Abstract
Phosphoinositide signal transduction pathways in nuclei use enzymes that are indistinguishable from their cytosolic analogues. We demonstrate that distinct phosphatidylinositol phosphate kinases (PIPKs), the type I and type II isoforms, are concentrated in nuclei of mammalian cells. The cytosolic and nuclear PIPKs display comparable activities toward the substrates phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate. Indirect immunofluorescence revealed that these kinases were associated with distinct subnuclear domains, identified as “nuclear speckles,” which also contained pre-mRNA processing factors. A pool of nuclear phosphatidylinositol bisphosphate (PIP2), the product of these kinases, was also detected at these same sites by monoclonal antibody staining. The localization of PIPKs and PIP2 to speckles is dynamic in that both PIPKs and PIP2 reorganize along with other speckle components upon inhibition of mRNA transcription. Because PIPKs have roles in the production of most phosphatidylinositol second messengers, these findings demonstrate that phosphatidylinositol signaling pathways are localized at nuclear speckles. Surprisingly, the PIPKs and PIP2 are not associated with invaginations of the nuclear envelope or any nuclear membrane structure. The putative absence of membranes at these sites suggests novel mechanisms for the generation of phosphoinositides within these structures.
INTRODUCTION
Phosphoinositide signaling pathways are present in nuclei (Divecha et al., 1993; Maraldi et al., 1994). The first evidence for a nuclear pathway was the identification of diacylglycerol, phosphatidylinositol (PI),1 and phosphatidylinositol phosphate kinase (PIPK) activities in nuclear envelopes (Smith and Wells, 1983). These same activities were later shown to be retained in Friend cell nuclei that had been carefully stripped of their nuclear envelopes with detergent (Cocco et al., 1987; Divecha et al., 1991). Since then, various enzymes necessary for PI signaling such as phosphoinositide-specific phospholipase C (PLC), protein kinase C (PKC) and inositol-phosphate phosphatases have been identified in the nuclear interior (Kuriki et al., 1992; Martelli et al., 1992; Asano et al., 1994; York et al., 1994; Balboa et al., 1995; Liu et al., 1996; Sun et al., 1997).
The functional significance of the nuclear PI cycle remains poorly understood. The intranuclear PIs constitute a fraction of the total cellular PIs, and their levels are reported to change independently of the plasma membrane phospholipids (Divecha et al., 1993). For instance, nuclear phosphatidylinositol bisphosphate (PIP2), but not total cellular PIP2, decreases as cells progress through S-phase of the cell cycle (York and Majerus, 1994). An increase in nuclear PLC activity and diacylglycerol levels was also reported at the G2-M transition (Sun et al., 1997). Furthermore, PLCβ translocates into nuclei and is activated upon insulin-like growth factor 1 stimulation of Swiss 3T3 cells (Divecha et al., 1991; Martelli et al., 1992). PKCα or PKCβ then enters the nuclei, suggesting that these kinases are effectors of nuclear PLC activity (Divecha et al., 1991). Cellular differentiation and the actions of cytokines such as interleukin 1α or interferon α have also been demonstrated to influence nuclear PI metabolism (Zini et al., 1996b; Divecha et al., 1997).
The spatial organization of the phosphoinositide signaling within the nucleus is not known. However, nuclei stripped of their envelope with detergent still retain phosphoinositides and enzymes that metabolize the phosphoinositides. This suggests that the phosphoinositides must remain associated with nonmembrane nuclear structures; potentially these phosphoinositides are in form of proteolipid complexes (Cocco et al., 1987; Divecha et al., 1991; Martelli et al., 1992). In NIH 3T3 cells and rat liver cells, diacylglycerol, PI, and PIP kinase activities are reported to be associated with the nuclear matrix using biochemical approaches (Payrastre et al., 1992). PLCβ and PKC appear to colocalize on the nuclear matrix by immunoelectron microscopy (Zini et al., 1993; Maraldi et al., 1994). These data again imply that the nuclear PI signaling functions as a component of the nuclear matrix, potentially in the absence of a lipid bilayer.
PIPKs synthesize phosphatidylinositol 4,5-bisphosphate (PI4,5P2) by phosphorylating phosphatidylinositol 4-phosphate (PI4P) (Loijens et al., 1996). Several human isoforms have been cloned, and the type I and type II subfamilies (PIPKIs and PIPKIIs) are each represented by multiple members (Boronenkov and Anderson, 1995; Divecha et al., 1995; Loijens and Anderson, 1996; Castellino et al., 1997; Ishihara et al., 1998; Itoh et al., 1998). In addition to PI4,5P2, PIPKIs can generate phosphatidylinositol 3,4-bisphosphate (PI3,4P2) and phosphatidylinositol 3,4,5-trisphosphate from phosphatidylinositol 3-phosphate (PI3P) (Zhang et al., 1997). Murine PIPKIα has also been reported to produce phosphatidylinositol 3,5-bisphosphate under certain conditions (Tolias et al., 1998), and the generation of putative phosphatidylinositol 3,5-bisphosphate has been correlated with osmotic stress in both yeast and mammalian cells (Dove et al., 1997). Recent studies indicate that PIPKIIs are preferentially PIP 4-kinases as they phosphorylate PI3P and phosphatidylinositol 5-phosphate to synthesize PI3,4P2 and PI4,5P2 by a novel pathway (Rameh et al., 1997). Thus, various PIPK isoforms produce partially overlapping subsets of PI second messengers, which have diverse effectors and cellular functions.
PIPK activity had previously been reported in nuclei (Payrastre et al., 1992). Here, we present evidence that multiple PIPK isoforms are present in the nucleoplasm and are concentrated at nuclear speckles containing mRNA-processing components. PIP2 was also detected at speckles, consistent with its production by PIPKs localized to those sites.
