Abstract
We isolated the barley stem rust resistance genes Rpg5 and rpg4 by map-based cloning. These genes are colocalized on a 70-kb genomic region that was delimited by recombination. The Rpg5 gene consists of an unusual structure encoding three typical plant disease resistance protein domains: nucleotide-binding site, leucine-rich repeat, and serine threonine protein kinase. The predicted RPG5 protein has two putative transmembrane sites possibly involved in membrane binding. The gene is expressed at low but detectable levels. Posttranscriptional gene silencing using VIGS resulted in a compatible reaction with a normally incompatible stem rust pathogen. Allele sequencing also validated the candidate Rpg5 gene. Allele and recombinant sequencing suggested that the probable rpg4 gene encoded an actin depolymerizing factor-like protein. Involvement of actin depolymerizing factor genes in nonhost resistance has been documented, but discovery of their role in gene-for-gene interaction would be novel and needs to be further substantiated.
Keywords: actin depolymerizing factor, barley, disease resistance domains, map-based cloning
Stem rust caused by the fungus Puccinia graminis (Pg) was historically one of the most significant foliar diseases of barley and wheat, with genetic resistance being the primary means of control. Durable resistance in barley has been achieved against many pathotypes for the past 60+ years by the widespread use of the single resistance gene, Rpg1 (1).
Barley can be attacked by Pg f. sp. tritici (Pgt), the wheat stem rust pathogen and Pg f. sp. secalis (Pgs), the rye stem rust pathogen. In cultivated barley, five genes are known to confer resistance to Pgt and three to Pgs (2, 3). Only Rpg1 has been cloned and characterized (4, 5). A virulent Pgt pathotype, designated QCC, was isolated from Midwestern barley cvs. containing Rpg1 in 1989 (6). A resistance gene identified in barley line Q21861 was designated rpg4. It acts in recessive manner to pathotype QCC and is temperature-sensitive (3, 7, 8). Besides rpg4, Q21861 carries Rpg5 providing resistance to Pgs isolate 92-MN-90 (3, 8). The Rpg5 gene, previously RpgQ, is dominant or semidominant in action and was reported to cosegregate with rpg4, although three exceptions were found in 769 F2 progeny (9). The rpg4 gene was mapped to the long arm of barley chromosome 7(5H) (10). More detailed mapping and identification of syntenic rice chromosome regions (11, 12) allowed the development of a physical map covering the presumed rpg4 gene region (13).
Advances in molecular techniques and tools have facilitated the cloning of numerous plant disease resistance genes (R genes) in the past two decades. R genes are grouped into different classes according to their protein domain structure (14). The largest group consists of the NBS-LRR family of R genes, which is characterized by an N-terminal nucleotide-binding site (NBS) and C-terminal leucine rich repeats (LRRs). Another class, with relatively few members, is the extracellular LRR and transmembrane (TM) domain containing genes conferring resistance to the fungus Cladosporium fulvum, the leaf mold pathogen of tomato. The rice genes Xa21 (15) and Xa26 (16) conferring resistance to the bacterial blight pathogen Xanthomonas oryzae are the only examples of receptor-like kinase genes consisting of an extracellular LRR, a TM, and a cytoplasmic serine/threonine protein kinase (S/TPK). Last, there is a class of R genes that consists of S/TPK domains. S/TPK R genes include the previously described barley Rpg1 gene (4); the tomato Pto gene, which confers resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (17); and the Arabidopsis PBS1 gene, which confers resistance to the bacterial pathogen Pseudomonas syringae pv. phaseolicola (18).
The S/TPK group of R genes is unique in that two of the members, Pto and PBS1, have been shown to require an NBS-LRR gene, Prf and RPS5, respectively, for resistance (19, 20). This demonstrates that NBS-LRR and protein kinases sometimes work together to provide resistance to plant pathogenic organisms.
Here, we report the cloning and preliminary characterization of two unique barley stem rust resistance genes Rpg5 and rpg4. Rpg5 encodes an R gene protein containing the NBS, LRR, and S/TPK domains in a single transcript. We validated the candidate Rpg5 gene by multiple allele sequencing and VIGS (21).
Available recombinants point to the rpg4 gene encoding an actin depolymerizing factor (Adf). Adfs play an important role in cytoskeleton organization and have been reported to be involved with nonhost disease resistance (22). Their role in gene-for-gene interaction would be novel and needs to be further substantiated.
Results
High-Resolution Genetic and Physical Mapping.
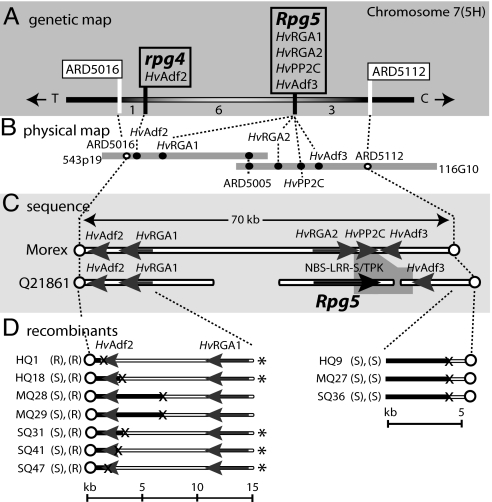
High-resolution genetic mapping identified the Rpg5/rpg4 containing region between RFLP markers ARD5112 and ARD5016. This region was delimited by 10 cross-overs, 7 in the HvAdf2 region and 3 between ARD5112 and HvAdf3 (Fig. 1 A and D). The recombinants around the HvAdf2 region separated rpg4 (resistance to Pgt pathotype QCC) from Rpg5 (resistance to Pgs isolate 92-MN-90) (Fig. 1D).
