Abstract
Small RNAs from plants are known to be highly complex and abundant, with this complexity proportional to genome size. Most endogenous siRNAs in Arabidopsis are dependent on RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) for their biogenesis. Recent work has demonstrated that the maize MEDIATOR OF PARAMUTATION1 (mop1) gene is a predicted ortholog of RDR2. The mop1 gene is required for establishment of paramutation and maintenance of transcriptional silencing of transposons and transgenes, suggesting the potential involvement of small RNAs. We analyzed small RNAs in wild-type maize and in the isogenic mop1-1 loss-of-function mutant by using Illumina's sequencing-by-synthesis (SBS) technology, which allowed us to characterize the complement of maize small RNAs to considerable depth. Similar to rdr2 in Arabidopsis, in mop1-1, the 24-nucleotide (nt) endogenous heterochromatic short-interfering siRNAs were dramatically reduced, resulting in an enrichment of miRNAs and transacting siRNAs. In contrast to the Arabidopsis rdr2 mutant, the mop1-1 plants retained a highly abundant heterochromatic ≈22-nt class of small RNAs, suggesting a second mechanism for heterochromatic siRNA production. The enrichment of miRNAs and loss of 24-nt heterochromatic siRNAs in mop1-1 should be advantageous for miRNA discovery as the maize genome becomes more fully sequenced.
Keywords: miRNA, mop1, rdr2, small RNA
The small RNAs found in a typical plant cell include a small number of highly abundant, mainly 21-nt microRNAs (miRNAs) and a large number of small interfering RNAs (mainly 24 nt heterochromatic siRNAs, or simply siRNAs) recognizing many diverse sequences. In addition, several additional subclasses of varying and in some cases overlapping functional importance have been described, including the trans-acting siRNAs (ta-siRNAs) (1–3), natural antisense siRNAs, a type thus far observed only under stress conditions (4, 5), and natural antisense miRNAs (6).
MicroRNAs have a variety of regulatory roles in development and stress responses (for review, see ref. 7). In addition, work in Arabidopsis has led to the hypothesis that transcription of repeats is performed by the plant-specific RNA polymerase IV (pol IV) followed by reverse transcription and cleavage by RDR2 and DCL3, respectively. These repeated sequences include transposons and retrotransposons in plants (8), and this series of events produces a complex set of heterochromatic siRNAs (for review, see ref. 9). The genome size of plants varies substantially among species mainly caused by variation in content of repeated DNA. The complexity of siRNAs is correspondingly greater in rice than in Arabidopsis (10), consistent with larger numbers of repeated sequences in rice.
The maize b1 locus is an excellent model for paramutation, a phenomenon in which alleles communicate in trans, resulting in meiotically heritable gene expression changes (11). Molecular work, combined with fine-structure recombination mapping, has demonstrated that this activity is mediated by tandem repeats at b1 that are required to establish and maintain its transcriptional silencing (12). Mutational analysis has identified genes required for paramutation, such as mop1 (13). In addition to effects on paramutation at multiple genes (13), mop1 mutants demonstrate pleiotropic effects, including reactivation of transcriptionally silenced Mutator transposons and transgenes (14, 15), as well as stochastic developmental phenotypes (13). Cloning of mop1 has shown that it encodes an RNA-dependent RNA polymerase predicted to be orthologous to the Arabidopsis RDR2 gene (16), suggesting RNA-mediated chromatin changes are associated with paramutation.
Prior work characterized the Arabidopsis small RNA population in several Arabidopsis small RNA biosynthetic mutants (17, 18). The rdr2 mutant was particularly informative because it was depleted for most repeat-associated, 24-nt heterochromatic small RNAs (19), resulting in enrichment for unusual RDR2-independent siRNAs and miRNAs, which enabled the identification of new Arabidopsis miRNAs. Because the large number of repeat-associated siRNAs makes it a major challenge to identify low-abundance miRNAs, we hypothesized that deep sequencing of small RNAs in the maize mop1 mutant could enrich for these and other sequences. Here, we report that the proportion of known miRNAs is higher in the mop1-1 mutant relative to wild type and 24-nt siRNA levels are proportionally reduced. However, we also found that the extent of the small RNA population of mop1-1 is unlike that of the Arabidopsis rdr2 mutant, and our data suggest the existence of a second heterochromatic siRNA mechanism in maize that produces ≈22-nt siRNAs.