MATERIALS AND METHODS
Antibodies
Recombinant human PIPKIIα (Boronenkov and Anderson, 1995) was expressed in Escherichia coli, purified, and coupled to CNBr-activated Sepharose. Using this matrix, rabbit polyclonal antibodies raised against human erythroid 53-kDa PIPKII (Bazenet et al., 1990) were affinity purified. Recombinant, His-tagged human PIPKIIβ (Castellino et al., 1997) was used to immunize rabbits, and the sera was affinity purified using PIPKIIβ coupled to Sepharose as above. Goat polyclonal anti-PIPKIIα antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) recognize peptides at the N terminus (N-19) or in the “insert” region (C-18) (Boronenkov and Anderson, 1995). The N-19 antibody specifically recognizes PIPKIIα and not PIPKIIβ by Western blotting (Boronenkov, Parker, and Anderson, unpublished observations). Production of anti-PIPKIα rabbit polyclonal antibodies that were affinity-purified using the full-length PIPKIα has been described (Zhang et al., 1997). An additional antibody pool against the unique C-terminal region of PIPKIα was isolated from the antisera, using the affinity column prepared from a hexahistidine-tagged fusion protein of PIPKIα residues 432–549.
The anti-PIP2 mAbs AM212 and AM2 were from Dr. Masato Umeda (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). The KT10 anti-PIP2 mAb and the KD2 anti-PIP mAb were kindly provided by Dr. Kyoko Fukami (University of Tokyo, Tokyo, Japan). The kt3g anti-PIP2 mAb was obtained from Perseptive Biosystems (Framingham, MA). Their specificities have been tested extensively by a variety of methods (Fukami et al., 1988; Fukami and Takenawa, 1989; Matuoka et al., 1988; Miyazawa et al., 1988). Human Sm reference serum was from the Centers for Disease Control (Atlanta, GA) (Hardin et al., 1982). The following antibodies were also used: SC35 mAb (American Type Culture Collection, Rockville, MD), FLAG M2 mAb (Kodak Eastman, New Haven, CT), B1C8 nuclear matrix protein mAb (Matritech, Cambridge, MA), β-actin AC-15 mAb (Sigma Chemical, St. Louis, MO), β-tubulin mAb (Amersham Life Sciences, Arlington Heights, IL), epidermal growth factor receptor (1005) rabbit polyclonal antibody (Santa Cruz Biotechnology), vimentin V9 mAb (Sigma), and glyceraldehyde-3-phosphate dehydrogenase mAb (BioDesign, Kennebunk, ME). mAb104 was a gift from Dr. Mark Roth (Fred Hutchinson Cancer Research Center, Seattle, WA), whereas a rabbit polyclonal antibody against an endoplasmic reticulum (ER)-located epoxide hydrolase was provided by Dr. Charles Kasper (University of Wisconsin, Madison, WI). Fluorescent dye-conjugated secondary antibodies and normal sera were from Jackson ImmunoResearch Laboratories (West Grove, PA), whereas HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology.
Cell Culture
Human transformed 2RA lung fibroblasts, human MG-63 osteosarcoma cells, human HeLa cells, and normal rat kidney NRK-49F fibroblasts were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and antibiotics (Life Technologies, Gaithersburg, MD). For inhibition of transcription, 2RA or NRK-49F cells in culture were treated with 10 μg/mL α-amanitin or 100 μM 5,6-dichlorobenzimidazole riboside (DRB; Sigma) for 4 h. At these concentrations, these inhibitors specifically cause inhibition of RNA polymerase II and induce reorganization of the nuclear speckles into larger and fewer structures (Spector et al., 1983; Carmo-Fonseca et al., 1992).
Immunofluorescence and Microscopy
For immunofluorescence studies, cells were grown on glass coverslips, which, if necessary, were coated with poly-l-lysine. Coverslips were rinsed in PBS, and then cells were fixed. A number of fixation methods were used. Cells were fixed for 15 min in PBS containing 4% formaldehyde at 24°C and permeabilized with 0.2% Triton X-100 in PBS for 7 min at 24°C. Cells were fixed with methanol at −20°C or dry ice for 10 min and washed with PBS. Cells were fixed with acetone at −20°C for 10 min and washed with PBS. After fixation the coverslips were washed in PBS, and they were blocked overnight at 4°C in BSA solution (PBS, pH 7.5, containing 3% BSA, 0.1% Tween 20, and 0.02% sodium azide). Where indicated, cells were preextracted with 0.2% Triton X-100 in PBS for 3 min on ice. All of the buffers were supplemented with 2 mM MgCl2. Incubation with primary antibodies in 3% BSA solution was for 1 h at 37°C. For methanol fixation, between 20 and 1 μg/ml primary polyclonal anti-PIPK antibody was used; for formaldehyde fixation between 10 and 1 μg/ml primary anti-PIPK antibody was used. Generally the AM212 mAb was used at 5 μg/ml, and anti-FLAG mAb was used at 10 μg/ml. Sm antiserum was used at a 1:600 dilution or for the detection of microspeckles as low as 1:6000 depending on which fixation was used. This was followed by labeling for 1 h at 24°C with fluorescent dye-conjugated secondary antibodies in 3% BSA solution supplemented with 10% normal goat serum. Biotin-conjugated concanavalin A (Con A; Vector Laboratories, Burlingame, CA) was used after methanol fixation like a primary antibody and was detected with Texas Red–conjugated streptavidin (Jackson ImmunoResearch). Coverslips were mounted on slides with PBS containing 90% glycerol, 0.1 g/mL 1,4-diazabicyclo(2.2.2.)octane (Eastman Kodak, Rochester, NY), and 1,4-phenylenediamine (Aldrich, Milwaukee, WI) and sealed using nail polish.
