Abstract
Lung disease is the leading cause of morbidity and mortality in cystic fibrosis (CF) patients. A modest number of bacterial pathogens have been correlated with pulmonary function decline; however, microbiological and molecular evidence suggests that CF airway infection is polymicrobial. To obtain a more complete assessment of the microbial community composition and dynamics, we undertook a longitudinal study by using culture-independent and microbiological approaches. In the process, we demonstrated that within complex and dynamic communities, the Streptococcus milleri group (SMG) can establish chronic pulmonary infections and at the onset of 39% of acute pulmonary exacerbations, SMG is the numerically dominant pathogen. We report the comprehensive polymicrobial community dynamics of a CF lung infection in a clinically relevant context. If a given organism, such as Pseudomonas aeruginosa, becomes resistant to antibiotic therapy, an alternative treatment avenue may mediate the desired clinical response by effectively managing the composition of the microbial community.
Keywords: Streptococcus milleri, oropharyngeal flora, normal microbiota, microbial communities
Pulmonary disease remains the leading cause of morbidity and mortality in cystic fibrosis (CF) (1). In CF, defective mucociliary clearance and impaired innate immunity lead to chronic pulmonary infections (2–4). CF airway disease is characterized by periods of stability punctuated by acute exacerbations in which overt immunological responses cause the majority of irreversible lung damage. Aside from respiratory viruses, which have been associated with up to one-third of pulmonary exacerbations (5, 6), the factors that lead to acute pulmonary exacerbation remain elusive. A small number of bacterial pathogens have been implicated in CF lung disease (7), with Pseudomonas aeruginosa being the most common, colonizing 80% of patients by early adulthood (8). However, both microbiological and molecular techniques have defined CF lower airway infections as polymicrobial (9–15).
In contrast to traditional microbial cultivation, culture-independent approaches offer a more complete view of lower airway microbiology (15). Terminal Restriction Fragment Length Polymorphism (T-RFLP) analysis is a common culture-independent technique for profiling polymicrobial communities. In adult CF patients, an average of 13.4 (±6.7) bacterial species are detectable by T-RFLP (11). Comprehensive microbial cultivation techniques substantiate that CF endobronchial secretions are complex polymicrobial specimens (13). However, the identity and clinical relevance of the majority of the bacterial species in CF airways remain enigmatic. It is clear that the reported microbial diversity in CF lower airways is not the consequence of sputum contamination by bacteria during passage through the oral cavity (12). Microbial communities in CF have been characterized from multiple patients at single time-points (cross-sectional studies), and perspectives on how the community structure changes within patients over time (longitudinal studies) have been limited.
In considering the polymicrobial nature of CF lung disease, the viridans streptococci are of interest because of their capacity to enhance P. aeruginosa pathogenicity by modulating virulence factor gene expression in the principal pathogen (13). Within the viridans streptococci, S. constellatus, S. intermedius, and S. anginosus (16, 17) are collectively referred to as the Streptococcus milleri group (SMG). The SMG are phenotypically diverse (16, 18, 19), however, they are grouped together based on molecular typing and some common biochemical properties (20). Clinically, the SMG are distinguished by their predilection for severe purulent infections (characterized by the accumulation of pus) (21–23). They are the etiologic agent in a number of life-threatening infections, including abscesses of the brain, liver, lung, and empyema (accumulation of pus in the pleural space) (23–32). SMG infections are thought to result from dissemination from local sites where the SMG are communal members of the microbiota (22, 33–35). They asymptomatically colonize the mouth, nasopharynx, gastro-intestinal tract, and genitourinary tract of 15–30% of the population (19). Although the SMG are a common cause of empyema (17, 36–38) and occasionally fatal community-acquired pneumonia (23, 28), its role in pulmonary infections remains largely unrecognized (17, 29). The confusion surrounding streptococcal taxonomy, the inability of the clinical microbiology laboratory to readily culture, identify, and enumerate SMG organisms, and their association with commensal microbiota have created an environment whereby the medical significance of these organisms is easily overlooked.
To determine whether the complete microbial community dynamics are relevant to the clinical course of CF lung disease, we investigated individual CF patients by using molecular (T-RFLP) and microbiological methods during and between periods of pulmonary exacerbations. The culture-independent approach revealed that, at the onset of acute pulmonary exacerbations, the community structure was highly similar and distinct from periods of clinical stability. The community profiling suggested that the SMG have the capacity to trigger pulmonary exacerbations. We confirmed that the clinical course of disease correlated to perturbations in the SMG population by developing a cultivation medium (McKay agar) to selectively follow the SMG population. At the onset of exacerbations, the SMG are the numerically dominant microorganism although not detected by standard sputum culture methods. McKay agar-culture surveillance revealed that SMG represents a very significant burden of disease in adult CF with 7 of 18 of hospitalizations over a 6-month period attributable to SMG. Antimicrobial therapy directed at the SMG proved more effective than anti-Pseudomonal therapy in these cases, and the clinical resolution of these exacerbations correlated with a decrease in SMG. We propose that the short list of clinically relevant bacterial species should be expanded to include the SMG and, furthermore, that the dynamic microbiome within CF airways must be considered to effectively guide therapy in the individual patient.
Results
Population Dynamics of the Microflora in CF Airways.