Fig. 1.
Genetic, physical, sequence, and recombinant characterization of the Rpg5/rpg4 locus. (A) High-resolution genetic map of the Rpg5 region based on 5,232 recombinant gametes. The Rpg5 flanking markers (ARD5016 and ARD5112) are designated above the white vertical bars. The candidate genes cosegregating with isolate 92-MN-90 (Rpg5) resistance and pathotype QCC (rpg4) resistance are inside the boxes labeled Rpg5 and rpg4, respectively. Numbers below the bar indicate the number of cross-overs. C and T represent centromere and telomere. (B) BAC physical map spanning the Rpg5/rpg4 locus. The two BAC clones (543P19 and 116G10) spanning the region are shown as horizontal gray bars. The white circles indicate the flanking genetic markers. Black circles indicate the position of Rpg5 and rpg4 candidate genes and the marker (ARD5005) used for chromosome walking. (C) Sequence annotation of the Rpg5 region. White horizontal bars represent sequenced regions from Morex and Q21861. White circles show the position of flanking markers. The black arrow represents the Rpg5 NBS-LRR-S/TPK gene. Gray arrows indicate annotated genes. The darker gray background between HvRGA2 and HvAdf3 indicates the region of colinearity breakdown. The scale is shown above in kilobases. (D) Recombinant sequence analysis. Horizontal bars represent sequence and/or genotyping from 10 lines with recombination defining the Rpg5/rpg4 region. Black indicates susceptible genotype, and white indicates the resistant Q21861 genotype. The “X” indicates the approximate point of recombination. The recombinant designations are labeled to the right with resistant (R) or susceptible (S), indicating the response to pathotype QCC (rpg4) and isolate 92-MN-90 (Rpg5), respectively. Gray arrows indicate candidate genes and the white circles represent flanking markers. Recombinant lines indicated with an asterisk have complete sequence analysis at the region of recombination. The scale is shown below in kilobases.
Physical mapping identified BAC clones 543P19 and 116G10 spanning the ARD5112 to ARD5016 region (Fig. 1B). Genetic and physical mapping confirmed that these BAC clones included the flanking markers and the Rpg5/rpg4 locus.
Identification and Analysis of Candidate Rpg5 and rpg4 Genes.
Sequence analysis of the cv. Morex BAC clones 543P19 and 116G10 identified a 70-kb region flanked by the RFLP markers ARD5016 and ARD5112. This region was annotated, and five candidate genes were identified, two encoding predicted R-like NBS-LRR proteins (HvRGA1 and HvRGA2), two actin depolymerizing factors (HvADF2 and HvADF3), and a protein phosphatase 2C protein (HvPP2C) (Fig. 1C).
PCR primers designed to the cv. Morex candidate genes amplified four of the five genes from the resistant line Q21861. The HvPP2C gene could not be amplified with Morex-specific PCR primers, suggesting a diverged gene or an indel event. The Q21861 noncolinearity region was amplified by using PCR chromosome walking (23) and sequenced revealing a S/TPK domain associated with the Rpg5 candidate gene HvRGA2 (Fig. 2). The S/TPK domain was absent from susceptible cvs. Morex, Steptoe, and Harrington, suggesting its involvement in Rpg5-mediated resistance.
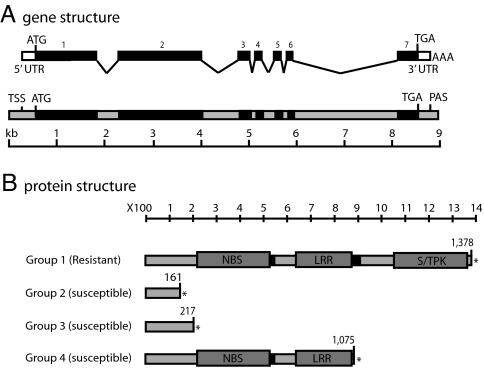
Fig. 2.