Digital images were acquired using an MRC-1024 laser scanning confocal microscopy system (Bio-Rad Laboratories, Hercules, CA) at the W.M. Keck Neural Imaging Laboratory (University of Wisconsin Medical School). For single fluorophore staining, a stack of the individual planar images with an 0.8-μm step was coaxially projected to obtain the final image using Confocal Assistant software (Bio-Rad). In the case of multiple fluorophores, sequential series of scans with a 0.2-μm step were acquired, and the corresponding individual thin optical sections were selected using NIH Image 1.59 software (National Institutes of Health, Bethesda, MD) and processed in Adobe Photoshop 4.0 (Adobe Systems, San Jose, CA).
Subcellular Fractionation and Western Blotting
To obtain total cell lysates, cultured cells were trypsinized, washed twice with cold PBS, and lysed in Triton lysis buffer (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 0.5% Triton X-100, 1 mM EDTA, 0.5 mM PMSF, 2 μg/ml leupeptin, and 10 trypsin inhibitor units/ml aprotinin). The nuclear isolation protocol was based on the method of Dignam et al. (1983) designed for preparation of splicing extracts and transcription factors. Harvested HeLa cells, obtained from the National Cell Culture Center (Minneapolis, MN), were washed and swollen in three packed-cell volumes of hypotonic buffer (10 mM HEPES, pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM PMSF, 2 μg/ml leupeptin, 10 trypsin inhibitor units/ml aprotinin, 2 μg/ml antipain, and 2 μg/ml chymostatin). All further buffers used for nuclear isolation contained 17 μg/ml calpain inhibitor I and 7 μg/ml calpain inhibitor II (Alexis, San Diego, CA) in addition to the inhibitors found in the hypotonic buffer. After 20 min on ice, the swollen cells were disrupted in a Dounce homogenizer with 20 strokes, and the completion of lysis was monitored by trypan blue exclusion staining. Nuclei were pelleted at 700 × g, and the supernatant was kept as the cytosolic fraction. Nuclei were then stripped in hypotonic buffer containing 0.8% Triton X-100 for 10 min on ice, spun at 700 × g, and washed several times in hypotonic buffer containing 25% glycerol and 0.5 mM DTT. This last step caused some leakage of the nuclear material. Stripped nuclei were treated with 350 mM KCl for 20 min at 4°C, disrupted by brief probe sonication (70 W, 2 × 10 s), and separated by 16,000 × g centrifugation (30 min) into the nuclear pellet (postextracted nuclei) and nuclear extract. The nuclear extract was dialyzed against hypotonic buffer with 20% glycerol and 0.5 mM DTT, and the resulting precipitate was removed by a 16,000 × g centrifugation (30 min). For some preparations, the nuclear extract was clarified with a 200,000 × g centrifugation for 1 h to remove any trace of membrane structures. A similar nuclear distribution of PIPKs was seen when stripped nuclei and nuclear matrix from NRK, HeLa, or 2RA cells were prepared by the method of Payrastre et al. (1992). The cellular fractions were transferred to an Immobilon-P polyvinylidene fluoride membrane (Millipore, Bedford, MA) and Western blotted as described previously (Zhang et al., 1997). The chemiluminescence was detected by film, with care being taken not to overexpose the film with the signal for any of the bands; the signals from individual protein bands were quantified by densitometric scans of the film from the Western blots with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The relative contents of the detected protein in subcellular fractions were then calculated by factoring in the amounts of total protein in every fraction. For the analysis of cross-contamination between the subcellular fractions, these Western blots were stripped with detergent, blocked, and reprobed with the antibodies specific for proteins restricted to specific subcellular fractions (as described in RESULTS). The relative amounts of these proteins in subcellular fractions were determined as outlined above for PIPKs.
Transfection of Epitope-tagged PIPKs
PIPKIIα and PIPKIIβ were N-terminally tagged with the FLAG epitope by subcloning their coding regions into pcDNA3-FLAG vectors provided by Dr. Jon Morrow (Yale University, New Haven, CT). These constructs were transiently transfected into 2RA cells using LipofectAMINE (Life Technologies) as previously described (Zhang et al., 1997).
Immunoprecipitations and Lipid Kinase Assays
Immunoprecipitations from HeLa nuclear extracts or cytosol were performed in hypotonic buffer supplemented with 150 mM NaCl and Nonidet P-40 at either 0.2% (PIPKIIα) or 0.5% (PIPKIα). Omnisorb cells (Calbiochem-Novabiochem, San Diego, CA) were used as the protein G matrix as previously described (Zhang et al., 1997). The PIPKIα antibody was used at 0.4 μg/100 μg protein for 2 h on ice, whereas the PIPKIIα N19 antibody was used at 1.2 μg/100 μg protein overnight at 4°C. Control experiments included mock immunoprecipitation in the absence of antibody and Western blotting a sample of the antibody used in the immunoprecipitation.
PIP kinase assays were performed as previously described (Zhang et al., 1997) using either 50 μM PI4P (Sigma) or PI3P dipalmitoyl ester (a gift from Dr. Glenn Prestwich, University of Utah, Salt Lake City, UT). The thin-layer chromatography plates were analyzed using a PhosphorImager and ImageQuant software (Molecular Dynamics). Radiolabeled lipids were subsequently scraped from the thin-layer chromatography plates, based on spots observed on the autoradiograph, and analyzed by scintillation counting (Zhang et al., 1997).
RESULTS
Type I and II PIPKs Localized to Nuclei in Cultured Cells
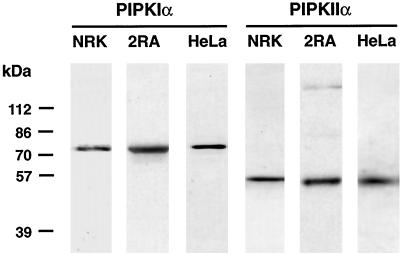
To characterize the intracellular distribution of the PIPKs and to provide tools to study the signaling pathways in which they participate, polyclonal antibodies were generated against two distinct PIPKs, the type I and type II isoforms. These antibodies immunoprecipitate their respective kinases from cell lysates and do not cross-react with kinases of the other type (Jenkins et al., 1994; Zhang et al., 1997). As shown in Figure 1, the antibodies specifically detected the 68-kDa PIPKIα and 53-kDa PIPKIIα in HeLa, NRK-49F, and 2RA cell lines. The PIPKIα antibodies did not cross-react with the closely related PIPKIβ (our unpublished results; Loijens and Anderson, 1996). The PIPKIIα antibodies appear to detect only the PIPKIIα (53 kDa by SDS-PAGE) by Western blotting cell lysates, because the PIPKIIβ migrates with a apparent size of ∼56 kDa. The PIPKIIα antibodies do weakly detect the homologous PIPKIIβ by Western blotting the E. coli–expressed protein.