Culture-independent approaches provide a comprehensive perspective of the polymicrobial nature of CF airways. Bypassing the requirement for microbial cultivation allows for the identification of organisms unable to grow by using standard culture conditions (such as anaerobes) or organisms obscured because of overgrowth of dominant community members (or fast growers) on standard selective mediums. To define the polymicrobial nature of CF airways, we chose to use T-RFLP as a means to follow the community structure in an individual CF patient (patient #1). T-RFLP was performed on prospectively collected sputum samples throughout an entire year, during periods of clinical stability that were punctuated by three pulmonary exacerbations requiring hospitalization. The analysis also included eight retrospective sputum samples dating back almost 5 years, two of which were collected at the time of hospital admission (Fig. 1).
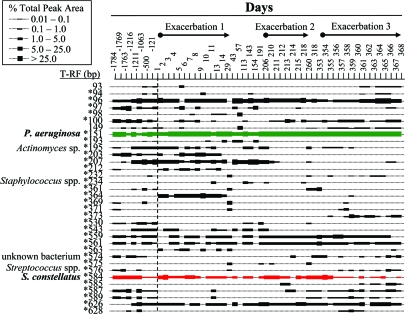
Fig. 1.
T-RFLP analysis of the microbial communities in sputum samples collected at admission to hospital for the first exacerbation and during the following year (days 1 to 368) and 5 retrospective samples (days −1784 to −121). The retrospective samples are shown left of the dashed line. The size in base pairs of each of the T-RFs detected during the 6-year period is shown on the left. * indicates T-RFs whereby bacterial species can be assigned by using in silico analysis. The abundance of each T-RF is plotted by using the legend. The names of the bacterial species cultivated from sputum are shown beside the T-RF for which they correspond. Green and red are the P. aeruginosa and S. constellatus T-RFs, respectively.
The patient was known to be chronically colonized by P. aeruginosa. A 151-bp terminal restriction fragment (T-RF) corresponding to P. aeruginosa represented 49% of the total collective area under all of the T-RF peaks from the complete 6-year T-RFLP dataset (54 total sputum samples). The other 51% of the total peak area could be organized into 91 different T-RFs [supporting information (SI) Fig. S1]. Thirty-four T-RFs were detected in 4 or more samples during the longitudinal analysis and present at levels >1% of the total peak area in at least 1 sample (Fig. 1). The T-RFs that did not satisfy the inclusion criteria are shown in Fig. S2. The composition of the microbial community is relatively simple compared with other microbial communities in the human body, such as the oral cavity and gastrointestinal and urogenital tracts. On average, 15 ± 5 unique T-RFs were detected from each sputum sample. Although T-RFLP has a number of caveats, it is a reliable tool for detecting compositional changes in communities of this complexity (39). Of the T-RFs detected in multiple samples, some were consistently present (for example, P. aeruginosa), and the presence of others fluctuated (detection was separated by months or even years) suggesting the presence of a highly dynamic population of microbes throughout the 6-year period (Fig. 1).
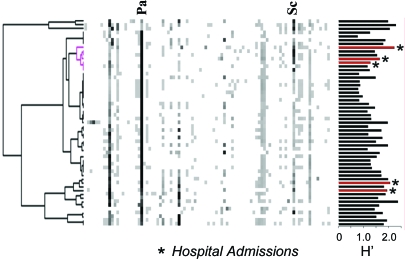
The rational for using a molecular approach to study CF microbiology was that it allowed us to determine whether the dynamics of the complete microbial consortia (represented by all of the detectable microbes) correlated with the clinical course of disease. To test the hypothesis that community structure and clinical status are interrelated, cluster analysis was used to statistically assess the T-RFLP profiles. This analysis indicated that the community composition at the time of hospital admission for the second and third exacerbation were highly similar (Fig. 2). Interestingly, the structure of these microbial communities was also very similar in composition to a retrospective hospital admission sample collected more than 5 years earlier. The clustering does not appear to be because of the diversity of the community (as measured by Shannon's diversity index) (40). Strikingly, the cluster of hospital admissions seems to be, in part, because of the abundance T-RF detected at 584 bp. We also observed an additional cluster representing the two additional hospital admission samples in the dataset (day 1 and day −1216) (Fig. 2). The relatedness among the composition of microbial communities at the onset of exacerbations suggests that community changes in CF airways may contribute to the transition from a chronic stable infection to an acute exacerbation infection.
Fig. 2.
Cluster analysis of longitudinal T-RFLP samples from Fig. 1. The calculated Shannon's diversity indices (H′) are plotted next to the corresponding T-RFLP profile. Those from sputums collected at the onset of pulmonary exacerbation are shown in red. The pink cluster highlights the relatedness of the microbial communities present upon admission to hospital.
Identification of the SMG as a Cause of Pulmonary Exacerbations in CF.
The strategy we used to identify organisms discerned with our culture-independent approach was to culture, identify (with 16S rRNA gene sequence), and experimentally determine the T-RFs for all differential colony types cultivated from patient #1 during the prospective year. Because of limitations associated with in silico-based microbial identification (39), we did not assign T-RF identities based on predictions. Using our approach, we could only assign a bacterial species to 13 of the 91 T-RFs (14.3%) (Fig. 1). Although we isolated 17 different organisms, 5 generated T-RFs identical to sizes of other cultured organisms. For example, Streptococcus sanguinis, Streptococcus parasanguinis, and Streptococcus salivarius all generate a 576-bp T-RF fragment.
