Abstract
Tumstatin is an angiogenesis inhibitor that binds to αvβ3 integrin and suppresses tumor growth. Previous deletion mutagenesis studies identified a 25-aa fragment of tumstatin (tumstatin peptide) with in vitro antiangiogenic activity. Here, we demonstrate that systemic administration of this tumstatin peptide inhibits tumor growth and angiogenesis. Site-directed mutagenesis identified amino acids L, V, and D as essential for the antiangiogenic activity of tumstatin. The tumstatin peptide binds to αvβ3 integrin on proliferating endothelial cells and localizes to select tumor endothelium in vivo. Using 3D molecular modeling, we identify a putative interaction interface for tumstatin peptide on αvβ3 integrin. The antitumor activity of the tumstatin peptide, in combination with bevacizumab (anti-VEGF antibody), displays significant improvement in efficacy against human renal cell carcinoma xenografts when compared with the single-agent use. Collectively, our results demonstrate that tumstatin peptide binds specifically to the tumor endothelium, and its antiangiogenic action is mediated by αvβ3 integrin, and, in combination with an anti-VEGF antibody it exhibits enhanced tumor suppression of renal cell carcinoma.
Keywords: angiogenesis, αvβ3 integrin, bevacizumab, tumstatin peptide
Tumor angiogenesis is a hallmark of tumor growth and metastasis (1). There are currently several antiangiogenic agents approved for human cancer therapy, but their effect is modest and short-acting, with therapy resistance generally developing within a few months. Thus, further research is required to improve the efficacy of antiangiogenic therapy and to identify genetic alterations that are likely to predict the therapeutic outcome for cancer patients.
We previously identified and characterized tumstatin as an endogenous antiangiogenic agent derived from the noncollagenous (NC1) domain of the α3 chain of type IV collagen (2). Tumstatin is an inhibitor of endothelial cell proliferation and suppresses the growth of various tumor types in mice (2, 3). It binds to endothelial cells via αvβ3 integrin, and this binding is speculated to be important for its antiangiogenic activity [see supporting information (SI) Table S1]. Integrins are a family of cell adhesion molecules. All integrins are heterodimeric transmembrane proteins, always consisting of an α and a β subunit, and αvβ3 integrin on endothelial cells is one such heterodimer (4). The heterodimer formation is essential for mediating outside-in cell signaling (5). The efficacy of tumstatin is profoundly increased in tumors above the size of 500 mm3, correlating with increased endothelial αvβ3 integrin expression at this stage (6). Although the C-terminal half of tumstatin exhibits direct tumor cell cytotoxicity, the antiangiogenic activity of tumstatin resides in the N-terminal half, restricted to amino acids 74–98 and termed the T7 peptide (3, 7). The T7 peptide (referred to as tumstatin peptide in this report) and the full length tumstatin protein have an equivalent antiproliferative effect on endothelial cells in vitro, and the inhibition of protein synthesis in endothelial cells is mediated by the tumstatin peptide (3, 8).
The current experiments were designed to identify the critical amino acids within tumstatin that confer antiangiogenesis activity and define the antiangiogenic and antitumor activity of tumstatin peptide. Additionally, we explored the endothelial binding characteristics of the tumstatin peptide in both the in vitro and in vivo settings. Combination of tumstatin peptide with anti-VEGF antibody was also explored for possible improvement in antitumor efficacy.
Results
Tumstatin Peptide Inhibits Tumor Growth.
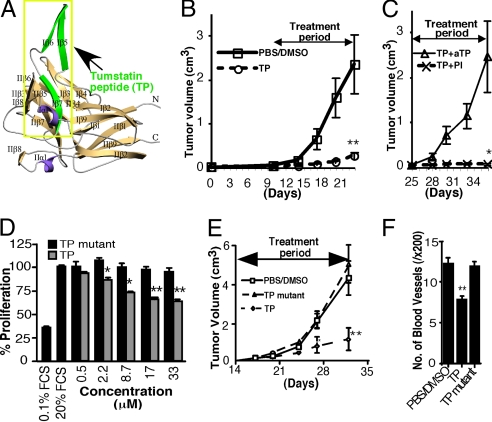
A putative structure representing tumstatin and containing the tumstatin peptide is shown in Fig. 1A. Tumstatin peptide significantly inhibits the growth of SCC-PSA1 teratocarcinomas, when compared with sham treatment (Fig. 1B). To test for tumstatin peptide specificity, an efficacy experiment was conducted by using a selective antitumstatin peptide antibody. The use of antitumstatin peptide but not the preimmune sera significantly reversed the effects of the tumstatin peptide (Fig. 1C), indicating that tumstatin peptide specifically inhibits tumor growth (9).