Figure 1.
anti-PIPKIα and anti-PIPKIIα polyclonal antibodies (10 μg/ml) selectively detected 68- and 53-kDa proteins, respectively, by Western blotting total lysates prepared from NRK-49F, 2RA, and HeLa cells.
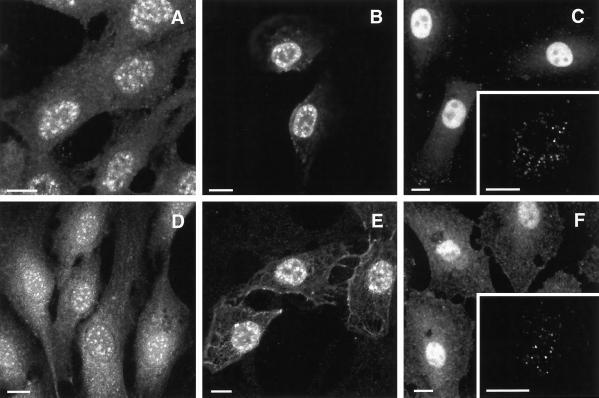
These affinity-purified, isoform-specific PIPK antibodies were used to determine the intracellular localization of the PIPKs in cultured NRK cells by indirect immunofluorescence. Cells fixed with methanol or acetone and stained with the type Iα or type IIα PIPK antibodies displayed intense nuclear staining for both isoforms (Figure 2, A and D, respectively). Moreover, the staining in nuclei was concentrated in distinct foci or nuclear speckles. This staining pattern was independent of fixation but was dependent on PIPK antibody concentration. When cells were fixed with 4% formaldehyde and stained with high concentrations of type I and II PIPK antibodies, diffuse nuclear staining with nucleolar exclusion was observed (Figure 2, C and F). Intermediate concentrations of PIPK antibodies (<5 μg/ml) emphasized the speckle pattern as in Figure 2, A and D, whereas at even lower antibody concentrations, the speckle structures became smaller and more numerous (Figure 2, C and F, insets), possibly representing the sites with the highest concentration of the PIP kinases within nuclei. These observations were reminiscent of the threshold effect reported previously when different dilutions of various antibodies toward splicing factors were used in immunofluorescence (Neugebauer and Roth, 1997). Similarly, we have observed the appearance of smaller dots in place of speckles when lower dilutions of Sm antiserum and SC35 antibody were used, either after formaldehyde or −20°C acetone fixations. To determine whether the diffuse nuclear staining at high antibody concentration represented a pool of soluble kinases, cells were mildly preextracted with detergent, followed by fixation and antibody staining. As shown in Figure 2, B and E, detergent-preextracted human 2RA fibroblast cells stained with high antibody concentration displayed a speckled pattern of nuclear staining with a reduction in the diffuse background staining. These data suggest that both detergent soluble and insensitive populations of the PIPKs were present within nuclei (also see below). This is consistent with previous biochemical findings demonstrating both detergent-soluble and -insoluble nuclear PIPK activities (Divecha et al., 1991; Payrastre et al., 1992). Furthermore, the PIPKIs and PIPKIIs associated with nuclear speckles were resistant to detergent extraction, suggesting a stable association of the kinases with these nuclear structures. This nuclear localization of PIPKIα and PIPKIIα was observed in a variety of transformed primate and rodent cell lines, transformed and nontransformed human cell lines, and neonatal mouse cardiomyocytes.
Figure 2.
Indirect immunofluorescence of NRK-49F rat fibroblasts indicated nuclear localization of PIPKIα (A–C) and PIPKIIα (D–F). The speckled staining in nuclei was observed with methanol fixation (A and D), or with a brief preextraction using 0.2% Triton X-100 and then fixation with formaldehyde (B and E). Strong diffuse nuclear staining upon formaldehyde fixation was obtained when anti-PIPK antibodies were used at 10 μg/ml (C and F). However, this picture at lower concentrations progressed to speckles and then resolved into a pattern of smaller dots when 0.5 μg/ml anti-PIPKIα or 1 μg/ml anti-PIPKIIα antibodies were used (C and F, insets). Insets, thin optical sections of magnified view of the nuclei of human MG-63 cells. Fixation and staining protocols are detailed in MATERIALS AND METHODS. Bar, 10 μm.
As controls, PIPKIα preimmune IgG and IgG from anti-PIPKIα sera depleted of PIPKIα-immunoreactive antibody species gave background signals by immunofluorescence. Furthermore, preincubation of the PIPKIIα antibody with an excess of denatured recombinant PIPKIIα abolished staining, and the nuclear staining was only weakly blocked when the closely related, denatured rPIPKIIβ was combined with the PIPKIIα antibody. No cross-reactivity between anti-PIPKI antibody and PIPKIIs (and vice versa) in immunofluorescence experiments was detected.