The first microbiological workup from patient #1 was conducted at the time of admission to the hospital for treatment of an acute pulmonary exacerbation (day 1). Sputum samples were collected at admission, before antimicrobial therapy and daily throughout the hospitalization. Sputum was cultured on standard solid media recommended for CF microbiology (MacConkey, Chocolate, Columbia 5% Sheep Blood, and Mannitol-Salts) and nonselective media [Brain Heart Infusion (BHI) and Trypticase Soy supplemented with Yeast extract (TSY)]. A numerically dominant organism (50-fold greater colony forming units than P. aeruginosa) was identified on BHI and TSY but notably absent on Columbia 5% Sheep Blood agar (CBA) plates (Fig. 3A). The organism was identified by 16S rRNA sequence as S. constellatus, a member of the SMG, and corresponds to the highly abundant 584-bp T-RF identified in the longitudinal study (Fig. 1), which was prevalent in the two clusters of hospital admission sputums (Fig. 2). Fig. 3B summarizes the T-RFLP data for S. constellatus during this first period of hospitalization.
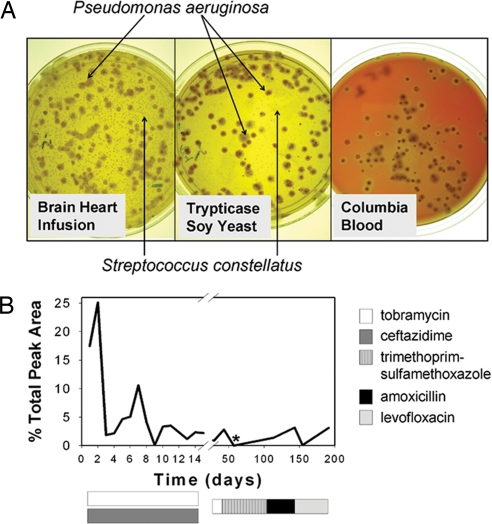
Fig. 3.
Detection of SMG by culture-dependent and culture-independent approaches. (A) Microbial cultivation of admission sputum in patient #1, pulmonary exacerbation 1, revealed that S. constellatus was the numerically dominant organism on BHI agar and TSY agar. CBA failed to grow S. constellatus (10−5 dilution plates shown) after 5 d at 37°C in 5% CO2. (B) The relative abundance of the 584-bp T-RF corresponding to S. contellatus is shown throughout patient #1's first pulmonary exacerbation (day 1–14) and period of clinical stability after discharge from hospital (day 15–200). * indicates the failure to detect S. constellatus by BHI culture.
Intravenous antibiotic treatment with tobramycin and ceftazidime was initiated on admittance. The patient remained hospitalized for 15 days and achieved only modest clinical remission. Inhaled tobramycin was continued for 1 month because the patient had not returned to baseline functional status. We detected S. constellatus in both samples produced during this time by using BHI culture and by T-RFLP (days 29 and 43) (Fig. 3B). The anti-Pseudomonal treatment was discontinued because of the limited clinical success and therapy, trimethoprim-sulfamethoxazole (TMP-SMX), directed at the S. constellatus was initiated. After 2 weeks, the patient reported feeling well for the first time since the onset of the exacerbation. No S. constellatus was detected by culture or by T-RFLP (day 57) (Fig. 3B).
The patient remained stable for more than a month during the TMP-SMX treatment, but on day 113, S. constellatus was the near numerically dominant organism by sputum culture and was detected by T-RFLP (day 113) (Fig. 3B). The lack of efficacy of TMP-SMX in suppressing S. constellatus suggested that resistance may have developed, which we later confirmed (Table S1). Treatment with amoxicillin and levofloxacin was modestly effective at reducing the S. constellatus, as indicated by plating and T-RFLP (day 154) (Fig. 3B). However, the limitations of the BHI cultivation practices for S. constellatus (the issue of P. aeruginosa overgrowth) prompted us to develop a selective medium for the identification and enumeration of SMG in sputum.
The use of selective mediums in the clinical laboratory for the isolation of respiratory pathogens from sputum is the foundational basis for guiding antibiotic therapy. The failure of the S. constellatus isolate to be cultured on CBA agar, the solid medium currently used for identification of such organisms, suggested that a proportion of SMG isolates may be escaping detection by using the current protocol for CF microbiology. To address this possibility, we developed a complex solid medium for the isolation of SMG, McKay Agar. With a push toward developing molecular diagnostics and culture-independent technologies, little has been done to change the traditional microbiologic practices for CF. McKay agar represents a medium for the identification of overlooked pathogens in CF. It is supplemented with colistin, oxolinic acid, and sulfadiazine to prevent the growth of principal CF pathogens (e.g., P. aeruginosa and S. aureus). Bromcresol purple is included as a colorimetric indicator to aid in distinguishing SMG colonies from other oropharyngeal isolates because SMG produce acid when grown on this medium. We validated McKay agar against a panel of SMG strains and observed that all SMG strains cultured with equal efficiencies on BHI and McKay but with varying efficiencies on CBA (unpublished data).