Rpg5 predicted gene structure from resistant and susceptible cultivars. (A) cDNA and genomic DNA structures with introns (gray), exons (black), and UTRs white shown to scale. Rpg5 predicted mRNA structure is shown above the genomic sequence with exons numbered above. ATG represents the start methionine codon and TGA represents the stop codon. AAA is the mRNA poly(A) tail, TSS is the transcription start site, and PAS is the polyadenylation signal. The scale below is shown in kilobases. (B) Organization of the predicted protein structure is shown to scale with predicted boundaries indicated by boxes. NBS denotes the nucleotide-binding site, LRR denotes 12 imperfect leucine-rich repeats, S/TPK denotes the serine threonine protein kinase domain, and the black bars represent predicted transmembrane domains. Numbers above indicate the last amino acid preceding the stop codon. Stop codons are represented by an asterisk. Cultivars or lines were placed into four groups. Group 1 (resistant) consists of Q21861 with a predicted intact and functional Rpg5 gene. Group 2 (susceptible) consists of the cultivars Morex and Steptoe. They have a cytosine-to-adenine conversion that introduces a stop codon resulting in a predicted truncated protein. In addition, they are missing the kinase domain coding region. Group 3 (susceptible) contains Golden Promise and MD2. They have a single cytosine insertion causing a frame-shift mutation that results in a stop codon and a predicted truncated protein. Group 4 (susceptible) consists of Harrington. Harrington is missing the kinase domain coding region and the second transmembrane domain.
To characterize the noncolinearity region further, a Q21861 λ library was developed and clone RSB762 identified. The RSB762 sequence contained the Rpg5 gene with 5 kb 5′ region and 7.5 kb extending 3′ into the noncolinearity region. Additional chromosome walking resulted in 15,708-bp Q21861 sequence. This completed the noncolinearity region except for a small retrotransposon block, estimated at ≈2 kb by restriction mapping, which we could not assemble because of its repetitive nature. The S/TPK domain was the only additional gene found in the Q21861 indel region.
An approximately 18-kb region between HvRGA1 and Rpg5 contained a large retrotransposon block in Morex. We were not able to assemble Q21861 contiguous sequence from this region. The partial sequences obtained were all repetitive and/or retrotransposon-like, with no evidence of additional genes. The 57.3-kb total Q21861 sequence identified the same candidate genes as in the susceptible Morex sequence, except that the HvPP2C gene was replaced by a protein kinase domain.
Comparison of Morex and Q21861 sequences suggested that HvRGA2 was the Rpg5 gene (nucleotide and amino acid numbers given are from the Q21861 Rpg5 sequence, starting with the first nucleotide in the translation start codon and the first amino acid). The Morex HvRGA2 consisted of typical NBS-LRR domains and a PP2C gene a short distance downstream (Fig. 1C). The Q21861 HvRGA2 gene consisted of NBS-LRR-S/TPK domains (Figs. 1C and 2). Furthermore, the susceptible cv. Morex genomic and cDNA sequence revealed a nucleotide substitution at position +483, resulting in a stop codon at amino acid position 161 and a predicted truncated protein (Fig. 2B, Group 2). This correlation of an apparently nonfunctional protein and a susceptible phenotype suggested that HvRGA2 could be the Rpg5 gene. Sequence analysis of the HvRGA2 alleles from the susceptible parents showed Steptoe to be similar to Morex, encoding a predicted truncated protein. MD2 and Golden Promise contained a single-nucleotide insertion (C337) causing a frame-shift mutation at amino acid position 114 resulting in a stop codon at aa position 217 (Fig. 2B, Group 3). The susceptible cv. Harrington HvRGA2 allele contained an amino acid sequence very similar to Q21861, but it was lacking the PK domain (Fig. 2B).
The remaining candidate genes were eliminated based on allele sequencing. The candidate gene HvRGA1 codes for an 895-aa (98.5-kDa) predicted NBS-LRR protein with highest homology to an Oryza sativa hypothetical NBS-LRR protein (GenBank accession no. EAY80410). Sequence analysis of HvRGA1 alleles revealed that the resistant parent Q21861 and the susceptible parent MD2 had identical HvRGA1 predicted amino acid sequences. The susceptible parent Harrington differed from Q21861 by only a single amino acid (C324R). The susceptible cvs. Morex and Steptoe were identical at the amino acid level and differed from Q21861 by five amino acids (S290A, K340N, A445G, N474D, and F586L). The single amino acid difference between Q21861 and Harrington occurred within the NBS domain but was not within one of the highly conserved sub domains. Two of the amino acid differences between Q21861 and Morex/Steptoe occurred within the NBS domain and three occurred within the predicted LRR region. The amino acid sequence identity between Q21861 and MD2 in addition to the very limited polymorphism between Q21861 and Harrington suggested that HvRGA1 was not Rpg5.
The HvAdf3 gene codes for a protein with the highest amino acid homology to a Lophopyrum elongatum actin depolymerizing factor-like gene (GenBank accession no. AAG28460). Allele sequencing from the mapping population parents (Steptoe, MD2, and Q21861) and cv. Morex showed that HvADF3 had no polymorphism at the amino acid level, suggesting that it is a highly conserved protein and not the Rpg5 gene.
The HvPP2C gene sequence from cv. Morex codes for a protein with the highest homology to an expressed O. sativa protein phosphatase 2C family protein (GenBank accession no. ABF99721). Specific HvPP2C primers (from Morex) failed to amplify Q21861, MD2 or cv. Golden Promise (susceptible), suggesting that either the HvPP2C gene was not present or was highly diverged. Southern blot analysis confirmed that HvPP2C was absent from Q21861 genome (data not shown). Sequence analysis of the λ clone RSB762 confirmed the absence of HvPP2C gene from this region of the resistant line Q21861.