Fig. 1.
Tumstatin peptide inhibits tumor growth and tumor angiogenesis. (A) Predicted secondary structure of tumstatin. The tumstatin peptide is indicated by the yellow box and highlighted in green. Secondary structure elements are colored beige (strands), purple (α-helices), and gray (coil) and labeled according to convention (9). (B and C) The effect of tumstatin peptide (TP) on SCC-PSA1 teratocarcinoma growth. Tumor growth curves show the mean tumor volume ± SEM, n = 13–15. From day 25, the tumstatin peptide-treated mice were given preimmune serum (PI) or a tumstatin peptide specific-antibody (aTP) together with tumstatin peptide, n = 4–5. *, P <0.05 compared with aTP+TP; **, P < 0.01 compared with PBS/DMSO. (D) The importance of tumstatin peptide sequence for antiendothelial activity. C-PAE cells were incubated with tumstatin peptide or tumstatin peptide mutant and cell proliferation was assessed. *, P < 0.05 and **, P < 0.01, compared with tumstatin peptide mutant at the same concentration. (E) The effect of tumstatin peptide, tumstatin peptide mutant and sham treatment on LLC tumor growth. Tumor growth curves show the mean tumor volume ± SEM, n = 8. *, P < 0.05 compared with control or tumstatin peptide mutant; **, P < 0.01 compared with control or tumstatin peptide mutant. (F) The effect of tumstatin peptide and tumstatin peptide mutant on microvessel density (per ×200 high-power field) in LLC tumors on day 32. **, P < 0.01 compared with control or tumstatin peptide mutant–ANOVA analysis.
Site-Directed Mutagenesis of Tumstatin Peptide Identifies L, V, and D as the Critical Amino Acids for Antiangiogenesis and Antitumor Activity.
Homology information derived from sequence comparison between tumstatin and the NC1 domain of α5 chain of type IV collagen, which lacks antiangiogenic activity (10), led to the identification of amino acids that might contribute to the antiangiogenic activity of tumstatin. Using rationale site-directed mutagenesis, we embarked on mapping the crucial amino acids within the tumstatin peptide and constructed seven different sequence variants. Within the 25 aa of tumstatin, the L, V, and D amino acids were found to be essential for the antiangiogenic activity (Table 1). In proliferation assays, the MIN mutant peptide (tumstatin mutant peptide) does not exhibit any activity, whereas the tumstatin peptide inhibits proliferation of endothelial cells by ≈50% (Fig. 1D). Next, we demonstrate that the tumstatin mutant peptide is incapable of inhibiting growth of syngenic Lewis lung carcinoma (LLC) tumors in mice, when compared with the tumstatin peptide (Fig. 1E). This suppression of tumor growth by the tumstatin peptide is also associated with a decrease in microvessel density (Fig. 1F and Fig. S1).
Table 1.
Site-directed mutagenesis of tumstatin peptides and their action on endothelial cell proliferation
| Tumstatin peptide | Amino acid sequence | Endothelial cell inhibition* |
|---|---|---|
| T7 | TMPFLFCNVNDVCNFASRNDYSYWL | ++++ |
| T7-mutant 1 (MIN) | TMPFMFCNINNVCNFASRNDYSYWL | - |
| T7-mutant 2 (MI) | TMPFMFCNINDVCNFASRNDYSYWL | - |
| T7-mutant 3 (IN) | TMPFLFCNINNVCNFASRNDYSYWL | + |
| T7-mutant 4 (N) | TMPFLFCNVNNVCNFASRNDYSYWL | + |
| T7-mutant 5 | TMPFLFCNVNDVCNFASRNDAK | - |
| T7-mutant 6 | TMPFLYCNPGDVCYYASRNDKSYWL | + |
| T7-mutant 7 | TLPFAYCNIHQVCHYAQRNDRSYWL | ++ |
*Percentage inhibition of endothelial cell proliferation (C-PAE cells), relative to tumstatin peptide, at 33 μM concentration. −, no activity; +, <25% of tumstatin peptide activity; ++, 25–50% of tumstatin peptide activity; +++, 50–75% of tumstatin peptide activity; ++++, maximal activity of tumstatin peptide (T7).