Results
Sequencing of Maize mop1-1 by Sequencing-by-Synthesis (SBS).
Arabidopsis rdr2 mutants have substantially reduced endogenous siRNAs and correspondingly enriched miRNAs (18, 19). We hypothesized that mop1-1 would be similar and that deep sequencing of small RNAs in mop1-1 would help identify additional low-abundance miRNAs from cereal crops. We generated libraries as described in ref. 8, by using immature ears of either wild-type or mop1-1 maize, an allele with a Mutator insertion in the middle of exon 4 (16). This tissue was selected because it is a rich source of RNA as many cells are actively dividing, prior work in Arabidopsis demonstrated flowers provided more complex siRNA profiles, and paramutation is maintained in this tissue.
We used Illumina's SBS technology for sequencing of the small RNA libraries, producing a total of 5.6 million sequences from two channels of wild type and 7.2 million from three channels of mop1-1 [supporting information (SI) Tables S1 and S2]. Sequencing was to a length of 27 nt; and after removing the 3′ adapter sequence from the signatures, only sequences longer than 15 nt (the vast majority) were retained. Because each library totaled >5 million sequences, we normalized the abundance of each signature to 5 million [units of transcripts per 5 million (TP5M)]. The abundance values ranged from 1 to 47,836 TP5M, with the highest value corresponding to miR168. A large majority of the signatures had very low abundance levels, with the average abundances in wild type of ≈1.3 TP5M and that in mop1-1 ≈2.3 of TP5M. This difference suggested that many low-abundance small RNAs are absent in mop1-1.
Our first indication that mop1-1 and rdr2 mutants are not fully equivalent came from an assessment of small RNA complexity. Our prior work with rdr2 using Massively Parallel Signature Sequencing (MPSS) demonstrated a reduction in small RNA sequence diversity in rdr2 of >80% compared with wild type (18). We confirmed these earlier MPSS data by resequencing rdr2 and wild-type Arabidopsis inflorescences with SBS, which showed an 85% reduction in distinct sequences in rdr2 (Tables S1 and S2), calculated before genome matching to make the analysis comparable with maize, for which the full genome sequence is not available. Small RNA diversity in mop1-1 was reduced, but to only 53% that of wild-type maize, 0.34 versus 0.65 proportion distinct sequences; Tables S1 and S2). This difference in reduction in sequence complexity in rdr2 versus mop1-1 prompted examination of the maize data in more detail.
Small RNA Sizes in the Maize Small RNA Libraries and the Maize Genome.
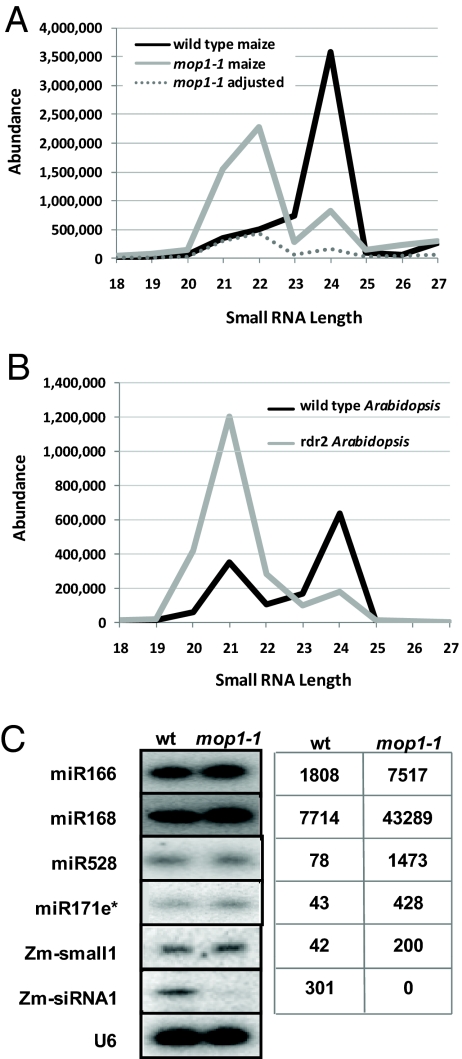
Compared with wild type, the small RNA size distribution in mop1-1 was substantially reduced in 24-nt siRNAs (Fig. 1A), consistent with the loss of heterochromatic siRNAs in rdr2 (18) (Fig. 1B). Almost as striking was an apparent enrichment of 21- and 22-nt small RNAs in mop1-1. The increase in 21-nt RNAs was similar to Arabidopsis rdr2 and dcl2/3/4 (17), but the prominent class of 22-nt small RNAs in mop1-1 was different. Deep sequencing in a genetic background lacking siRNAs would be expected to enrich for miRNAs (predominantly 21 nt) and produce an elevation in their abundances that is simply the result of the loss of siRNAs. We examined several known maize miRNAs in the sequences from the mop1-1 mutant compared with wild type, revealing an average enrichment of miRNAs in the mop1-1 mutant of 5.3-fold (Fig. 1 C and Table 1). To control for the loss of siRNAs, we performed an RNA gel blot with RNA from wild type and mop1-1 loaded to reflect equal levels of the U6 spliceosomal RNA, which is unlikely to be altered in a mop1 mutant. When this RNA gel blot was hybridized with probes of two highly abundant miRNAs, no difference was observed in their levels between wild type and mop1-1 (Fig. 1C). Therefore, we adjusted the abundances of all small RNAs in the mop1-1 sequences by dividing by 5.3, the average overall enrichment. With these miRNA-normalized levels, there remained a 22-nt peak of abundance in mop1-1 similar that of wild-type maize (Fig. 1A), suggesting an unusual class of small RNAs compared with Arabidopsis (Fig. 1B).
Fig. 1.
Small RNA sizes and abundances in wild-type and mop1-1 or rdr2 libraries. (A) Plot comparing the total abundance of maize small RNA sequences versus their size for wild type and mop1-1. The dashed gray line indicates the abundances of the mop1-1 sample adjusted downward based on the average 5.3-fold enrichment of miRNAs observed in C. (B) Plot comparing the total abundance of Arabidopsis small RNA sequences versus their size for wild-type and rdr2 inflorescences. (C) (Left) RNA gel blot analysis of miRNA abundances in wild type and mop1-1. Total RNA was normalized based on levels of the U6 spliceosomal RNA. Zm-small1 is a putative new miRNA candidate described in Results. The Zm-siRNA is a randomly selected 24-nt small RNA from a high-density siRNA cluster. (Right) Original SBS abundance of each miRNA. Measured across the known maize miRNAs, these raw abundances were 5.3-fold higher in the mop1-1 data.
Table 1.
Registered maize miRNA families matched by small RNAs from immature ears
| miRNA | mop1-1 | Wild type |
|---|---|---|
| miR156a/b/c/d/e/f/g/h/i | 1,170 | 98 |
| miR156j | 148 | 10 |
| miR156k | 38 | 1 |
| miR159a/b | 9,986 | 3,052 |
| miR159c/d | 55 | 23 |
| miR160a/b/c/d/e | 147 | 166 |
| miR160f | 1 | 1 |
| miR162 | 2 | 1 |
| miR164a/b/c/d | 61 | 18 |
| miR166a | 129,603 | 22,941 |
| miR166b/c/d/e/f/g/h/i | 9,629 | 1,471 |
| miRNA166j/k | 23,636 | 4,462 |
| miR166l/m | 60,735 | 9,285 |
| miR167a/b/c/d | 42,729 | 3,242 |
| miR167e/f/g/h/i | 1,554 | 112 |
| miR168a/b | 128,980 | 17,608 |
| miR169a/b | 49 | 10 |
| miR169c | 21 | 1 |
| miRNA169f/g/h | 8 | 3 |
| miR169i/j/k | 17 | 5 |
| miR171a/b | 1 | 0 |
| miR171d/e/i/j | 203 | 53 |
| miR171f | 1 | 0 |
| miR171h/k | 3 | 2 |
| miR172a/b/c/d | 87 | 14 |
| miR172e | 1,249 | 197 |
| miR319a/b/c/d | 32 | 9 |
| miR393 | 7 | 2 |
| miR394a/b | 45 | 33 |
| mi396a/b | 92 | 12 |
| miR399a/c | 3 | 1 |
| miR399e | 8 | 2 |
| miR408 | 11 | 6 |
Values indicate raw abundances for perfect matches to known maize miRNAs with matches precisely aligned to the annotated 5′ end of the miRNA (some matching small RNAs were slightly longer and were also counted if they perfectly matched).