Genetic mapping identified seven recombination events between the markers ARD5016 and HvRGA1, a physical region of 12 kb containing HvAdf2 and HvRGA1 (Fig. 1D). The sites of recombination were identified to within ≈200-bp intervals by sequencing and SNP analysis. Recombinants with the susceptible cvs. HvAdf2 allele were resistant to the stem rust isolate 92-MN-90, eliminating HvAdf2 from consideration as the Rpg5 gene (Fig. 1D). However, the six recombinants differentiating rpg4 (Pgt pathotype QCC resistance) from Rpg5 (Pgs isolate 92-MN-90 resistance) and an additional recombinant occurring distal of HvAdf2 (HQ1) identified HvAdf2 as the probable rpg4 gene.
HvAdf2 codes for a 147 aa (16.2 kDa) actin-depolymerizing factor-like protein with highest homology to the O. sativa actin-depolymerizing factor 4 expressed gene (GenBank accession no. ABF99587.1). Sequence analysis of the HvAdf2 alleles revealed that the resistant parent Q21861 and susceptible parent Steptoe and cv. Morex differed by three amino acids (Q39H, A101T, and S135G). However, the pathotype QCC susceptible parent Harrington had an HvAdf2 gene identical to the Q21861 allele at the amino acid level.
Structure and Expression of the HvRGA2 (Rpg5) Gene.
RT-PCR analysis of HvRGA2 showed it was expressed at the mRNA level in all parents tested, indicating that transcription did not correlate with resistance. However, PCR primers designed to amplify the junction between the LRR domain and the S/TPK domain produced an RT-PCR product only from Q21861, MD2, and Golden Promise [supporting information (SI) Table S1]. Although MD2 and Golden Promise contain the intact NBS-LRR-S/TPK transcript, both MD2 and Golden Promise alleles were shown to contain a frame-shift mutation within the N-terminal region of the gene resulting in a stop codon at amino acid position 217 and a putative truncated protein (Fig. 2B, Group 3).
To obtain HvRGA2 (Rpg5) transcription start site (TSS) primers were designed from the genomic sequence in 100 bp (Table S1) increments. They were used in RT-PCRs to delimit the TSS to within 100 bp at position −408 to −346 bp.
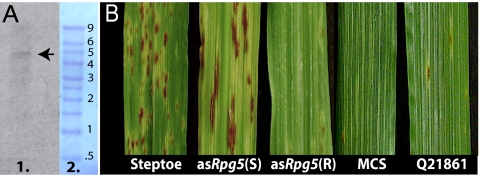
HvRGA2 (Rpg5) encodes an apparently functional NBS-LRR-S/TPK gene containing seven exons transcribed into a predicted 4.4-kb mRNA coding for a 1,378-aa (151.6-kDa) predicted protein (Fig. 2). The transcript size was confirmed by Northern blot analysis, which showed a single hybridizing band at ≈4.8 kb (Fig. 3A).
Fig. 3.
Northern analysis of Rpg5 and virus-induced gene silencing phenotypes. (A) Northern blot analysis of Rpg5 transcript. Lane 1 is Q21861 poly(A) mRNA with the Rpg5 specific probe hybridizing to a single band at ≈4.8 kb, indicated by the arrow to the right. Lane 2 is a methylene blue stain of the RNA marker after transfer to membrane. Numbers to the right indicate molecular mass of Millennium Marker (Ambion) bands. (B) Barley seedlings inoculated with antisense Rpg5 or MCS VIGS constructs and challenged with Pgs isolate 92-MN-90. Approximately 30% of the inoculated seedlings showed a typical susceptible reaction to the Pgs 92-MN-90 fungal infection. These are marked asRpg5(S), whereas seedlings that failed to show a reaction to the fungal infection are marked asRpg5(R). A representative seedling is shown for each group. The MCS antisense (BSMV-MCS) treated plant shows typical resistant reaction to Pgs 92-MN-90. The seedling leaves marked Steptoe and Q21861 are the rust-susceptible and resistant virus uninoculated controls, respectively.
The S/TPK domain contains all nine conserved amino acids (24), suggesting a functional kinase. The kinase domain had the highest homology to an O. sativa putative S/TPK (GenBank accession no. EAZ08788) but also showed significant similarity to the known R gene Pto (36% amino acid identity and 53% amino acid similarity). The Rpg5 S/TPK domain was very homologous to the unknown function Rpg1 gene family members ABC1041 and ABC1063 with 60% and 61% amino acid identity, respectively, and 76% amino acid similarity (25).
The NBS-LRR region contained the 4 NBS conserved subdomains and 12 imperfect LR repeats. This region had the highest homology to an O. sativa hypothetical protein (GenBank accession no. EAY98635). The Rpg5 NBS-LRR is very similar to the rice gene Pi-ta (40% amino acid identity and 54% amino acid similarity) conferring resistance to the rice blast fungus (26).
The TMPRED program (www.ch.embnet.org/software/TMPRED_form.html) predicted two transmembrane domains, one on the C-terminal side of the NBS domain and the other on the C-terminal side of the LRR domain (Fig. 2B, Group 1).
Virus-Induced Gene Silencing of the Rpg5 Gene.
