholar]
- 43.Horvat-Switzer RD, Thompson AA. HOXA11 mutation in amegakaryocytic thrombocytopenia with radio-ulnar synostosis syndrome inhibits megakaryocytic differentiation in vitro. Blood Cells Mol Dis. 2006;37:55–63. doi: 10.1016/j.bcmd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Gruss P, Kessel M. Axial specification in higher vertebrates. Curr Opin Genet Dev. 1991;1:204–210. doi: 10.1016/s0959-437x(05)80071-1. [DOI] [PubMed] [Google Scholar]
- 45.Boulet AM, Capecchi MR. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development. 2004;131:299–309. doi: 10.1242/dev.00936. [DOI] [PubMed] [Google Scholar]
- 46.Patterson LT, Pembaur M, Potter SS. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- 47.Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L, Pu Y, Hepps D, Danielpour D, Prins GS. Posterior Hox gene expression and differential androgen regulation in the developing and adult Rat prostate lobes. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Gu X, Vander Velden K. DIVERGE: Phylogeny-based analysis for functional–structural divergence of a protein family. Bioinformatics. 2005;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- 51.Pohnke Y, Kempf R, Gellersen B. CCAAT/enhancer-binding proteins are mediators in the protein kinase A-dependent activation of the decidual prolactin promoter. J Biol Chem. 1999;274:24808–24818. doi: 10.1074/jbc.274.35.24808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.