We wanted to determine whether the genomic loci generating the various classes of small RNAs in mop1-1 were the same as wild type. Because the maize genome was not yet fully sequenced, we used as a surrogate two large contigs comprising ≈14 Mb; this was 7.8 Mb from the short arm of Chr. 1 and 6.6 Mb from the long arm of Chr. 9, which represent an ancient duplication within the maize genome (20). Unfortunately, no maize centromeres or pericentromeric locations are yet available for analysis because the small RNA populations in these regions are distinctive in both Arabidopsis and rice. Approximately 200,000 different small RNAs matched each contig from each genotype (Table S3), averaging approximately seven locations per small RNA. The maize small RNAs are on track to match more locations than in either Arabidopsis or rice (Fig. S1), even using <1% of the maize genome.
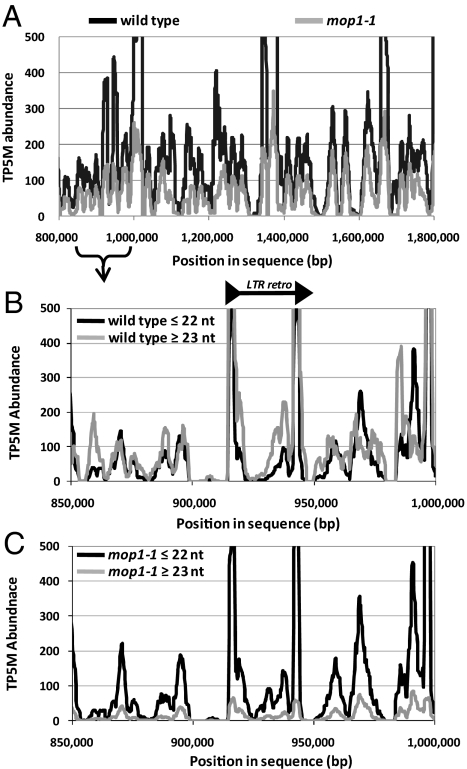
An analysis of the distribution of small RNAs and their abundances indicated that nearly all genomic regions with matching small RNAs in wild-type maize also had matching small RNAs in mop1-1 (Fig. 2A). This trend was different from Arabidopsis, which has a small number of discrete loci generating small RNAs in wild type that are dramatically reduced in rdr2 (18). As an example, we show a 250-kb region of maize chromosome 1 with the size distribution of small RNAs matching this region from wild type and mop1-1 (Fig. 2 B and C). This analysis showed the dramatic decrease of small RNAs ≥23 nt in mop1-1 but retention of the 22-nt class.
Fig. 2.
Chromosomal distribution of small RNAs from immature ears. (A) Small RNAs were matched to a segment of maize chromosome 1, as described in Results. The x axis indicates the location on the contig, and the y axis indicates the abundance of the small RNA in TP5M, using miRNA-normalized abundances. The lines are representative of a moving average of 10 windows of 10,000 bp. The curly brace indicates the region expanded in B and C. (B and C) Sliding-window calculation of the sum of abundance of small RNAs matched to each location in a 250-kb region of the contig. The x axis indicates the location on the contig, and the y axis indicates the sum of abundance in TP5M. The LTR retrotransposon is marked, including the terminal repeats (black arrowheads). The values were calculated separately for wild type (B) and mop1-1 (C), and for small RNAs ≤22 nt and ≥23 nt.
This analysis was expanded to include different types of genomic sequences within the two large contigs. Based both on the ratios of different sequences and of hits-normalized-abundances, there were substantially fewer small RNAs in all genomic regions for small RNAs ≥23 nt (Table S4 and Fig. 2 B and C) in mop1-1 compared with wild type, although there were still 24-nt siRNAs observed in mop1-1. All classes of sequences showed an apparent enrichment level in the 22-nt size class of ≈5-fold in mop1-1, an effectively unchanged level when accounting for sampling effects (i.e., the average 5.3-fold increase of miRNAs described above).