The BSMV vector (21) was used to posttranscriptionally silence the Rpg5 gene with its antisense RNA. Approximately 30% (average of three independent experiments) of the Q21861 BSMV-asRpg5 treated seedlings showed a conversion from incompatible reaction to compatible, whereas all of the control BSMV-MCS-treated plants remained resistant to isolate 92-MN-90 (Fig. 3B). The BSMV-MCS control demonstrated that the BSMV-VIGS vector itself did not induce susceptibility to fungal infection.
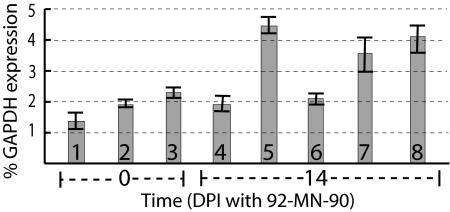
Quantitative analysis (qRT-PCR) of the Rpg5 mRNA was carried out on samples taken at the time of fungal infection (designated TP0) and at 14 d after fungal infection (designated TP14). At TP0, the BSMV-asRpg5 construct (Fig. 4, bar 1) showed significant mRNA reduction with reference to the BSMV uninoculated control (Fig. 4, bar 3) and BSMV-MCS inoculated control (Fig. 4, bar 2). At TP14, the uninoculated uninfected control (Fig. 4, bar 4) showed an Rpg5 mRNA level comparable to the TP0 control (Fig. 4, bar 3), but the TP14 BSMV uninoculated control (Fig. 4, bar 5) showed much higher mRNA levels which are comparable to the BSMV-MCS inoculated control (Fig. 4, bar 8) and the BSMV-asRpg5 inoculated plants that did not show resistance phenotype (Fig. 4, bar 7). Only the BSMV-asRpg5 inoculated seedlings showing a susceptible phenotype (Fig. 4, bar 6) had highly reduced Rpg5 mRNA levels compared with the uninoculated and MCS inoculated seedlings (Fig. 4).
Fig. 4.
Expression of the Rpg5 gene after BSMV-VIGS induced gene silencing and infection with isolate 92-MN-90 given as percentage of GAPDH gene expression. Bars represent BSMV-VIGS data from plants sampled at either time point 0 or 14 d postfungal infection (DPI). Sample 1 is inoculated with BSMV-asRpg5, 2 is inoculated with BSMV-MCS, and 3 is the uninoculated control all sampled at TP0. Sample 4 is the untreated control, 5 is the virus uninoculated control, 6 is inoculated with BSMV-asRpg5 sample taken from seedlings showing susceptible fungal reaction, 7 is inoculated with BSMV-asRpg5 sample taken from seedlings failing to show susceptible fungal reactions, and 8 is inoculated with BSMV-MCS, all sampled at TP14. The plants were examined for disease reaction at 14 DPI. The tissue analyzed by qRT-PCR was taken at TP0 or TP14 d after inoculation with isolate 92-MN-90.
The data suggested that the Rpg5 mRNA may be induced by infection with isolate 92-MN-90. A time-course experiment showed that there is a strong induction of the Rpg5 mRNA at day 1 (12.7%, relative to a GAPDH control) and a second smaller peak at days 8 (3.5%) through 11 (3.2%). This response, however, was also observed in mock and uninoculated controls and therefore is not fungus infection specific (data not shown). We conclude that the Rpg5 mRNA levels fluctuate because of environmental or developmental reasons, but the exact cause and pattern remain to be investigated. However, the Rpg5 mRNA silencing by the BSMV-asRpg5 VIGS inoculation must be made with a control sample taken at the same time point under identical environmental conditions.
Discussion
High-resolution mapping identified a 70-kb region on chromosome 7(5H) encompassing the Rpg5 and rpg4 genes. Within this locus, we identified five candidate genes. Multiple allele sequencing and recombinant characterization eliminated four genes, leaving only the HvRGA2 as the candidate Rpg5 gene (Fig. 1). Allele sequencing and VIGS confirmed the candidate Rpg5 gene. This is the first report of an R-gene encoding a disease-resistance protein composed of NBS-LRR-S/TPK domains. A search of the Oryza and Arabidopsis genomes did not identify any genes with this novel three-domain structure. The protein also contains two predicted transmembrane domains, suggesting a potential membrane-bound protein. The location of the transmembrane domains, if functional, suggests that the RPG5 LRR domain may reside outside the cell and act as the pathogen receptor, whereas the NBS and PK domains are intracellular and propagate the disease resistance signaling.
An NBS-LRR domain is present in a majority of plant disease resistance proteins identified to date (14). Another class of resistance proteins is represented by S/TPKs such as Pto, PBS1, and RPG1 (14, 18). The tomato Pto and the Arabidopsis PBS1 S/TPKs require function of the NBS-LRR genes Prf and RPS5, respectively, for disease resistance (18, 19). The Rpg5 gene, identified here, is the only reported disease resistance gene where all three domains are present in a single protein. The S/TPK of the Rpg5 gene is clearly required for disease resistance, as indicated by the susceptible cv. Harrington allele that appears to have an intact and expressed NBS-LRR protein but lacks the protein kinase domain. The presence of all three domains in a single protein may facilitate studying their interactions.