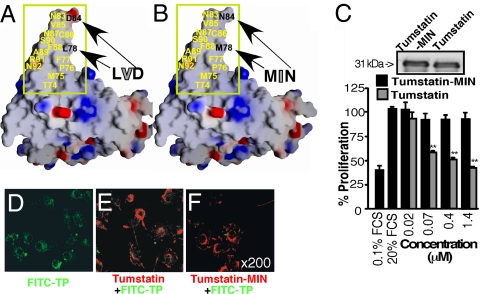
Next, we used full-length tumstatin to specifically mutate amino acids L, V, and D within the tumstatin peptide region and replaced them with M, I, and N amino acids respectively (to mimic the NC1 domain of the α5 chain of type IV collagen) (Fig. 2 A and B). As shown in the 3D model of tumstatin, these amino acids are part of the putative exposed tumstatin peptide region of the protein (Fig. 2 A and B). Using 293 embryonic kidney epithelial cells, we expressed the recombinant tumstatin and recombinant tumstatin-MIN (mutant) proteins and used the proteins in endothelial proliferation assays. Recombinant tumstatin inhibits endothelial proliferation by almost 60%, whereas the recombinant tumstatin mutant does not exhibit this activity, further supporting the hypothesis that the L, V, and D amino acids are critical for the antiangiogenesis activity of tumstatin (Fig. 2 C and Inset).
Fig. 2.
Identification of amino acids responsible for the anti-angiogenesis property of tumstatin. (A and B) Electrostatic surface representation of the tumstatin and the tumstatin mutant protein homology models. Surface charges are shown as: blue, positive; and red, negative. The tumstatin peptide region is indicated by the yellow rectangle, and the point-mutated residues are indicated by arrows. In the tumstatin peptide mutant and tumstatin mutant protein, amino acids L (78), V (82) and D (84) were replaced with amino acids M, I and N, respectively. The V to I mutation is not visible in this orientation of the surface model (indicated by gray letters). (C) The importance of tumstatin peptide sequence for the anti-endothelial activity of tumstatin. Cell proliferation in C-PAE cells incubated with tumstatin or tumstatin mutant protein was assessed. **, P < 0.01, compared with tumstatin mutant protein at the same concentration. (Inset) Recombinant production of tumstatin and tumstatin mutant protein. Using standard Western blot procedure, 500 ng of protein was run in each lane on a 15% acrylamide gel, and an anti-FLAG antibody was used with anti-mouse IgG linked to horseradish peroxidase (Sigma), as primary and secondary antibody, respectively. Lane 1, tumstatin mutant; lane 2, tumstatin. (D–F) Tumstatin peptide (TP) binding to endothelial cells preexposed to tumstatin or tumstatin mutant protein. C-PAE cells were incubated with FITC-tumstatin peptide (D), tumstatin (E), or tumstatin mutant protein (F) before addition of FITC-tumstatin peptide. An anti-FLAG antibody was used to visualize tumstatin and tumstatin mutant protein. Green: FITC-tumstatin peptide; red: anti-FLAG (tumstatin and tumstatin mutant protein). Confocal microscopy.
Endothelial Cells Have a Binding Site for Tumstatin Peptide.
To address the capacity of tumstatin peptide to bind endothelial cells, we synthesized a FITC-conjugated tumstatin peptide (FITC-tumstatin peptide). FITC-tumstatin peptide and tumstatin peptide inhibit endothelial cell proliferation at comparable levels, demonstrating that the antiangiogenic activity of tumstatin peptide is not compromised when conjugated to FITC (Fig. S2).
To analyze the binding capacity of tumstatin peptide, endothelial cells were incubated with recombinant full-length tumstatin or tumstatin mutant protein followed by FITC-tumstatin peptide. Although tumstatin peptide by itself exhibited extensive binding to endothelial cells, the binding was significantly reduced when preincubation was performed with recombinant full-length tumstatin (Fig. 2 D–F). These results demonstrate that endothelial cells can bind tumstatin peptide, and that tumstatin peptide and full-length tumstatin protein share a binding site on the endothelial cells. However, endothelial cells also partially bind to the recombinant tumstatin mutant protein (Fig. 2F), demonstrating that the mutations within the tumstatin peptide sequence disrupt the activity, but not the entire binding capacity of the protein to the proliferating endothelium. Furthermore, antitumstatin antibodies inhibit the attachment of endothelial cells to tumstatin peptide-coated culture plates in a dose-dependent manner, when compared with control antibodies (Fig. S3). This result demonstrates that endothelial cells have a binding site for the tumstatin peptide.
Tumstatin Peptide Binds to αvβ3 Integrin on Proliferating Endothelial Cells.