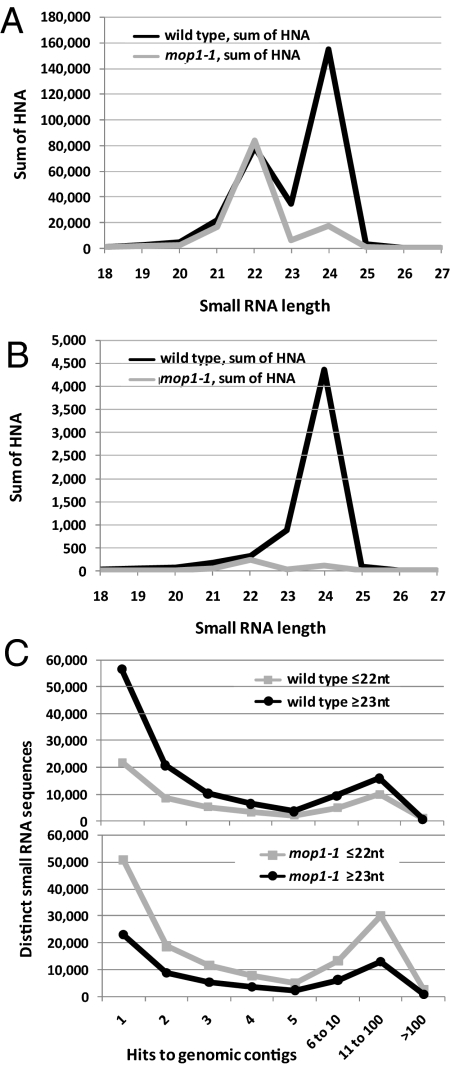
When we compared relatively high-copy-number sequences like LTR retrotransposons with lower-copy repeats such as DNA transposons, we observed that there was a much more substantial loss of 23/24-nt small RNAs in the mop1-1 mutant at the low-copy repeats compared with the high-copy repeats (Figs. S2 and S3). When analyzed across all LTR retrotransposons and DNA transposons on the contigs, it was also clear that the ratio of 22- to 24-nt small RNAs is quite different between these classes of elements in wild type, with a much larger proportion of 22-nt small RNAs matching high-copy LTR retrotransposons (Fig. 3 A and B). When we plotted distinct small RNAs versus their hits in the two maize contigs, we observed a substantially different ratio of low- to high-hit small RNAs in mop1-1 versus wild type (Fig. 3C and Table S5), particularly for small RNAs ≥23 nt, consistent with a decrease in low-hit 23/24-nt small RNAs in mop1-1. In light of these maize repeat-associated siRNA results, we further analyzed the Arabidopsis rdr2 SBS data for hits within repeats. As noted (Fig. 1), there is a greater reduction in rdr2 small RNAs relative to wild-type Arabidopsis compared with mop1-1 relative to wild-type maize. In addition, in rdr2 the remaining siRNAs associated with both retroelements and DNA transposons were predominantly 21 nt (Fig. S4). Comparing the Arabidopsis 22-mers in wild type and rdr2 reveals that 26% of the 22-mers are from retroelements in wild-type Arabidopsis, but the loss-of-function mutation in rdr2 dramatically reduces the 22-mers to <1%; most of the remaining 22-mers are miRNA- or ta-siRNA-related.
Fig. 3.
Sizes and hits for repeat- or contig-mapped maize small RNAs. (A) Size distribution of maize small RNAs mapping to LTR retrotransposons, predominantly high-copy elements, on the Chr. 1 and 9 contigs. The LTR retrotransposons were identified by RepeatMasker as gypsy- or copia-type elements and spanned 6,670,685 bp on the two contigs. The y axis indicates the sum of the hits-normalized-abundance (HNA), calculated by dividing the normalized abundance (in TP5M) for each small RNA by the number of genomic locations to which the small RNA maps (the “hits”). The mop1-1 abundances have been reduced by 5.3-fold (normalized by miRNA levels). (B) Same calculations as in A, but performed for predominantly low-copy DNA transposons identified by RepeatMasker, including hAT, hAT-Ac, En-Spm, Harbinger, or MuDR elements. These spanned 85,149 bp on the two contigs. (C) Plots of distinct, contig-matching small RNAs versus their hits. Data represent only small RNAs matched to the Chr. 1 and Chr. 9 maize contigs. The values were calculated separately for wild type (Upper) and mop1-1 (Lower) and for small RNAs ≤22 nt and ≥23 nt. A comparison of the ratios using different ranges for low versus high hits is in Table S5.