The Pto and Prf proteins have been reported to act coincidentally with one another for detecting and eliciting a disease-resistance response to the pathogen (27). Given this close interaction between the PK and NBS-LRR proteins, it is not surprising that a gene has been found that combines the two domains in one protein.
The Rpg5/rpg4 locus also confers resistance to pathotype QCC. The HvAdf2 gene was identified as the probable rpg4 gene based on analysis of the genetic recombinants. Interestingly, whereas we found recombinants that expressed resistance to isolate 92-MN-90 but susceptibility to pathotype QCC, the reciprocal recombinants were not observed. In general, resistance to pathotype QCC corresponded with resistance to isolate 92-MN-90 in studies of a large number of domesticated and wild barleys (B.S., unpublished data). The observation that the QCC-susceptible cv. Harrington has an expressed Adf2 gene identical to the resistant Q21861 allele at the amino acid level also suggested that additional factors may be involved. One hypothesis is that the recessive rpg4 gene functions as a pathotype QCC-specific disease-resistance gene only in the presence of another functional disease resistance gene, perhaps Rpg5.
ADFs are critical in remodeling the actin cytoskeleton during normal plant development and under biotic and abiotic stress. They are typically small proteins that function in rapid recycling of actin monomers (28). Actin functions in cytoskeleton organization, which coordinates essentially all aspects of plant growth (29). Actin microfilament polymerization has been shown to be involved in nonhost disease resistance. For example, using cytochalasin E, an inhibitor of actin microfilament polymerization and Arabidopsis defense-related mutants eds1, pad4, and nah4, it was shown that nonhost resistance to the wheat powdery mildew fungus Blumeria graminis f. sp. tritici in Arabidopsis largely depends on actin cytoskeleton dynamics and function of the EDS1 gene (30). Similar observations have been documented in other systems.
The posttranscriptional gene silencing of Rpg5 by VIGS showed significant reduction of Rpg5 transcript at both sampling time points 0 d (seedlings pooled) and 14 d (susceptible and resistant plants sampled separately). Approximately 30% of the BSMV-asRpg5 plants resulted in a susceptible reaction to the rust fungus. At the 14-d time point, the level of silencing observed corresponded to the phenotype observed with the susceptible plants showing significant reduction in the Rpg5 mRNA, whereas the resistant plants had mRNA level similar to the controls (Fig. 4). The time-course experiments indicated that Rpg5 mRNA levels were variable over time. However, induction by fungal infection could not be established because of similar observations in mock and uninoculated controls. The variable Rpg5 mRNA levels were surprising, because previous experiments with Rpg1 showed low but steady mRNA levels with or without fungal infection (31, 32).
In summary, we have identified two types of plant disease-resistance genes. The Rpg5 gene combines features of NBS-LRR type disease-resistance gene with the S/TPK domain, suggesting that this gene may function both in pathogen perception and signal transduction. The candidate gene for rpg4 needs further verification, but, if confirmed, it would show involvement of actin cytoskeleton in race-specific disease resistance.
Materials and Methods
Plant Materials.
Progeny from the crosses Steptoe/Q21861, Harrington/Q21861, and MD2/Q21861 were used for genetic mapping. Q21861 is the source of the stem rust-resistance genes Rpg5 and rpg4 (3, 9). Steptoe and Harrington are barley cultivars, and MD2 is a genetic stock with multiple dominant mutations. All three are susceptible to stem rust. Plants were grown in the greenhouse with day/night temperatures of 18°C/14°C, respectively. Metal halide lights supplemented a 16-/8-h light/dark photoperiod.
Molecular Markers.
RFLP markers were generated as described (13) or by designing PCR primers based on the cv. Morex or line Q21861 sequence (this study). Primers are described in Table S1.
Genetic and Physical Mapping.
High-resolution mapping at the Rpg5/rpg4 locus used 50 recombinants selected from 5,232 gametes between the flanking markers Aga5 and ABG391. These 50 recombinants were reduced to 10 between the flanking markers ARD5016 and ARD5112. BAC physical maps were generated as described (13).
Sequencing and Sequence Analysis.
The cv. Morex BAC clones (543P19 and 116G10) forming a contig across the Rpg5/rpg4 region were sequenced at the Institute for Genomics Research (TIGR). All other cultivar DNA was sequenced with the BigDye terminator system on an ABI 373 DNA sequencer (Applied Biosystems) at the Laboratory for Biotechnology and Bioanalysis, Washington State University, Pullman. The λ clone RSB762 was subcloned into the NotI site of pBluescript (Stratagene) and sequenced by using the EZ::TNTM<KAN-2> Insertion Kit (Epicentre). All PCR-generated fragments were either directly sequenced or cloned into pGEM-T Easy vector (Promega) and sequenced by using the EZ::TNTM<KAN-2> Insertion Kit (Epicentre). For direct sequencing, gel slices were placed in plugged tips (Rainin Instrument) and frozen at −20°C. After complete thawing, the tip was centrifuged at 4,000 × g for 10 min. The eluate was extracted two times with 24:1 chloroform:isoamyl alcohol and precipitated with 0.3 M NaOAc and 2.5 volumes 95% EtOH. Primers and clones are described in Table S1.