Previous studies have demonstrated that tumstatin binds to αvβ3 integrin on endothelial cells, in an Arg-Gly-Asp (RGD)-independent manner (7). Endothelial cells show similar attachment capacity to culture plates precoated with either native tumstatin or tumstatin peptide and tumstatin mutant protein or tumstatin mutant peptide (Fig. S4). Preincubation of endothelial cells with an αvβ3 integrin antibody inhibited the attachment to both peptide and full-length tumstatin protein, whereas preincubation with α2 integrin subunit antibody did not inhibit binding to endothelial cells (Fig. S4). These results indicate that tumstatin binds to αvβ3 integrin on endothelial cells via the tumstatin peptide subunit. Using confocal microscopy, we further confirmed that FITC-tumstatin peptide colocalizes with αvβ3 integrins on the endothelial cell surface (Fig. S5). Finally, the SSC-PSA1 teratocarcinoma cell line was found to lack expression of the αv and β3 integrin subunits, and the αvβ3 integrin dimer (Fig. S6), excluding tumor cell cytotoxicity as the reason for tumor growth inhibition (Fig. 1B).
We demonstrate that FITC-tumstatin peptide binds only to proliferating endothelial cells and not to a 100% confluent culture of endothelial cells that are contact-inhibited and express VE cadherin on the cell surface (Fig. S7). When endothelial cells are proliferating (40% confluency), FITC-tumstatin peptide binding is robust and extensively colocalized with αvβ3 integrin on the cell surface (Fig. S7). Upon reaching 100% confluency, the expression of αv integrin subunit and FITC-tumstatin peptide binding becomes negligible (Fig. S7). These results suggest that proliferating endothelial cells specifically express αvβ3 integrin and such robust expression disappears when the endothelial cells cease to proliferate at 100% confluency, a time point at which the expression of endothelial cell adhesion molecule, VE cadherin, is the highest. Concomitantly, the binding of FITC-tumstatin peptide decreases as the cells reach 100% confluency. Collectively, these results suggest that the capacity of tumstatin peptide to bind proliferating endothelial cells correlates with the expression of αvβ3 integrin. Furthermore, colocalization studies with FITC-tumstatin peptide and tumor vascular markers (CD31 and entactin) demonstrate that tumstatin peptide binds only to a subset of the endothelial cells (Fig. S8). BrdU labeling (to assess for cells in a proliferative state) reveals that the tumstatin peptide mostly binds to proliferating tumor endothelium (Fig. S8).
Next, we examined the binding capacity of FITC-tumstatin peptide to blood vessels during pathological and physiological (repair-associated) angiogenesis in mice. We demonstrate that FITC-tumstatin peptide binds to angiogenic vessels of LLC tumors and also angiogenic vessels associated with Matrigel plug assays (Fig. S9). Interestingly, FITC-tumstatin peptide did not bind to angiogenic vessels associated with liver regeneration and wound healing in mice (Fig. S9). This observation is in accordance with our previous observation that, whereas tumor and Matrigel plug-associated angiogenic vessels express αvβ3 integrin in a subset of their neovasculature, the physiological repair-associated, angiogenic vessels do not express αvβ3 integrin (6).
FITC-tumstatin peptide was used as a therapeutic in mice with LLC tumors (see above). Therefore, we evaluated whether FITC-tumstatin peptide localizes to any other vasculature in tumor-bearing mice, after systemic administration. Examination of >35 different tissue locations in these mice reveals that FITC-tumstatin peptide specifically binds to tumor-associated angiogenic vessels and not to normal organ vasculature in the mice (Fig. S9).
The ability of FITC-tumstatin peptide to bind to tumor angiogenic vessels of tumstatin deficient (collagen IV-α3−/−) and heterozygous (collagen IV-α3+/−) mice with LLC tumors was compared (Fig. S10). Absence of endogenous tumstatin in the collagen IV-α3-deficient mice leads to significantly more FITC-tumstatin peptide localization when compared with the tumors in the heterozygotes. These results indicate that in heterozygous mice, endogenous tumstatin attaches to some of the neovessels and contributes to physiological control of tumor angiogenesis, whereas in mice lacking tumstatin, all putative αvβ3 integrin-binding sites are free and available for increased FITC-tumstatin peptide attachment.