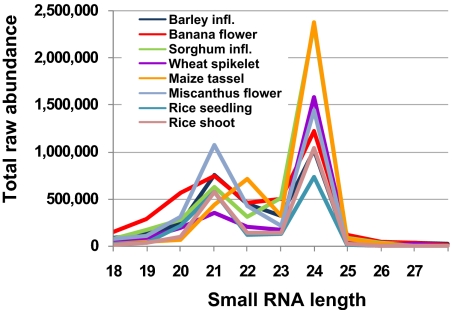
We next asked whether the 21/22-nt class of heterochromatic siRNAs from maize is found in other monocot species, by examining small RNA libraries produced from wild-type flowers or parts of flowers from wheat, sorghum, banana, barley, miscanthus, and from maize tassels. Only maize shows a larger 22-nt peak relative to the 21-nt peak (Fig. 4), and this peak is observed in independent maize libraries from different inbred backgrounds and tissues. The reduction in 23-nt small RNAs in the maize tassel (Fig. 4) versus ears (Fig. 1A) may reflect a different ratio of the 21/22 versus 23/24 class of siRNAs or a specific reduction in 23-nt siRNAs in the tassel; perhaps these two classes are differentially regulated in these tissues. Although the size profile of maize is unusual within the monocots, a recent study has demonstrated that pine has predominantly 21-nt, not 24-nt, siRNAs perhaps caused by the lack of DCL3 (21).
Fig. 4.
Size distribution of small RNAs from diverse monocot species. A plot comparing the total abundance of small RNA sequences versus their size for monocot species was sequenced as part of a comparative sequencing project (http://smallrna.udel.edu).
MicroRNA-Derived Small RNAs Identified in the Wild-Type and mop1-1 Libraries.
The deep SBS dataset allowed us to quantify the accumulation of specific miRNA families, although without replication, this quantitative analysis represents a rough estimate. Approximately half of the known miRNAs are expressed >5-fold in mop1-1 compared with wild type, including miR166a and miR168a/b (before correcting for the 5.3-fold difference described above; Table 1), the most abundant miRNAs in both the wild-type and mop1-1 immature ear libraries, consistent with their developmental roles.
From the SBS data, miR160 was less abundant in mop1-1 compared with wild type, and miR156 and miR166 were of particularly high abundance in mop1-1 (Table 1). Examples of other miRNAs with unusual characteristics are miR162, miR399, and several members of the miR171 family, which had a very low accumulation. One speculation is that the different accumulation levels are caused by a secondary level of control by a siRNA-mediated pathway, which is altered in mop1-1. Eleven known miRNAs were never observed in either wild-type or mop1-1 libraries from either ears or tassels; this could indicate that these miRNAs are not expressed in the tissues or conditions sampled, some of these are not bona fide miRNAs, or sequence-based biases in cloning and/or sequencing contributed to their absence.
To identify novel miRNA candidates, we examined small RNAs mapping to the two maize contigs with characteristics of known plant miRNAs described in ref. 18, by using several criteria such as the dense clustering of the small RNAs signatures and the position of the signatures relative to annotated genes to identify new miRNAs. Within just this 14-Mb maize sequence, small RNAs mapping to the hairpin structures of two known miRNAs (miR528 and miR171e), and one new miRNA candidate (Zm-small1), were identified. The abundances of these small RNAs were confirmed by RNA gel blots (Fig. 1C). miR528 has been described only in rice (22). Interestingly, the abundant small RNA that mapped to miR171 corresponds to the miRNA* and was expressed at a level twice as high as the mature miRNA (Fig. 1C). Zm-small1 is a strong candidate for a novel miRNA because it is unaffected in mop1-1 compared with wild type, is 21 nt in length, does not map to an siRNA cluster, and has a relatively high abundance. Although the prevalence of 22-nt siRNAs complicates the analyses, the enrichment of miRNAs and loss of 24-nt heterochromatic siRNAs in mop1-1 should be advantageous for miRNA discovery.
Discussion
The comparison of the maize mop1-1 and Arabidopsis rdr2 small RNA profiles was informative for both miRNA and siRNA characterization. The rdr2 mutant was a powerful genotype in which to identify new miRNAs. In wild-type maize, known miRNAs are <10% of the small RNAs, whereas in the mop1-1 mutant, miRNAs represented nearly 40% of total small RNAs. Thus, the mop1-1 enrichment should provide greater sensitivity to low-abundance miRNAs, providing substantial utility for miRNA discovery. Even in a relatively small portion of the maize genome we have identified new miRNA candidates. With the pending availability of the complete maize genome, we anticipate that these data will prove useful for the discovery of additional maize miRNAs. However, achievement of a level of siRNA reductions in maize similar to that in rdr2 in Arabidopsis may require additional mutations combined with mop1-1.