Contigs were assembled by using Vector NTI Advance 9.0 contig express (Invitrogen). The cultivar sequence comparisons were done by using Vector NTI alignX and the National Center for Biotechnology Information bl2seq function (www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). All sequences were analyzed with the BLASTX and BLASTN algorithms.
Library Construction.
High-molecular-mass Q21861 genomic DNA was isolated by using a modified CTAB method (33). Genomic DNA (≈20 μg) was partially digested with 0.1 units of Sau 3AI for 1 hour to yield a majority of 15- to 23-kb fragments. DNA was dephosphorylated with 0.1 units of calf intestine alkaline phosphatase for 30 min and separated in 1.2% low melting agarose. DNA fragments 9–23 kb were excised from the gel and recovered by β-agarase treatment. Genomic fragments were ligated into the Lambda Dash II vector (Stratagene) predigested with BamHI and packaged using the Gigapack III XL packaging extract (Stratagene).
RNA Isolation and Northern Blot.
Isolation of RNA, Northern blot analysis, and hybridization were as described (25).
RT-PCR.
Approximately 1 μg of total RNA was used for RT-PCRs by using M-MLV Reverse Transcriptase (Promega) under the manufacturer's recommended conditions. RT-PCR fragments were directly sequenced after elution from agarose gels as described for direct sequencing.
3′ and 5′ RACE.
3′ RACE was carried out by using the 3′ RACE System (Invitrogen) following the manufacturer's recommended conditions. 5′ RACE was carried out by using the FirstChoice RLM-RACE Kit (Ambion) following the manufacturer's recommended conditions and SMART technology (34). The gene specific primers used for RACE are described in Table S1.
VIGS and qRT-PCR.
Barley plants for VIGS experiments were grown in the growth chamber in plastic pots with a day/night temperature of 20°C +/−1°C and 18°C +/− 1°C, respectively. A 20-/4-h light/dark photoperiod was provided by cool fluorescent tubes (525 uE/m2s).
Rpg5 was silenced by using VIGS as described (21). A 314-bp Rpg5 cDNA fragment (+2,033 to 2,346 bp) was generated by PCR and ligated in antisense (as) orientation into BSMV-VIGS infectious clone (pγPDS4as) digested with PacI and NotI (21). The BSMV-asRpg5 construct was cotranscribed with the α and β genomes of the tripartite BSMV virus by using the mMessage mMachine T7 kit (Ambion), and the T7 promoter. RNA was inoculated onto Q21861 barley plants at the two-leaf stage. The seedlings were inoculated with isolate 92-MN-90 urediniospores at 0.025 mg per plant mixed with a talc carrier ≈11–12 days after virus infection. After inoculation, the plants were misted and placed in the dark under high humidity conditions for 22 h, then exposed to light and misted periodically. After 4 h, the misting was stopped, and the leaves were left to dry slowly. When the leaf surfaces were completely dry, plants were moved to the growth chambers at 20°C and 80% relative humidity. The plants were scored for compatibility or incompatibility at 14 days postfungal infection
The negative control contained a 121-bp antisense fragment of the multi cloning site (MCS) from pBluescript K/S (Stratagene). The MCS sequence did not hybridize to barley genomic DNA at low stringency conditions, indicating no homologous regions in the barley genome. Q21861 and Steptoe were used as the resistant and susceptible virus uninoculated controls. qRT-PCR was performed on Rotor-Gene 2000 thermocycler (Corbet Research) using the QuantiTect SYBR green PCR system (Qiagen). The Rpg5 primers used are described in Table S1 and GAPDH primers were described (35). Tissue samples for qRT-PCR were collected at 0 or 14 d after fungal infection. Tissue samples for the time course experiment were collected at 0, 1, 3, 5, 8, 11, and 14 days after fungal inoculation.
Supplementary Material
Acknowledgments.
This research was supported by U.S. Department of Agriculture National Research Initiative Grant No.2004-35301-14635. This is Scientific Paper No. 0402-08 from the College of Agricultural, Human, and Natural Resource Sciences Research Center, Washington State University, Project 0196.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU883792 (Rpg5 cDNA from barley line Q21861); EU878778 (genomic sequence from barley line Q21861); EU812563 (genomic sequence from barley line Morex); EU881932–EU881935 (HvAdf2 genomic sequence from barley lines Golden Promise, Harrington, MD2, and Steptoe, respectively); EU883581–EU883583 (HvRGA1 genomic sequence from barley lines Harrington, MD2 and Steptoe, respectively); EU883787–EU883790 (HvRGA2 genomic sequence from barley lines Golden Promise, Harrington, MD2, and Steptoe, respectively); and EU883791 (HvAdf3 genomic sequence from barley line Steptoe)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0807270105/DCSupplemental.