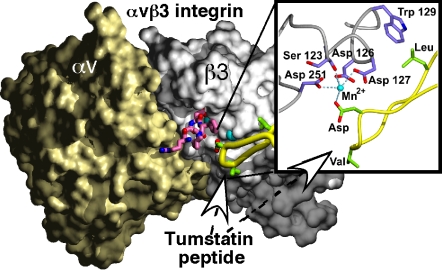
The crystal structure of the extracellular domain of αvβ3 integrin and its complex with the common RGD peptide ligand motif was solved a few years ago (11). The crystal structure of the NC1 domain hexamer of type IV collagen from human placenta is also reported (12). We used the combined information to generate a 3D homology model of the putative binding site of tumstatin and tumstatin peptide to αvβ3 integrin (Fig. 3). The T7 peptide region of tumstatin protrudes out of the putative structure of tumstatin and interacts with a surface groove on the β3 domain of αvβ3 integrin, and this potential binding site is distinct from the RGD peptide attachment locus (11). In our model, the negatively charged D (84) side chain of tumstatin peptide interacts with a positively charged manganese ion (Fig. 3 Inset). Mutation of this residue to N (D→N) would be expected to disrupt this interaction. Substitution of L (78) with a bulkier M side chain (L→M) would also have a disruptive effect on the metal-binding site by steric interaction with D (126) (on the β3 subunit of αvβ3 integrin), which coordinates the manganese atom. In contrast, it is difficult to determine what effect, if any, the conservative mutation of leucine to isoleucine (V→I) at position 78 will have on the interaction with the β3 integrin subunit.
Fig. 3.
Tumstatin interaction with αvβ3 integrin. 3D modeling of the potential interaction between tumstatin and αvβ3 integrin and surface diagram of the ligand binding site. The αv and the β3-domains of αvβ3 integrin are shown as beige and gray surfaces, respectively. The noncompetitive RGD peptide is a magenta stick model. The tumstatin peptide region of tumstatin is shown in yellow, and the LVD amino acids of tumstatin peptide are green sticks. The manganese atoms interacting with the RGD and tumstatin peptides are shown as cyan spheres. The surfaces were calculated with the program GRASP (13) and used as input for Povscript+ (14). The final images were ray-traced in POV-Ray (www.povray.org). (Inset) Diagram illustration of the tumstatin peptide interaction with the β3 integrin subunit of αvβ3 integrin. The amino acids of the β3 integrin subunit that interact with the manganese atom are shown as sticks colored blue, whereas the backbone of β3 integrin is depicted in gray. The tumstatin peptide is shown in yellow, with the LVD amino acids colored green. D (84) of tumstatin peptide is shown to interact with the manganese atom in the β3 subunit of αvβ3 integrin.
Combination of Tumstatin Peptide with Anti-VEGF Antibody Provides Enhanced Antitumor Activity.
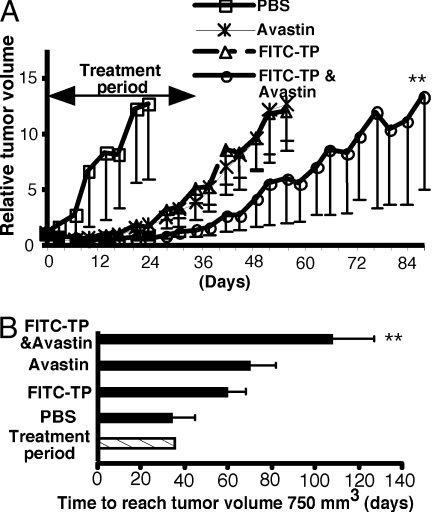
The 786 human renal cell carcinoma xenografts on athymic nude mice were used to assess combination treatment of tumstatin peptide with an anti-VEGF antibody (Fig. 4 A and B). The mice were administered tumstatin peptide and/or an anti-VEGF antibody (bevacizumab, Avastin) for 5 weeks to assess the long-term effects and toxicity associated with this treatment regimen. The mice tolerated the combined therapy, without overt toxicity or weight loss (data not shown). In the 786 tumors, tumstatin peptide or anti-VEGF antibody suppressed tumor growth but without statistical significance, whereas the combination of the two demonstrated a significant tumor growth inhibition, when compared with other treatment arms (Fig. 4B). These results provide strong preclinical evidence for a possible combined efficacy of tumstatin peptide and anti-VEGF antibody.
Fig. 4.
Improved antitumor efficacy of combined therapy with anti-VEGF antibody and tumstatin peptide. (A) The effect of combination antiangiogenic therapy in 786 human renal cell carcinoma. Tumor growth curves show the mean relative tumor volume ± SEM, n = 5–7. The tumor volume when treatment was started is normalized on the y axis. **, P < 0.02, compared with the control group. (B) The time for 786 human renal cell carcinomas to reach a tumor volume of 750 mm3. Bars depict the mean ± SEM, n = 5–7. **, P < 0.02, compared with the control group.