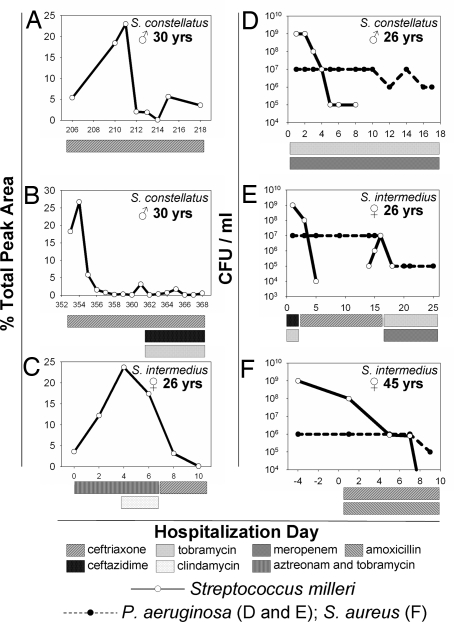
On day 206, patient #1 was admitted with a second pulmonary exacerbation. Sputum collected before antibiotic therapy grew S. constellatus as the numerically dominant organism by McKay agar culture (Fig. S3). The patient was treated with i.v. ceftriaxone, a drug with no anti-Pseudomonal activity. S. constellatus responded to antibiotic treatment, the patient's clinical symptoms resolved, and lung function returned to baseline. SMG was no longer detectable by sputum cultivation within 6 d, and the relative proportion of the T-RF corresponding to S. constellatus began to decrease after 5 d of therapy (Fig. 4A). On discharge, the patient was prescribed doxycycline as a prophylactic strategy to prevent relapse infection with S. constellatus. The utility of McKay agar culture was highlighted during this therapy because the SMG detection limit increased 100-fold, revealing S. constellatus at levels by quantitative sputum culture that where not measurable on all other alternative media, including BHI because of P. aeruginosa overgrowth.
Fig. 4.
Examples of pulmonary exacerbations caused by the SMG. The relative abundance of the T-RF corresponding to the S. constellatus during patient #1's second (A) and third (B) pulmonary exacerbation and the S. intermedius exacerbation described in detail in the text (C) are shown. (D–F) Additional examples of cases where McKay agar culture detected SMG as the numerically dominant pathogen at the time of hospital admission (day 1). The SMG species identified in each exacerbation and the patients' sex and age are shown in the top right of each image. Antibiotic therapy is described in the legend. SMG levels are always shown with a bold solid line. Exacerbations (A–E) were also associated with P. aeruginosa (bold dashed line) and exacerbation (F) was associated with S. aureus.
The patient had a third pulmonary exacerbation on day 353, and again S. constellatus was detected as the numerically dominant organism by McKay culture. Intravenous ceftriaxone was administered during the first 8 d of hospitalization. S. constellatus levels rapidly fell below the P. aeruginosa levels, and the S. constellatus T-RF rapidly decreased in relative abundance (Fig. 4B). Ceftriaxone treatment was continued for an additional 7 d along with ceftazidime and tobramycin. During this time, S. constellatus was only sporadically detectable by T-RFLP (Fig. 4B). The antibiotic treatment regimen resulted in the resolution of acute exacerbation symptoms.
Throughout the year, P. aeruginosa populations remained stable as measured by quantitative microbiology and T-RFLP despite aggressive anti-Pseudomonal therapy at various times (Figs. 3B and 4 A and B). S. constellatus levels, however, reflected the disease state. The organism was the numerically dominant organism only at the onset of each exacerbation, and resolution of symptoms correlated with a decrease in numbers. Microbiological data on this patient supports the T-RFLP results (41).
To further support our hypothesis that pulmonary exacerbation was triggered by changes in the microbial community structure resulting in a resurgence of the resident SMG population, we used pulsed-field gel electrophoresis (PFGE) to confirm that the S. constellatus isolates from patient #1 shared a common lineage. We subjected 4 isolates separated by 354 days to PFGE. All four isolates represented the numerically dominant organisms cultured upon admission to hospital, and the isolate that emerged with TMP-SMX resistance produced identical PFGE profiles (Fig. S4). This result confirms that the S. constellatus population dynamics seen in patient #1 were because of fluctuations in levels of a single chronically colonized S. constellatus strain.
SMG Behaves as Pathogens in Other CF Patients.
After the results obtained with patient #1, we investigated whether the SMG were escaping conventional identification in other patients. Remarkably, in the next CF patient admitted for an acute pulmonary exacerbation, the numerically dominant organism grew on McKay agar and was identified as S. intermedius, another SMG. Once again, the organism was not detected by using conventional CF sputum microbiology protocols. It took 4 d from the time of admission to confirm the identity of the organism, during which time she was treated empirically for P. aeruginosa with i.v. aztreonam and tobramycin. She did not clinically respond to the treatment and required supplemental oxygen. After the identification of S. intermedius, clindamycin was added to her treatment regimen, but after 3 d, there was no symptomatic improvement, and S. intermedius remained the numerically dominant organism in her sputum. We subsequently confirmed clindamycin resistance (data not shown). Ceftriaxone treatment was started, and the clinical response was immediate; symptoms subsided, pulmonary function improved, and the patient was discharged from hospital after only 5 d of treatment. The resolution of the acute exacerbation symptoms correlated with a >1,000-fold drop in S. intermedius numbers and no change in P. aeruginosa bacterial load (41). T-RFLP analysis revealed an increase in the relative abundance of the 584-bp T-RF, corresponding to S. intermedius, during treatment with aztreonam, tobramycin, and clindamycin. This may reflect S. intermedius becoming a more dominant member of the community as other susceptible organisms are removed. After initiating ceftriaxone treatment, the percent total peak area of the S. intermedius T-RF dropped 100-fold (Fig. 4C).