Endogenous siRNAs corresponding to transposons, retrotransposons, centromere repeats, and pseudogenes virtually disappear in the Arabidopsis rdr2 background whereas in the mop1-1 mutant, the removal of the 23/24-nt siRNAs leaves a 21/22-nt class of putative siRNAs associated predominantly with high-copy repeats. This difference is unexpected given that the genes appear orthologous. There is no evidence from the current maize genome sequence that there is another RDR closely related to RDR2 (www.chromdb.org), and the mop1-1 mutation is a loss-of-function allele, suggesting that an additional layer of small RNA complexity exists in maize. A comparison among wild-type libraries from other monocots did not show a similarly large proportion of 22-nt siRNAs. This finding is true even for small RNA populations from other large-genome cereals such as wheat and barley, which also lack the 22-nt small RNA peak, suggesting that this siRNA population may be a unique characteristic of maize. It will be interesting to examine the impact of loss-of-function mutations in orthologs of rdr2/mop1 in other monocots to dissect further their small RNA populations.
Several potential mechanisms could explain the biogenesis of the 22-nt class of siRNAs. An additional Dicer copy could work with other components of the siRNA biogenesis pathway to generate 22-nt siRNAs. In contrast to metazoan and fungal genomes that encode just one or two Dicers, plants have many more Dicer-like genes (23). The rice genome encodes six Dicer-like genes, two more than Arabidopsis. The additional genes are duplications of DCL2 and DCL3; the latter cleaves RDR2-generated dsRNAs into 24-nt siRNAs. Dicer-like enzymes such as DCL2 and DCL4 function to prevent viral infection in plants; DCL2 is capable of generating 22-nt small RNAs from double-stranded viral RNA (24). Without the complete genome sequence, it is difficult to know exactly how many Dicer-like genes exist in maize. However, at least five DCL genes have been identified in maize; two most similar to DCL3 and one each most similar to DCL1, DCL2, and DCL4 (www.chromdb.org). There could be partial redundancy of the RDR2(MOP1)/DCL3 pathway with the RDR6/DCL4 pathway, leading to the production of both RDR2/MOP1-dependent and RDR6-dependent 21/22-nt siRNAs. It is also possible that the dsRNA template for generating the 22-nt siRNAs is produced by an uncharacterized RDR or in an RDR-independent manner. For example, natural trans-antisense RNAs could be synthesized from the high-copy-number retrotransposons. Confirmation of additional pathways in maize awaits the completion of the maize genome, additional mutant screens, and biochemical analyses.
A large proportion of the maize genomic sequence examined contains high-copy retrotransposons, which were associated with dense clusters of small RNAs. In contrast, most regions immediately flanking the predicted maize genes were devoid of small RNAs, as were the genes themselves (Fig. S3), except when those genes contained tandem repeats or DNA transposons where siRNAs were also concentrated. This pattern in maize of “siRNA islands” interspersed between almost every gene was not obvious with our Arabidopsis and rice small RNA data probably because their relatively compact genomes have fewer intergenic heterochromatic repeats. The maize small RNA data may serve as a useful “filter” when combined with genomic annotation to identify potential genes, noncoding RNAs, and regulatory sequences.
The mop1 mutants have striking, albeit stochastic pleiotropic, developmental phenotypes relative to wild type, including stunting and flowering defects (13). These visible “mutant” phenotypes produced in the mop1-1 background suggest that if there are multiple siRNA-based regulatory pathways they have little overlap and redundancy. Several potential mechanisms could contribute to these phenotypes. There may be specific miRNAs or siRNAs whose miss-regulation in mop1 mutants causes developmental phenotypes. One example in Arabidopsis is the large effect on the expression of a key flowering gene, FLC, through siRNA-mediated heterochromatin formation of a nearby transposon (25). Another nonmutually exclusive possibility is that some phenotypes are generated by mutations caused by the insertion of transposons reactivated in the mop1-1 mutant. The small RNA data demonstrated that the relatively lower-copy repeats have far fewer ≈22-nt putative siRNAs, which may lead to reactivation of low-copy-repeat sequences when the ≈24-nt pathway is disrupted in mop1-1. Consistent with this, Mutator transposons can be reactivated after multiple generations in the absence of MOP1 (14, 26).