Discussion
Tumstatin is a member of a class of proteins and factors known as endogenous angiogenesis inhibitors (15). In this report, we demonstrate that the antiangiogenic activity of tumstatin resides within the T7 peptide region of this protein fragment and is further associated with amino acids L, V, and D. Several mutagenesis experiments were designed to validate that the L, V, and D amino acids are important for the antiangiogenic activity of tumstatin. The L, V, and D amino acids of tumstatin peptide are critical for the activity mediated via αvβ3 integrin on proliferating endothelium. The tumstatin peptide binds specifically to proliferating endothelial cells on the tumor-associated vasculature, and the inhibition of angiogenesis and tumor growth is associated with the expression of αvβ3 integrin on tumor endothelial cells. Using 3D modeling, we examined how tumstatin may interact with its integrin receptor, but the exact binding site and interaction characteristics must await cocrystallization of tumstatin and αvβ3 integrin. Collectively, our study provides biochemical data to support the notion that tumstatin is an endogenous angiogenesis inhibitor that requires αvβ3 integrin for its action (6). Furthermore, our data suggest that a therapeutic regime combining tumstatin with currently approved antiangiogenic agents, such as the anti-VEGF antibody bevacizumab (Avastin), may provide enhanced tumor growth suppression and delay in cancer progression.
Malignant neoplasms have multiple signaling pathways that are up-regulated simultaneously (16, 17). Therefore, blocking just one signaling axis likely results in up-regulating or further escalating other pathways (18–20). This is one possible explanation for the lack of long-term efficacy of angiogenesis inhibitors, such as anti-VEGF antibodies, in the clinic. In this regard, ≈80% of renal cell carcinoma patients respond to anti-VEGF and IFN-α combination treatment, with disease stabilization. Nevertheless, only 10–20% of the patients exhibit an objective tumor regression (21, 22). Tumstatin is directly cytotoxic to the tumor endothelium, inhibiting protein synthesis (8), whereas bevacizumab indirectly affects the tumor vasculature by blocking the VEGF ligand secreted by various cell types within the tumor (23). Additionally, blocking VEGF signaling causes vascular normalization within malignant tumors, and could increase the amount of tumstatin peptide reaching the tumor via improved tumor perfusion (24). Therefore, adding tumstatin peptide in a combination therapy mixture is likely a viable strategy to prolong the therapeutic response of bevacizumab.
Identification of the critical amino acids responsible for the antiangiogenesis action of tumstatin provides us an opportunity to evaluate for possible SNPs involving the L, V, and D amino acids in a general human population and cancer patients. Variations in the tumstatin gene sequence could result in compromised endogenous angiogenesis inhibition, leading to enhanced rate of tumor progression, as demonstrated for endostatin (25, 26). Such genetic screening for SNPs might identify individuals who would benefit from supplemental recombinant tumstatin or tumstatin peptide to control the rate of tumor growth.
Materials and Methods
In Vivo Tumor Trials.
For the in vivo tumor experiments, one million SCC-PSA1 teratocarcinoma (SP), LLC, or 786 human renal cell carcinoma (786-O) cells were injected s.c. on the back of SV129 (SP), C57/BL6 (LLC), and Nu/Nu (786-O) mice, respectively. The LLC tumors were further grown on both collagen IV-α3 deficient (−/−) and heterozygous (+/−) littermate control mice. Further information is listed in SI Methods.
Production of Recombinant Tumstatin, Tumstatin Mutant, and Synthetic Peptides.
Two sets of primers were created for two individual PCR amplifications. The first set of primers was designed to amplify a sequence from 40 to 255 bp of tumstatin and to mutate the amino acids L (78), V (82), and D (84) to MIN in the antiangiogenic tumstatin peptide region. The PCR was started 40 bp into the gene sequence to delete several amino acids from the N terminus of tumstatin, which are not part of the antiangiogenic NC1 domain (2). The second set of primers was designed to amplify a sequence from 205 to 735 bp, thus creating the second half of the tumstatin molecule.
Products from the above two PCRs were used as templates for one-third PCR where the forward primer of the first PCR and the reverse primer of the second PCR were combined. This third PCR was designed to obtain a mixed pool of products containing no mutations (tumstatin, 40–735 bp) or a tumstatin MIN mutant (tumstatin-MIN, 40–735 bp). The PCR products were cloned into a PCEP-PU vector modified to contain a BM40 signal sequence and a FLAG-tag. This construct was used to transfect 293 human embryonic kidney cells. Positive clones were selected for puromycin resistance, as described in ref. 2. Supernatant collected from these cells was purified by using an anti-FLAG column and eluted with FLAG peptide. Fractions were collected, dialyzed, and concentrated for Western blot analysis and use.
The tumstatin peptide, FITC-conjugated tumstatin peptide and tumstatin peptide mutants were synthesized and purified by HPLC at the Tufts University Core Facility (Boston).