Further culture surveillance with McKay agar revealed that a high proportion of hospital admissions in our adult patient population are attributable to SMG. In addition to the exacerbations described in detail, 7 of 18 acute pulmonary exacerbations in our adult population over a 6-month period were associated with SMG (3 are shown in Fig. 4 D–F). All of these SMG strains were missed by using conventional approaches.
Discussion
We followed the microbial dynamics during and between pulmonary exacerbations in CF by using both molecular and culture-based approaches. We demonstrated that a comprehensive perspective on the microbial populations present in sputum can explain the clinical course of lung disease in these patients. When the SMG became a dominant member of the microbial community, clinical intervention was required. Treating the SMG as primary pathogens resulted in effective resolution of pulmonary exacerbations and a return to clinical stability. We have recently had similar findings with two cases of idiopathic chronic bronchiectasis exacerbations unrelated to CF. In these cases, no pathogen was detected by standard clinical microbiology, but McKay agar detected significant levels of SMG. Moreover, SMG-directed therapy resolved the symptoms (unpublished data). These results suggest that the SMG may represent a more significant respiratory pathogen than recognized. We recently undertook a retrospective review of the impact of SMG on CF, which revealed that the SMG is a significant cause of additional pulmonary exacerbations and invasive disease (41). There has been only one brief report of SMG in CF lung disease (42). SMG were also identified by Harris et al. (by a culture-independent approach because the SMG failed detection by routine culture) (15) and Tunney et al. (14), but these studies did not correlate the microbiology with the clinical course of disease.
The absence of any further reported cases of SMG pulmonary infections in CF is likely not because it is uncommon, but rather because SMG organisms frequently do not grow on traditional laboratory media and, if grown, are often dismissed as clinically irrelevant normal microbiota. The fastidious nature of SMG cultivation is highlighted by the failure to grow half of all SMG isolates for antibiotic susceptibility testing (24, 43, 44). McKay agar offers a solution to this problem and provides superior recovery efficiencies over blood agar culture, which is of utmost importance when culturing SMG from nonsterile specimens such as sputum.
The mechanisms of SMG pathogenicity are not well understood. SMG strains can produce hyaluronidase, DNase, ribonuclease, chondroitin sulfatase, gelatinase, and collagenase, all enzymes that may contribute to tissue disruption and abscess formation (45–50). It is worthwhile to consider that the mechanisms of pathogenicity might rely on the polymicrobial context of CF airways. It is common to isolate the SMG from mixed infections with strict anaerobes (23, 32, 51, 52) or with P. aeruginosa (23, 32, 52). In a mouse model of acute pneumonia, polymicrobial infections combining S. constellatus with Prevotella resulted in a 6-fold increase in mortality relative to either organism alone (29). Strikingly, several of the patients described in this work have a T-RF of the predicted size of Prevotella spp. The retrospective T-RFLP analysis indicates that this T-RF, along with the SMG T-RF, has been present for the last 6 years in patient #1. Given that certain CF oropharyngeal (OF) species have the capacity to modulate P. aeruginosa gene expression through microbe-microbe interactions (13), it will be interesting to determine whether there is a role for SMG–P. aeruginosa interactions in promoting pulmonary exacerbations.
To date, we have isolated all three species of SMG from patients in our adult CF patient population with no evidence for a particular epidemic strain. We are continuing our SMG surveillance in the adult population and are expanding to include the pediatric CF population and patients with idiopathic chronic bronchiectasis. To further our understanding of SMG pathogenesis, we are sequencing the genomes of CF and invasive isolates of S. constellatus, S. intermedius, and S. anginosus.
The life expectancy of CF patients continues to increase because of the benefits of current multifaceted clinical interventions. The consequence this increased longevity has on the microbiology of CF airways has not been established. It remains a possibility that the SMG have gone unrecognized in CF lung disease, not only because of culture and identification inadequacies, but because this group of pathogens has emerged as the result of changes in the CF patient demographics. P. aeruginosa adaptation during chronic colonization in CF has been associated with a loss of virulence determinants (53). This is seemingly contradictory to the increasing exacerbation frequency and decline in lung function in these patients. Our results suggest that other pathogens, such as SMG, may play a significant role as pathogens in the adult CF population. The description of SMG with pathogenic relevance to CF lung disease may be but one of many examples of how the OF can serve as a reservoir to elusive pathogens. Why is it so common for CF patients to clinically improve during antibiotic therapy when there are no indications of a bacteriologic response in the perceived principal pathogens? The dynamic nature of the OF population is a plausible explanation. During antibiotic treatment, changes in the composition of the OF communities are currently not evaluated. The removal of OF species during antibiotic treatment that behave as true pathogens, or that mediate their pathogenic effects through microbe-microbe interactions, may be the mechanism for the resolution of pulmonary exacerbations in such patients. Advancing the understanding of the role of polymicrobial dynamics in CF lung disease provides the clinician with alternate therapeutic targets. If a given organism, such as P. aeruginosa, becomes resistant to antibiotic therapy, an alternative treatment avenue may mediate the desired clinical response by effectively managing the composition of the microbial community in such a way that removes bacterial species responsible for exacerbating the state of a principal pathogen.