Biochemical Fractionation Also Demonstrated That the PIPKs Are Nuclear Enzymes
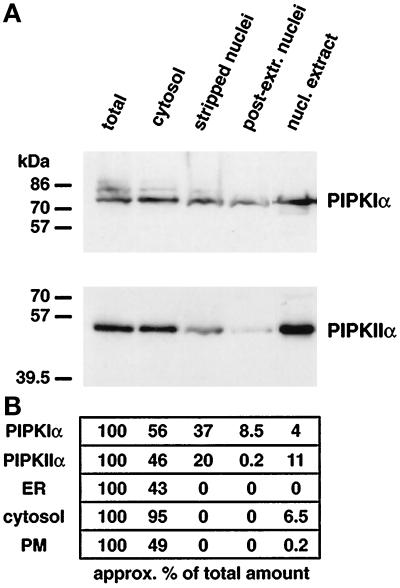
To provide further evidence for the nuclear localization of PIPKs, HeLa, NRK, and 2RA cells were separated into subcellular fractions by established methods (Dignam et al., 1983; Payrastre et al., 1992; York et al., 1994). As representative of these experiments, a HeLa cell fractionation is shown in Figure 3, which is based on the method of Dignam et al. (1983), for isolation of nuclei. Cell fractions with equal protein loads were Western blotted with antibodies against PIPKIα and PIPKIIα (Figure 3A), and the relative amounts of the kinases in each fraction were quantified (Figure 3B) by Western blotting (see MATERIALS AND METHODS). Shown are the total cell lysate, crude cytosol, which contains cytosol and membranes, and membrane-stripped nuclei. The nuclei have been extracted with 0.8% Triton X-100, a treatment that completely removes the nuclear envelope and soluble nuclear material (Divecha et al., 1991; Vann et al., 1997). The detergent extraction step resulted in the removal of PIPKs from nuclei, and this likely represents the soluble PIPK described above. However, a large fraction of both PIPKI and PIPKII proved to be resistant to detergent extraction. This observation also suggested the presence of two pools of PIPKs within nuclei and correlated well with the above immunofluorescence results and the biochemical data showing that kinase activities are retained by nuclei (Divecha et al., 1991; Payrastre et al., 1992). Relative to the total lysate, ∼37% of the cellular PIPKIα and 20% of PIPKIIα was quantified to be tightly retained in nuclei stripped of their envelopes (Figure 3B). When stripped nuclei were then extracted with high ionic strength, the majority of the PIPKs were removed from nuclei and were present in the nuclear extract. Preparation of stripped nuclei and nuclear matrix of NRK, HeLa, or 2RA cells by the method of Payrastre et al. (1992) gave a similar nuclear distribution for the PIPKs (our unpublished data).
Figure 3.
Subcellular fractionation of HeLa cells showed that nuclei contained PIPKIα and PIPKIIα. HeLa cells were disrupted and separated into nuclear and crude cytosolic (cytosol) fractions by low-speed sedimentation. The nuclei were membrane stripped with 0.8% Triton X-100, washed, and pelleted (stripped nuclei). Stripped nuclei were then treated with 0.35 M KCl and separated into soluble (nuclear extract) and insoluble (postextracted nuclei) fractions by high-speed centrifugation. Equal amounts of protein from each fraction were used to do Western blots for both PIPKIα and PIPKIIα (A). The purity of the cellular fractions was assessed by reprobing the Western blots (B) with antibodies toward proteins from the ER, cytosol, and plasma membrane (PM), as described in MATERIALS AND METHODS.
The subcellular fractions were also assayed for the presence of proteins that reside in the ER, plasma membrane and cytosolic fractions by quantitative Western blotting blotting (see MATERIALS AND METHODS). The stripped nuclei isolated by our fractionation method did not contain sizable quantities of plasma membrane, ER, or cytosolic contamination, as monitored by epidermal growth factor receptor, epoxide hydrolase, and glyceraldehyde-3-phosphate dehydrogenase immunoreactivity, respectively (Figure 3B). The resultant nuclear extract did contain minor amounts of actin and tubulin but was free of intermediate filaments as monitored by vimentin immunoreactivity, consistent with previous reports (Payrastre et al., 1992).
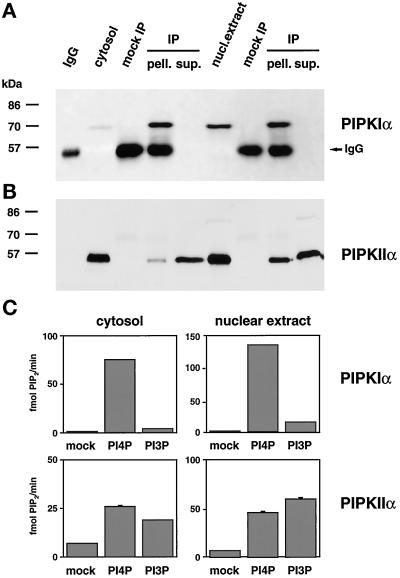
To examine the activity of the nuclear PIPKs, type Iα and type IIα enzymes were immunoprecipitated from either crude cytosol or nuclear extract (Figure 4). The PIPKIα antibody quantitatively removed all PIPKIα from either the cytosol or nuclear extract (Figure 4A), whereas only a fraction of PIPKIIα was immunoprecipitated by the N-19 peptide antibody in each case (Figure 4B). These immunoprecipitated kinases were then tested for activity toward PI4P and PI3P (Figure 4C). The substrate preferences observed for the given PIPK isoform in the cytosolic and nuclear extract fractions were indistinguishable. PIPKIα preferred PI4P over PI3P, whereas PIPKIIα had almost equal preference for both substrates, as had been previously reported (Zhang et al., 1997). As a control, mock immunoprecipitations demonstrated that no PIPKs were nonspecifically isolated.
Figure 4.