Cell Proliferation Assay.
Bovine pulmonary arterial endothelial cells (C-PAE cells) were stimulated by using VEGF and bFGF and incubated with tumstatin protein, tumstatin mutant protein, tumstatin peptide, or tumstatin peptide mutants for 48 h, after which the number of viable cells was counted. For further details, see SI Methods.
In Vitro Competitive Cell Binding of Tumstatin and Tumstatin Mutant Protein vs. FITC-Tumstatin Peptide.
C-PAE cells were preincubated with 30 μg/ml of full-length tumstatin or tumstatin mutant protein for 20 min at 37°C, before adding 30 μg/ml of FITC-tumstatin peptide to the medium and incubating for further 20 min. As a positive control, C-PAE cells were incubated with only FITC-tumstatin peptide for 20 min. The cells were thereafter fixed in acetone. Tumstatin binding was visualized by using a mouse anti-FLAG antibody (Sigma) for 1 h at room temperature, followed by a rhodamine-conjugated anti-mouse IgG secondary antibody (Jackson Immunoresearch) for 1 h at room temperature and analyzed by confocal microscopy.
Cell Attachment Assays.
C-PAE cells were preincubated with 1, 5, 10, or 50 μg/ml of polyclonal antitumstatin peptide or antitumstatin antibody, or control rabbit IgG (preimmune serum) for 15 min at room temperature, before plating them in 96-well plates precoated 2 h with tumstatin peptide (50 μg/ml) or bovine serum albumine (BSA). The cells were thereafter incubated for 45 min to allow cell attachment. The percentage of cell attachment was detected by methylene blue staining and calculated based on optical density at a wave length of 655 nm. For further details, see SI Methods.
In a separate experiment, using the same protocol as above, C-PAE cells were preincubated with 10 μg/ml of a mouse monoclonal antibody to human α2 integrin subunit (clone P1E6), αvβ3-integrin (clone LM609), or control mouse IgG for 15 min (all antibodies: Chemicon) before plating them in 96-well plates precoated overnight with 50 μg/ml of recombinant protein (tumstatin or tumstatin mutant) or synthetic peptide (tumstatin peptide or tumstatin peptide mutant). The percentage of cell attachment was determined as described above.
In Vitro Binding of Tumstatin Peptide and Dependence on Cell Proliferation Status.
C-PAE cells were grown to 40% or 100% confluency on eight-chamber slides (Lab-Tek). After preincubation with FITC-tumstatin peptide, the cells were fixed and then incubated with a monoclonal mouse anti-human αv integrin subunit (Chemicon) or a polyclonal goat anti-human VE-cadherin (Santa Cruz Biotechnology) primary antibody. Subsequently, the immunoreaction was detected by using rhodamine-conjugated secondary antibodies (Jackson Immunoresearch), and the sections were analyzed by confocal microscopy. For further details, see SI Methods.
For comparative integrin labeling of SP and C-PAE cells, the above staining procedure was employed using monoclonal mouse anti-human αv integrin subunit or αvβ3 integrin antibodies (Chemicon), hamster anti-mouse β3 integrin subunit antibody (BD PharMingen), and corresponding fluorescent IgG secondary antibodies (Jackson Laboratories).
Molecular Modeling.
The crystal structure of the [(α1)2α2]2 NC1 [Protein Data Bank (PDB) ID 1LI1] (residues 2–229 of chain A) was used to generate a homology model for tumstatin by using the program MODELLER 9v3 (27). The model with the lowest value of the object function was chosen for blind-docking simulations performed by using the fast Fourier transformation methodology as implemented in the program GRAMM (28). The homology model for tumstatin was chosen as the ligand, and the β3 domain from the crystal structure of αvβ3 integrin in complex with the RGD-peptide (PDB ID 1L5D) as the receptor.
BrdU and FITC-Tumstatin Peptide Distribution Experiments.
Animals were killed 90 and 45 min after i.v. injection of BrdU and FITC-tumstatin peptide, respectively. LLC tumors (from collagen IV-α3 (−/−) and (+/−) mice), Matrigel plugs, regenerating liver tissue and skin wound tissue was collected and processed for microscopy. For further details, see SI Methods.
Statistics.