Materials and Methods
Microbiology Techniques.
Sputum samples were collected from patients in accordance with ethical guidelines in sterile containers (retrospective samples were stored at −80°C). Sputum was sheared by passage through a 1-cc syringe (without a needle) and serially diluted in Brain Heart Infusion Broth (BD), and 100 μl of the 10−3 and 10−5 dilutions were cultured on standard media (BD): MacConkey agar, Pseudomonas Isolation agar, Mannitol Salt agar, Chocolate agar, CBA, BHI agar, and TSY agar (Trypticase Soy agar with 3 g/L yeast extract). Plates were incubated at 37°C with 5% CO2 for a minimum of 5 d. Individual colonies were purified by serial passage three times. Bacterial strains were identified by PCR amplification and sequencing of part of the 16S rRNA gene by using primers 8f and 926r (54). Identification to the species level corresponds to >97% identity to the closest match in the RDP database (http://rdp.cme.msu.edu/).
Susceptibilities to antibiotics were determined by modified Kirby-Bauer disk diffusion methods according to the Clinical Laboratory Standards Institute, formerly National Committee on Clinical Laboratory Standards guidelines (55).
Pulsed-field gel electrophoresis was performed as described (56) by using ApaI. Fragments were separated on a Chef Mapper Electrophoresis Cell (Bio-Rad) in 0.5X TBE at 4°C and 6 V/cm with switch times ramped from 5 to 35 seconds.
McKay Agar.
One liter of McKay agar was made by combining the following ingredients and pH adjusting to 7.2 before autoclave sterilization: 13.3 g Nutrient Broth (BD), 5 g dextrose, 10 g Yeast extract (Difco), 5 g Tryptone (BD), 2 g K2HPO4, 40 ml salt solution (NaHCO3 10 g/L, NaCl 2 g/L, K2HP04 1 g/L, KH2PO4 1g/L, MgSO4·7H20 0.5 g/L, and CaCl2·2 H20 0.25 g/L), 1 ml Tween 80, 1 mg Crystal Violet, 60 mg Bromcresol Purple, 10 μg Vitamin K, 0.05 g Hemin, and 15 g/L Bacto Agar (Difco). Sterilized media was supplemented with 20 ml l-arginine (2.5% w/vol), sulfadiazine, colistin sulfate, and oxolinic acid, added to a final concentration of 500 μg/ml, 10 μg/ml, and 5 μg/ml, respectively.
Terminal Restriction Fragment Polymorphism Analysis.
Total DNA was prepared from sputum by using a Bead Beater (Biospec) as described (11) without addition of Sputasol. A total of 20 ng of DNA was used as template in a PCR with primers 8f (5 ′-AGAGTTTGATCCTGGCTCAG-3 ′) labeled with 6FAM (ABI Biosciences) and 926r (5 ′-CCGTCAATTCCTTTRAFTTT-3 ′) (54) by using described reaction conditions (11). Every sputum sample was subjected to three independent PCRs, and the resulting products were pooled and purified by using a DNA Clean and Concentrator 5 column (Zymo Research). Two hundred nanograms of each purified PCR product was digested with 20 units of CfoI (Roche), according to the manufacturer's instructions, at 37°C for at least 6 h. Approximately 5 ng of digested PCR product were injected into an ABI 3730 Genetic Analyzer, and fragment analysis was done by using the GeneMapper software package (ABI Biosystems); LIZ1200 (ABI Biosystems) was used as a size standard. The total area under the peak for a given T-RFLP profile was totaled, and each T-RF was represented as a percentage of the total peak area (57). T-RFs were assigned to corresponding organisms cultivated from the sputum samples by performing the fragment analysis protocol on 16S amplified from purified colonies. The in silico analysis method for T-RF prediction has been described (58). Shannon's diversity index was calculated as described (40). T-RFLP profiles were clustered by using Cluster and visualized with TreeView (59).
Supplementary Material
Acknowledgments.