Nuclear and cytosolic PIPKs exhibited similar kinase activities toward PI4P and PI3P as substrates. PIPKIα (A) and PIPKIIα (B) could be selectively immunoprecipitated from HeLa cytosol or nuclear extract. (A) PIPKIα was immunoprecipitated from either 200 μg of cytosol or 100 μg of nuclear extract with the rabbit anti-PIPKIα polyclonal antibody, followed by Western blotting with the PIPKIα C-terminal isoform-specific antibody. (B) The goat N19 peptide PIPKIIα isoform-specific antibody was used to immunoprecipitate PIPKIIα from 130 μg of cytosol or 65 μg of nuclear extract. PIPKIIα was then detected by blotting with the rabbit anti-PIP5KIIα polyclonal antibody. (C) PIPKIα and PIPKIIα, immunoprecipitated from 400 μg of cytosol or 200 μg of nuclear extract, were assayed for lipid kinase activity toward either PI4P or PI3P. The assays shown are representative of two or three immunoprecipitations from two different nuclear preparations. As controls, mock immunoprecipitations (mock IP) were performed in the absence of the antibody, and a sample of the antibody used for the immunoprecipitation was Western blotted (IgG).
PIPKs Associated with Nuclear Speckles Containing mRNA-processing Factors
By immunofluorescent staining, both PIPKIα and PIPKIIα displayed a detergent-resistant subnuclear localization suggesting a compartmentalization of the enzymes in the nucleus (Figure 2). This punctate pattern was reminiscent of nuclear speckle staining commonly observed for splicing factors (Lamond and Earnshaw, 1998); cells were double labeled with anti-PIPK antibodies and human Sm sera, an autoimmune antibody that recognizes an epitope in small nuclear RNA-binding proteins (Hardin et al., 1982). In Figure 5, the PIPKs colocalized identically with the Sm-positive nuclear speckles in methanol-fixed NRK cells. This is indicated by the yellow color resulting from overlaying thin optical sections of the FITC signal (green) from the PIPK antibodies and with the Texas Red signal from the Sm antibodies. This colocalization was also observed in 2RA fibroblasts that had been preextracted and fixed with formaldehyde, and when cells were examined with antibodies specific for other proteins found in speckles such as SC35, mAb104, and B1C8 (our unpublished results). These antibodies are specific for components of the mRNA processing machinery (Fu and Maniatis, 1990, Roth et al., 1990) or nuclear matrix (Wan et al., 1994).
Figure 5.
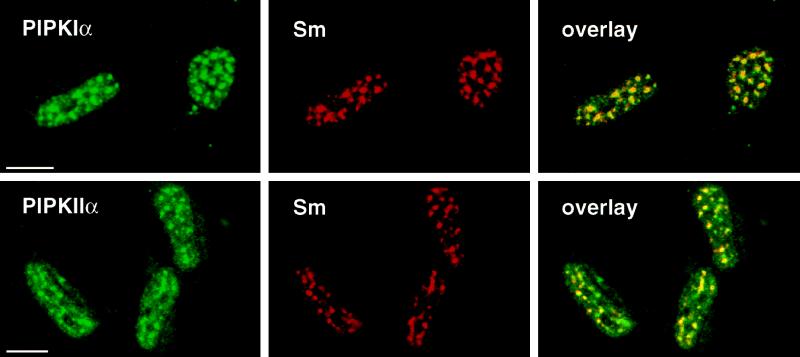
PIPKs colocalized with components of the mRNA-processing machinery in nuclear speckles. Methanol-fixed rat NRK fibroblasts were double-labeled with anti-PIPKIα or anti-PIPKIIα polyclonal antibodies and human Sm antiserum (recognizes components of small nuclear RNA-binding proteins). Thin optical sections obtained by confocal scanning laser immunofluorescence microscopy are shown. Colocalization is represented by yellow in the overlays. Bar, 10 μm.
To provide conclusive evidence that specific PIPK isoforms were nuclear and associated with speckles, PIPKIIα and the homologous PIPKIIβ were epitope-tagged and transiently expressed in cultured cells. The overexpressed, FLAG-tagged PIPKIIs localized to nuclei giving a diffuse staining (Figure 6, top row). Staining with the corresponding PIPKII antibodies indicated that the transfected kinases were expressed at levels substantially higher than the untransfected cells around them, suggesting that overexpression had obscured or saturated the speckle association. Indeed, preextraction of the cells with 0.2% Triton X-100 revealed the kinases to exhibit a nuclear speckle pattern, which colocalized with Sm staining (Figure 6, bottom row). These results, although reflecting an overexpression situation, were consistent with the distribution of endogenous nuclear PIPKs to the detergent-soluble and -insoluble compartments. Importantly, PIPKIIα antibodies did not detect overexpressed PIPKIIβ, but anti-PIPKIIβ antibodies detected overexpressed PIPKIIα and PIPKIIβ. However, PIPKIIβ antibodies did not give a strong signal with untransfected cells. This provided evidence that PIPKIIα antibodies were isoform specific for immunofluorescence staining, whereas the PIPKIIβ antibodies detect both PIPKII isoforms, but only when they are overexpressed. The staining with the FLAG antibody clearly indicated that overexpressed PIPKIIβ localized to the nucleus (Figure 6). These combined data indicate that both PIPKIIα and PIPKIIβ nuclear localize and associate with nuclear speckles.
Figure 6.
Epitope-tagged PIPKIIα and PIPKIIβ localized to nuclei and nuclear speckles when expressed in cultured fibroblasts. Indirect immunofluorescence was performed on formaldehyde-fixed human 2RA fibroblasts transiently overexpressing FLAG epitope-tagged PIPKIIα or PIPKIIβ. Double labeling with anti-PIPKIIα or PIPKIIβ antibodies and anti-FLAG M2 antibodies showed primarily diffuse nuclear localization of the expressed kinases (top row). Localization of overexpressed kinases to the speckles was revealed by a very brief preextraction of the cells with 0.2% Triton X-100 (bottom row) and staining with anti-FLAG antibody. Speckles were costained with the human Sm serum. Bar, 10 μm.