In contrast to mop1 mutants, the Arabidopsis rdr2 mutant has no such dramatic phenotypes. This is surprising because heterochromatic siRNAs are substantially diminished in rdr2, which one might have suspected would lead to stochastic activation of transposons and the generation of epialleles, such as reported for the ddm1 in Arabidopsis that dramatically reduces DNA methylation within repeats (27). However, it has been suggested that the rdr2 mutation causes a decrease in production of the demethylase ROS1, leading to hypermethylation at CG sites and additional protection against the activation of transposons (28). A more complete understanding of transposon reactivation in rdr2 and mop1 mutants will await larger-scale studies of DNA methylation in these mutants. With the impending availability of the complete maize genomic sequence, it should be possible to use our data to identify and characterize specific regulatory loci that are altered in mop1 mutants, which could contribute to understanding the stochastic developmental phenotypes appearing in mop1 mutants.
Experimental Procedures
Plant Growth and RNA Gel Blot Analysis.
Small-ear libraries are from maize variety K55 (wild type and mop1-1), and the small-tassel library was prepared from B73. The mop1-1 allele is described in ref. 13. Tissues were harvested from plants grown outdoors in Tucson, AZ, 68 days after germination; young tassel small ears containing ovules were 2–4 cm long. The data for the plant species in Fig. 4 were obtained from the comparative sequencing project described on http://smallrna.udel.edu, where details on the libraries are available.
Low-molecular-weight RNA gel blots, radiolabeled probes for specific small RNAs, and hybridization/wash conditions were described in ref. 8. Blot hybridization analysis was performed as described in ref. 29; the blot shown is representative of two independent experiments.
SBS Data Generation and Analysis.
For all libraries, total RNA was isolated by using TRIzol reagents (Invitrogen). SBS sequencing was performed by Illumina using small RNA libraries constructed as described (8, 30). Adapter sequences were identified and removed by using a Perl script. Small RNAs shorter than 15 nt were removed. We compared SBS data to maize contigs (20) and assigned signatures to each location with a perfect match. The number of matches was recorded as “hits.” Repeat analysis was performed as described in ref. 8 by using a combination of programs including RepeatMasker (www.repeatmasker.org/), Einverted, and Etandem (31).
Supplementary Material
Acknowledgments.
We thank Mayumi Nakano for her work on the web interface and Rosa Jaime-Frias for growing the maize plants. This work was supported by National Science Foundation Grants 0439186 and 0548569 (to P.J.G. and B.C.M.), MCB-0235329 (to V.L.C), 0321437 (to B.C.M.), and 638525 (to B.C.M. and P.J.G.); by National Institutes of Health Grant DP1 OD000575 (to V.L.C); and by U.S. Department of Agriculture Grant 2007-01991 (to P.J.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE12173 (series identifier), GPL7071 (platform identifier), and GSM306487 and GSM306488 (sample identifiers)]. The raw and normalized SBS data are also available at http://mpss.udel.edu/maize; this web site allows users to query data based on physical location, gene identifiers, or by sequence.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808066105/DCSupplemental.
References
- 1.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katiyar-Agarwal S, et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc Natl Acad Sci USA. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 8.Lu C, et al. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 9.Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 10.Nobuta K, et al. An expression atlas of rice mRNA and small RNA. Nat Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- 11.Chandler VL. Paramutation: From maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorweiler JE, et al. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisch D, Carey CC, Dorweiler JE, Chandler VL. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc Natl Acad Sci USA. 2002;99:6130–6135. doi: 10.1073/pnas.052152199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinnis KM, Springer C, Lin Y, Carey CC, Chandler V. Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics. 2006;173:1637–1647. doi: 10.1534/genetics.106.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 17.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing, and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 18.Lu C, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruggmann R, et al. Uneven chromosome contraction and expansion in the maize genome. Genome Res. 2006;16:1241–1251. doi: 10.1101/gr.5338906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolgosheina EV, et al. Conifers have a unique small RNA silencing signature. RNA. 2008;14:1508–1515. doi: 10.1261/rna.1052008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, et al. Loss-of-function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margis R, et al. The evolution and diversification of Dicers in plants. FEBS Lett. 2006;580:2442–2450. doi: 10.1016/j.febslet.2006.03.072. [DOI] [PubMed] [Google Scholar]
- 24.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, He Y, Amasino R, Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodhouse MR, Freeling M, Lisch D. The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics. 2006;172:579–592. doi: 10.1534/genetics.105.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 28.Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007;145:1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.