We thank the CF patients for their invaluable contributions and the Southern Alberta Cystic Fibrosis Clinic for patient care and participation in sample sputum sample gathering. This work was supported by a Canadian Cystic Fibrosis Foundation grant (to M.G.S.). M.G.S. is supported as an Alberta Heritage Foundation for Medical Research Scientist and Canada Research Chair in Microbial Gene Expression. C.D.S. is supported by an Alberta Heritage Foundation for Medical Research studentship and a Canada Graduate Scholarship from Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804326105/DCSupplemental.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsui H, et al. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell. 1998;95:1005–1015. doi: 10.1016/s0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 3.Goldman MJ, et al. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 4.Smith JJ, Travis SM, Greenberg EP, Welsh MJ. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 5.Ong EL, et al. Infective respiratory exacerbations in young adults with cystic fibrosis: Role of viruses and atypical microorganisms. Thorax. 1989;44:739–742. doi: 10.1136/thx.44.9.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hordvik NL, et al. Effects of acute viral respiratory tract infections in patients with cystic fibrosis. Pediatr Pulmonol. 1989;7:217–222. doi: 10.1002/ppul.1950070406. [DOI] [PubMed] [Google Scholar]
- 7.Gilligan PH, Kiska DL, Appleman MD. Cumitech: Cystic fibrosis microbiology. In: Appleman MD, editor. Vol 43. Washington, D.C.: Am Soc Microbiol; 2006. [Google Scholar]
- 8.Rajan S, Saiman L. Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect. 2002;17:47–56. doi: 10.1053/srin.2002.31690. [DOI] [PubMed] [Google Scholar]
- 9.Sibley CD, Rabin H, Surette MG. Cystic fibrosis: A polymicrobial infectious disease. Future Microbiology. 2006;1:53–61. doi: 10.2217/17460913.1.1.53. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GB, et al. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2003;41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers GB, et al. characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers GB, et al. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J Clin Microbiol. 2006;44:2601–2604. doi: 10.1128/JCM.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 14.Tunney MM, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 15.Harris JK, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiley RA, Fraser H, Hardie JM, Beighton D. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group.”. J Clin Microbiol. 1990;28:1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiley RA, Beighton D, Winstanley TG, Fraser HY, Hardie JM. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): Association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243–244. doi: 10.1128/jcm.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ball LC, Parker MT. The cultural and biochemical characters of Streptococcus milleri strains isolated from human sources. J Hyg (Lond) 1979;82:63–78. doi: 10.1017/s002217240002547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poole PM, Wilson G. Occurrence and cultural features of Streptococcus milleri in various body sites. J Clin Pathol. 1979;32:764–768. doi: 10.1136/jcp.32.8.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruoff KL. Streptococcus anginosus (“Streptococcus milleri”): The unrecognized pathogen. Clin Microbiol Rev. 1988;1:102–108. doi: 10.1128/cmr.1.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edmiston CE, Jr, et al. Streptococcus milleri group (Streptococcus anginosus): Recovery from intra-abdominal and soft tissue sites. Ann Clin Lab Sci. 1991;21:56–61. [PubMed] [Google Scholar]
- 22.Parker MT, Ball LC. Streptococci and aerococci associated with systemic infection in man. J Med Microbiol. 1976;9:275–302. doi: 10.1099/00222615-9-3-275. [DOI] [PubMed] [Google Scholar]
- 23.Porta G, et al. Thoracic infection caused by Streptococcus milleri. Eur Respir J. 1998;12:357–362. doi: 10.1183/09031936.98.12020357. [DOI] [PubMed] [Google Scholar]
- 24.Belko J, Goldmann DA, Macone A, Zaidi AK. Clinically significant infections with organisms of the Streptococcus milleri group. Pediatr Infect Dis J. 2002;21:715–723. doi: 10.1097/00006454-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Chua D, Reinhart HH, Sobel JD. Liver abscess caused by Streptococcus milleri. Rev Infect Dis. 1989;11:197–202. doi: 10.1093/clinids/11.2.197. [DOI] [PubMed] [Google Scholar]
- 26.Gossling J. Occurrence and pathogenicity of the Streptococcus milleri group. Rev Infect Dis. 1988;10:257–285. doi: 10.1093/clinids/10.2.257. [DOI] [PubMed] [Google Scholar]
- 27.Hardwick RH, Taylor A, Thompson MH, Jones E, Roe AM. Association between Streptococcus milleri and abscess formation after appendicitis. Ann R Coll Surg Engl. 2000;82:24–26. [PMC free article] [PubMed] [Google Scholar]
- 28.Mirick GS, Thomas I, Curen EC, Horsfall FL. Studies of non-haemolytic streptococcus isolated from the respiratory tract of human beings. J Exp Med. 1944;80:391–440. doi: 10.1084/jem.80.5.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinzato T, Saito A. The Streptococcus milleri group as a cause of pulmonary infections. Clin Infect Dis 21 Suppl. 1995;3:S238–S243. doi: 10.1093/clind/21.supplement_3.s238. [DOI] [PubMed] [Google Scholar]
- 30.Velghe A, Van den NN, Janssens W, Smeets P, Vogelaers D. Streptococcus milleri-sepsis with lung and brain abscesses. Acta Clin Belg. 2004;59:369–372. doi: 10.1179/acb.2004.054. [DOI] [PubMed] [Google Scholar]