Polyphosphoinositides Were Also Present at Nuclear Speckles
Because PIPKs were associated with nuclear speckles, it was plausible that polyphosphoinositides could be produced at these same sites. Several monoclonal antibodies have been generated against PI4,5P2 that have been extensively characterized (Fukami et al., 1988; Fukami and Takenawa, 1989; Matuoka et al., 1988; Miyazawa et al., 1988). These antibodies were used to localize PI4,5P2 within nuclei by indirect immunofluorescence. Of the anti-PIP2 antibodies tested, the AM212 mAb (Miyazawa et al., 1988) intensely stained nuclear speckles in all cell lines examined. The pattern of staining with the AM212 antibody is shown for 2RA cells in Figure 7. In the top and bottom panels, PIP2 antibody staining colocalized with PIPKIα and PIPKIIα at speckles. When these cells were triple labeled for the kinases, AM212, and Sm, all signals were present at the same nuclear speckles.
Figure 7.
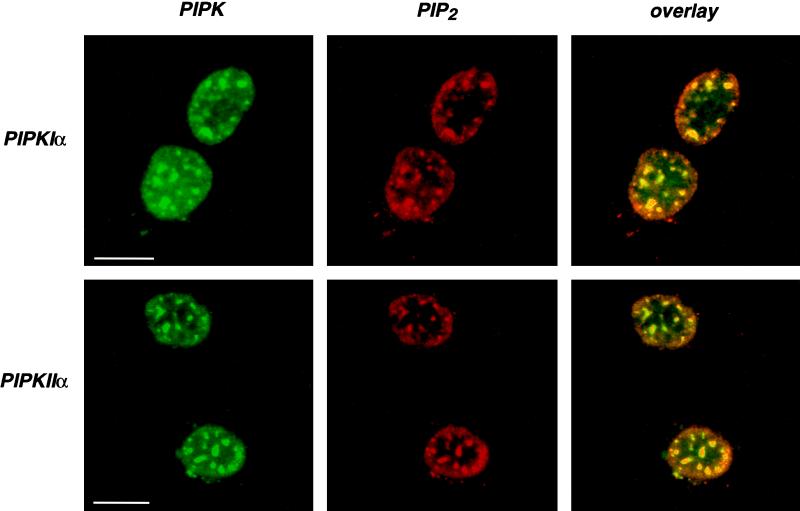
PIPKs colocalized with PIP2 in nuclear speckles. Prepermeabilized, formaldehyde-fixed human 2RA fibroblasts were double labeled with anti-PIPKIα or anti-PIPKIIα polyclonal antibodies and anti-PIP2 mAb AM212 (Miyazawa et al., 1988). Thin optical sections were obtained by confocal laser scanning microscopy. Colocalization is represented by yellow in the overlays. Bar, 10 μm.
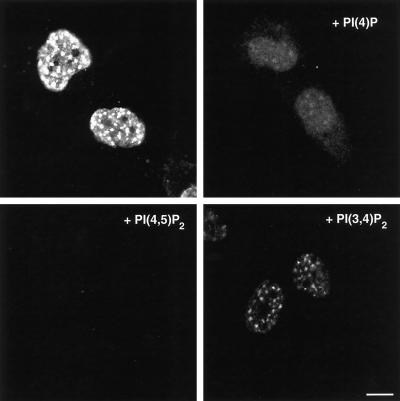
To characterize the specificity of the PIP2 antibody staining, the antibody was preincubated with various polyphosphoinositides and phospholipids. An excess of PI4,5P2 abolished the staining, whereas PI4P and PI3,4P2 at the same concentration were only partially inhibitory (Figure 8). Preincubation with PI or other phospholipids had no effect on antibody staining, and this was consistent with the characterized specificity of the AM212 PIP2 antibody (Miyazawa et al., 1988). Moreover, an intense signal was detected after formaldehyde fixation whether the cells were preextracted with Triton X-100, indicating that the PIP2 antibody staining was resistant to detergent. Methanol-fixed cells also retained nuclear speckle staining by the AM212 mAb (see below); however, the signal intensity was substantially reduced. This suggested that methanol fixation may have extracted some of the PIP2, as would be expected for a phospholipid. Thus, immunofluorescence indicated that a pool of polyphosphoinositides was present at nuclear speckles that could be either substrates or products of the PIPKs also associated with speckles.
Figure 8.
The specificity of the nuclear PIP2 signal was demonstrated by complete inhibition of staining after preincubation of the AM212 mAb with excess of PI4,5P2 liposomes but only partial inhibition by PI4P or PI3,4P2. Bar, 10 μm.
PIPKs and Polyphosphoinositides Were Not Associated with Known Intranuclear Membrane Structures
Invaginations of the nuclear envelope project deep within the nucleus and in some cases traverse it (Fricker et al., 1997). These membrane structures, when visualized by laser confocal microscopy in thin optical sections, would appear similar to nuclear speckles. Figure 9 shows these structures (denoted by arrows) in 2RA fibroblasts stained with the biotin-conjugated lectin Con A. Con A specifically binds mannose residues in the lumen of the ER and the nuclear envelope. Triple labeling with Con A (Figure 9, C and G), the AM212 mAb (Figure 9, B and F), and either PIPKIα (Figure 9A) or PIPKIIα (Figure 9E) antibodies revealed that PIPKs and PIP2 were not associated with these nuclear membrane structures.
Figure 9.

PIPKs and PIP2 were not associated with invaginations of the nuclear envelope. Invaginations of the nuclear envelope can transverse nuclei (Fricker et al., 1997) and produce a series of dots (arrows) seen here in thin optical sections of methanol-fixed human 2RA fibroblasts labeled with biotin-conjugated Con A (a lectin that binds mannose residues of nuclear envelope glycoproteins). Thin optical sections of the triple-labeling with anti-PIPKIα (A) or anti-PIPKIIα (E) polyclonal antibodies, Con A (C and G) and anti-PIP2 mAb AM212 (B and F) are shown. The overlay of PIPK and Con A staining patterns is shown in yellow (D and H). Bar, 10 μm.