We used the nonpaired Student's t test in comparison between the means. The Mann–Whitney test was used where a normal distribution of the data was not evident. ANOVA was used to determine statistical differences between more than two groups. A P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We wish to dedicate this manuscript to the late Dr. Judah Folkman for his constant support. We thank Drs. Akulapalli Sudhakar and Malin Sund for experimental help provided to H.S. during the course of this study. This work was primarily supported by National Institutes of Health Grant DK62987 and partially by National Institutes of Health Grants DK55001, DK61688, AA13913, and CA12550 and funds from the Department of Medicine for the Division of Matrix Biology at Beth Israel Deaconness Medical Center. H.P.E. was supported by grants from the University of Bergen, Norway; the Fulbright Association; and Helse Vest, Norway. H.S. was supported by the Stop and Shop Pediatric Brain Tumor Foundation.
Footnotes
Conflict of interest statement: Beth Israel Deaconess Medical Center has licensed the intellectual property surrounding tumstatin to EAI Corporation and is also an equity owner along with R.K.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807055105/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Maeshima Y, et al. Distinct antitumor properties of a type IV collagen domain derived from basement membrane. J Biol Chem. 2000;275:21340–21348. doi: 10.1074/jbc.M001956200. [DOI] [PubMed] [Google Scholar]
- 3.Maeshima Y, et al. Extracellular matrix-derived peptide binds to alpha (v) beta(3) integrin and inhibits angiogenesis. J Biol Chem. 2001;276:31959–31968. doi: 10.1074/jbc.M103024200. [DOI] [PubMed] [Google Scholar]
- 4.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Hamano Y, et al. Physiological levels of tumstatin, a fragment of collagen IV alpha3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via alphaV beta3 integrin. Cancer Cell. 2003;3:589–601. doi: 10.1016/s1535-6108(03)00133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeshima Y, Colorado PC, Kalluri R. Two RGD-independent alpha vbeta 3 integrin binding sites on tumstatin regulate distinct anti-tumor properties. J Biol Chem. 2000;275:23745–23750. doi: 10.1074/jbc.C000186200. [DOI] [PubMed] [Google Scholar]
- 8.Maeshima Y, et al. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science. 2002;295:140–143. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- 9.Siebold B, Deutzmann R, Kuhn K. The arrangement of intra- and intermolecular disulfide bonds in the carboxyterminal, non-collagenous aggregation and cross-linking domain of basement-membrane type IV collagen. Eur J Biochem. 1988;176:617–624. doi: 10.1111/j.1432-1033.1988.tb14321.x. [DOI] [PubMed] [Google Scholar]
- 10.Petitclerc E, et al. New functions for non-collagenous domains of human collagen type IV. Novel integrin ligands inhibiting angiogenesis and tumor growth in vivo. J Biol Chem. 2000;275:8051–8061. doi: 10.1074/jbc.275.11.8051. [DOI] [PubMed] [Google Scholar]
- 11.Xiong JP, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 12.Than ME, et al. The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link. Proc Natl Acad Sci USA. 2002;99:6607–6612. doi: 10.1073/pnas.062183499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 14.Fenn TD, Ringe D, Petsko GA. Povscript+: A program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- 15.Sund M, Zeisberg M, Kalluri R. Endogenous stimulators and inhibitors of angiogenesis in gastrointestinal cancers: Basic science to clinical application. Gastroenterology. 2005;129:2076–2091. doi: 10.1053/j.gastro.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 16.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 17.Relf M, et al. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 1997;57:963–969. [PubMed] [Google Scholar]
- 18.Miller KD, Sweeney CJ, Sledge GWJ. The Snark is a Boojum: The continuing problem of drug resistance in the antiangiogenic era. Ann Oncol. 2003;14:20–28. doi: 10.1093/annonc/mdg033. [DOI] [PubMed] [Google Scholar]
- 19.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, et al. Vascular remodeling marks tumors that recur during chronic suppression of angiogenesis. Mol Cancer Res. 2004;2:36–42. [PubMed] [Google Scholar]
- 21.Yang JC, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med. 2003;349:427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escudier B, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: A randomised, double-blind phase III trial. Lancet. 2007;370:2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 23.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 24.Batchelor TT, et al. AZD2171, a Pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iughetti P, et al. A polymorphism in endostatin, an angiogenesis inhibitor, predisposes for the development of prostatic adenocarcinoma. Cancer Res. 2001;61:7375–7378. [PubMed] [Google Scholar]
- 26.Lourenco GJ, et al. A high risk of occurrence of sporadic breast cancer in individuals with the 104NN polymorphism of the COL18A1 gene. Br Cancer Res Treat. 2006;100:335–338. doi: 10.1007/s10549-006-9259-z. [DOI] [PubMed] [Google Scholar]
- 27.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Tovchigrechko A, Vakser IA. Development and testing of an automated approach to protein docking. Proteins. 2005;60:296–301. doi: 10.1002/prot.20573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.