- 31.Molina JM, et al. Clinical and bacterial features of infections caused by Streptococcus milleri. Scand J Infect Dis. 1991;23:659–666. doi: 10.3109/00365549109024289. [DOI] [PubMed] [Google Scholar]
- 32.Van der AP. Clinical significance of Streptococcus milleri. Eur J Clin Microbiol. 1985;4:386–390. doi: 10.1007/BF02148688. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs JA, Pietersen HG, Stobberingh EE, Soeters PB. Bacteremia involving the “Streptococcus milleri” group: Analysis of 19 cases. Clin Infect Dis. 1994;19:704–713. doi: 10.1093/clinids/19.4.704. [DOI] [PubMed] [Google Scholar]
- 34.Murray HW, Gross KC, Masur H, Roberts RB. Serious infections caused by Streptococcus milleri. Am J Med. 1978;64:759–764. doi: 10.1016/0002-9343(78)90514-4. [DOI] [PubMed] [Google Scholar]
- 35.Salavert M, et al. Seven-year review of bacteremia caused by Streptococcus milleri and other viridans streptococci. Eur J Clin Microbiol Infect Dis. 1996;15:365–371. doi: 10.1007/BF01690091. [DOI] [PubMed] [Google Scholar]
- 36.Clarridge JE, III, Osting C, Jalali M, Osborne J, Waddington M. Genotypic and phenotypic characterization of “Streptococcus milleri” group isolates from a Veterans Administration hospital population. J Clin Microbiol. 1999;37:3681–3687. doi: 10.1128/jcm.37.11.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto N, et al. Trends in antimicrobial susceptibility of the Streptococcus milleri group. J Infect Chemother. 2002;8:134–137. doi: 10.1007/s101560200023. [DOI] [PubMed] [Google Scholar]
- 38.Maskell NA, et al. U.K. Controlled trial of intrapleural streptokinase for pleural infection. N Engl J Med. 2005;352:865–874. doi: 10.1056/NEJMoa042473. [DOI] [PubMed] [Google Scholar]
- 39.Hartmann M, Widmer F. Reliability for detecting composition and changes of microbial communities by T-RFLP genetic profiling. FEMS Microbiol Ecol. 2008;63:249–260. doi: 10.1111/j.1574-6941.2007.00427.x. [DOI] [PubMed] [Google Scholar]
- 40.Magurran AE. Ecological Diversity and Its Measurements. Princeton: Princeton Univ Press; 1988. p. 179. [Google Scholar]
- 41.Parkins MD, Sibley CD, Surette MG, Rabin HR. The Streptococcus milleri group - An unrecognized cause of disease in cystic fibrosis: A case series and literature review. Pediatr Pulmonol. 2008;43(5):490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 42.Cade A, Denton M, Brownlee KG, Todd N, Conway SP. Acute bronchopulmonary infection due to Streptococcus milleri in a child with cystic fibrosis. Arch Dis Child. 1999;80:278–279. doi: 10.1136/adc.80.3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ball JL, Malhotra RM, Leong P, Bacon AS. The importance of recognizing Streptococcus milleri as a cause of orbital cellulites. Eye. 2000;14(5):814–815. doi: 10.1038/eye.2000.226. [DOI] [PubMed] [Google Scholar]
- 44.Bourgault AM, Wilson WR, Washington JA. Antimicrobial susceptibilities of species of viridans streptococci. J Infect Dis. 1979;140:316–321. doi: 10.1093/infdis/140.3.316. [DOI] [PubMed] [Google Scholar]
- 45.Jacobs JA, Stobberingh EE. Hydrolytic enzymes of Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius in relation to infection. Eur J Clin Microbiol Infect Dis. 1995;14:818–820. doi: 10.1007/BF01691002. [DOI] [PubMed] [Google Scholar]
- 46.Shain H, Homer KA, Beighton D. Degradation and utilization of chondroitin sulphate by Streptococcus intermedius. J Med Microbiol. 1996;44:372–380. doi: 10.1099/00222615-44-5-372. [DOI] [PubMed] [Google Scholar]
- 47.Unsworth PF. Hyaluronidase production in Streptococcus milleri in relation to infection. J Clin Pathol. 1989;42:506–510. doi: 10.1136/jcp.42.5.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pulliam L, Porschen RK, Hadley WK. Biochemical properties of CO2-dependent streptococci. J Clin Microbiol. 1980;12:27–31. doi: 10.1128/jcm.12.1.27-31.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall R, Kaufman AK. Production of deoxyribonuclease, ribonuclease, coagulase, and hemolysins by anaerobic gram-positive cocci. J Clin Microbiol. 1981;13:787–788. doi: 10.1128/jcm.13.4.787-788.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steffen EK, Hentges DJ. Hydrolytic enzymes of anaerobic bacteria isolated from human infections. J Clin Microbiol. 1981;14:153–156. doi: 10.1128/jcm.14.2.153-156.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piscitelli SC, Shwed J, Schreckenberger P, Danziger LH. Streptococcus milleri group: Renewed interest in an elusive pathogen. Eur J Clin Microbiol Infect Dis. 1992;11:491–498. doi: 10.1007/BF01960802. [DOI] [PubMed] [Google Scholar]
- 52.Han JK, Kerschner JE. Streptococcus milleri: An organism for head and neck infections and abscess. Arch Otolaryngol Head Neck Surg. 2001;127:650–654. doi: 10.1001/archotol.127.6.650. [DOI] [PubMed] [Google Scholar]
- 53.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Committee for Clinical Laboratory Standards. Wayne, PA: NCCLS; 2000. Methods for dilution antimicrobial susceptibility testing for bacteria that grow aerobically. [Google Scholar]
- 56.Bartie KL, Wilson MJ, Williams DW, Lewis MA. Macrorestriction fingerprinting of “Streptococcus milleri” group bacteria by pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:2141–2149. doi: 10.1128/jcm.38.6.2141-2149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers GB, et al. Bacterial activity in cystic fibrosis lung infections. Respir Res. 2005;6:49. doi: 10.1186/1465-9921-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent AD, Smith DJ, Benson BJ, Triplett EW. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl Environ Microbiol. 2003;69:6768–6776. doi: 10.1128/AEM.69.11.6768-6776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.