References
- 1.Steffenson BJ. Analysis of durable resistance to stem rust in barley. Euphytica. 1992;63:153–167. [Google Scholar]
- 2.Sun Y, Steffenson BJ. Reaction of barley seedlings with different stem rust resistance genes to Puccinia graminis f sp tritici and P g f. sp. secalis. Can J Plant Pathol. 2005;27:80–89. [Google Scholar]
- 3.Jin Y, Steffenson BJ, Miller JD. Inheritance of resistance to pathotypes QCC and MCC of Puccinia graminis f sp tritici in barley line Q21861 and temperature effects on the expression of resistance. Phytopathology. 1994;84:452–455. [Google Scholar]
- 4.Brueggeman R, et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nirmala J, et al. Subcellular localization and functions of the barley stem rust resistance receptor-like serine/threonine-specific protein kinase Rpg1. Proc Natl Acad Sci USA. 2006;103:7518–7523. doi: 10.1073/pnas.0602379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roelfs AP, Casper DH, Long DL, Roberts JJ. Races of Puccinia graminis in the United States in 1989. Plant Dis. 1991;75:1127–1130. [Google Scholar]
- 7.Jin Y, Steffenson BJ, Fetch TGJ. Sources of resistance to pathotype QCC of Puccinia graminis f sp tritici in barley. Crop Sci. 1994;34:285–288. [Google Scholar]
- 8.Steffenson BJ, Jin Y, Rossnagel BG, Rasmussen JB, Kao K. Genetics of multiple disease resistance in a doubled-haploid population of barley. Plant Breeding. 1995;114:50–54. [Google Scholar]
- 9.Sun Y, Steffenson BJ, Jin Y. Genetics of resistance to Puccinia graminis f sp secalis in barley line Q21861. Phytopathology. 1996;86:1299–1302. [Google Scholar]
- 10.Borovkova IG, et al. Identification of molecular markers linked to the stem rust resistance gene rpg4 in barley. Phytopathology. 1995;85:181–185. [Google Scholar]
- 11.Han F, et al. Synteny with rice: Analysis of barley malting quality QTL and rpg4 chromosome regions. Genome. 1998;41:373–380. [Google Scholar]
- 12.Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A. Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4 using rice as an intergenomic cloning vehicle. Plant Mol Biol. 1997;35:187–195. [PubMed] [Google Scholar]
- 13.Druka A, et al. Physical mapping of barley stem rust resistance gene rpg4. Mol Gen Genet. 2000;264:283–290. doi: 10.1007/s004380000320. [DOI] [PubMed] [Google Scholar]
- 14.Martin GB, Bogdanove AJ, Sessa G. Understanding the function of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 15.Song W-Y, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- 17.Martin GM, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 18.Swiderski MR, Innes RW. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001;26:101–112. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- 19.Salmeron JM, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 20.Warren RF, Merritt RFPM, Holub E, Innes RW. Identification of three putative signal transduction genes involved in R-gene specified disease resistance in Arabidopsis. Genetics. 1999;152:401–412. doi: 10.1093/genetics/152.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holzberg S, Brosio P, Gross C, Pogue GP. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002;30:315–327. doi: 10.1046/j.1365-313x.2002.01291.x. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi Y, Yamada M, Kobayashi I, Kunoh H. Actin microfilaments are required for the expression of nonhost resistance in higher plants. Plant Cell Physiol. 1997;38:725–733. [Google Scholar]
- 23.Cottage A, Yang A, Maunders H, de Lacy RC, Ramsay NA. Identification of DNA sequences flanking T-DNA insertions by PCR-walking. Plant Mol Biol Rep. 2001;19:321–327. [Google Scholar]
- 24.Hanks S, Quinn A, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 25.Brueggeman R, Drader T, Kleinhofs A. The barley serine/threonine kinase gene Rpg1 providing resistance to stem rust belongs to a gene family with five other members encoding kinase domains. Theor Appl Genet. 2006;113:1147–1158. doi: 10.1007/s00122-006-0374-3. [DOI] [PubMed] [Google Scholar]
- 26.Bryan GT, et al. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2033–2045. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mucyn TS, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remedios DC, et al. Actin binding proteins: Regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–479. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 29.Wasteneys GO, Galway ME. Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu Rev Plant Biol. 2003;54:691–722. doi: 10.1146/annurev.arplant.54.031902.134818. [DOI] [PubMed] [Google Scholar]
- 30.Yun B-W, et al. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 2003;34:768–777. doi: 10.1046/j.1365-313x.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 31.Rostoks N, Steffenson BJ, Kleinhofs A. Structure and expression of the barley stem rust resistance gene Rpg1. Physiol Mol Plant Pathol. 2004;64:91–101. [Google Scholar]
- 32.Zhang L, Castell-Miller C, Dahl S, Steffenson B, Kleinhofs A. Parallel expression profiling of barley stem rust interactions. Funct Integr Genom. 2008;8:187–198. doi: 10.1007/s10142-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 33.Kleinhofs A, et al. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- 34.Tabansky I, Nurminsky DI. Mapping of transcription start sites by direct sequencing of SMARTTM RACE products. Biotechnology. 2003;34:482–486. doi: 10.2144/03343bm06. [DOI] [PubMed] [Google Scholar]
- 35.Horvath H, et al. Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc Natl Acad Sci USA. 2003;100:364–369. doi: 10.1073/pnas.0136911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.