Association of PIPKs and PIP2 with Nuclear Speckles Was Dynamic and Dependent on Transcriptional Activity
Treatment of cells with the transcriptional inhibitor α-amanitin at concentrations that specifically inhibit RNA polymerase II causes reorganization of nuclear speckles containing splicing factors into fewer and larger speckles as detected by the Sm sera or antibodies specific for other splicing factors (Carmo-Fonseca et al., 1992). Likewise, treatment with the transcriptional inhibitor DRB (Spector et al., 1983) causes reorganization of Sm speckles into larger dots or a scattered array of small dots (Davis et al., 1993). As shown in Figure 10, inhibition of RNA polymerase II in 2RA cells with α-amanitin (A) or NRK cells with DRB (B) caused the expected changes in Sm staining (compare with Figure 5). Triple labeling of treated cells indicated that the intranuclear distributions of PIPKIα (A), PIPKIIα (B), and PIP2 followed changes in Sm staining, demonstrating a physical and dynamic association of these signaling molecules with the speckles. The results were similar for both kinases with either treatment and independent of the fixation method used. In addition, expressed PIPKIIα and PIPKIIβ similarly reorganized when transfected 2RA cells were treated with α-amanitin (our unpublished results).
Figure 10.
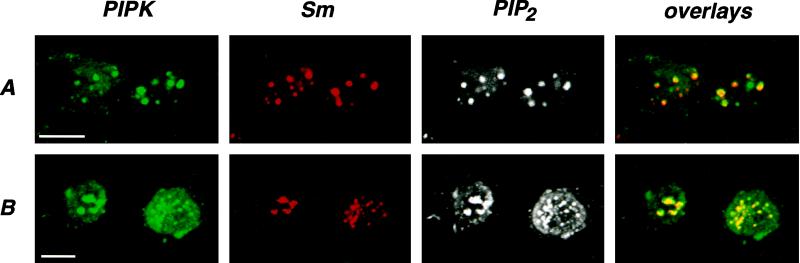
Association of PIPKs and PIP2 with nuclear speckles is dynamic. Treatment of 2RA cells with a 10 μg/ml concentration of the transcriptional inhibitor α-amanitin for 4 h caused reorganization of splicing-related nuclear speckles as detected by Sm antiserum into a few large dots (A, compare with Figure 5). Likewise, treatment of NRK cells with a 100 μM concentration of the transcriptional inhibitor DRB for 4 h caused reorganization of Sm speckles into large dots or a scattered array of small dots (B). Cells were triple labeled to show the changes in PIPKIα (A), PIPKIIα (B), and PIP2 distribution upon treatment with the inhibitors. The cells were fixed with 4% formaldehyde after 0.2% Triton X-100 preextraction. The PIPK and Sm staining patterns were overlaid (yellow) to demonstrate colocalization. Bar, 10 μm.
DISCUSSION
PIPK activities are present in many subcellular fractions including the plasma membrane, cytosol, endoplasmic reticulum, cytoskeleton and nuclei (Loijens et al., 1996). The discovery of at least six mammalian PIPK isoforms (Boronenkov and Anderson, 1995; Divecha et al., 1995; Loijens and Anderson, 1996; Castellino et al., 1997; Ishihara et al., 1998; Itoh et al., 1998) may partially explain the wide distribution of PIPKs in mammals. Previously, PIPK activity, together with PLC activity, has been reported in isolated nuclei (Cocco et al., 1987; Divecha et al., 1991) and associated with a biochemically defined structure called the inner nuclear matrix (Payrastre et al., 1992). However, the exact nature of PIPK enzymes involved and the properties of the compartment containing the phosphoinositide signaling enzymes were not defined. In this study, the immunofluorescence and fractionation experiments both suggest that the nucleus contains a substantial proportion of the total PIPKIα and PIPKIIα found in cells. The localization of PIPKs to a specific subcellular site is an important step toward understanding compartmentalization and function of phosphoinositide signaling pathways. Our results argue for nuclear PIPKs being present in two pools: a soluble pool, extractable by detergent; and a second pool, which was more tightly associated with nuclei. The latter was shown to be localized to structures called nuclear speckles.
Nuclei are highly ordered organelles composed of multiple subdomains with specific functions (Fakan et al., 1984; Nickerson et al., 1995; Fricker et al., 1997; Lamond and Earnshaw, 1998). One of these subdomains, consisting of interchromatin granule clusters, is observed with electron microscopy and hypothesized to be a site of assembly or storage of factors required to synthesize pre-mRNAs (Jackson et al., 1993; Spector, 1996; Misteli et al., 1997; Singer and Green, 1997). These structures contain small ribonucleoproteins, mRNA-splicing factors, and a hyperphosphorylated form of RNA polymerase II (Mortillaro et al., 1996). Perichromatin fibrils are at the periphery of the interchromatin granule clusters and have been proposed to be the sites of transcription and splicing (Huang and Spector, 1996; Pombo and Cook, 1996). The assemblies of multiple proximal interchromatin granule clusters are thought to correspond to the 20–40 intensely stained nuclear speckles above a diffuse background signal when immunofluorescence microscopy is performed with probes to a variety of splicing factors (Spector et al., 1991; Neugebauer and Roth, 1997). Resistance of this staining to extraction with nonionic detergents or treatment with DNase I suggests association of speckle components with a nuclear scaffold (Nickerson et al., 1995). The large number of replication, splicing, transcriptional, and other assemblies found in and around the nuclear speckles suggests that these must be sites that generate signals or are impacted upon by signal transduction. Protein kinases and phosphatases are known to reside at nuclear speckles and to regulate speckle morphology (Gui et al., 1994; Colwill et al., 1996; Misteli and Spector, 1996).
Several studies have identified PLCβ, PIP2, and PKC at the periphery of interchromatin granule clusters by immunoelectron microscopy of in situ nuclear matrix preparations from different cell types (Zini et al., 1993; Maraldi et al., 1994, 1995). The localization of PIPKs to speckles suggests that speckles may also be centers for nuclear PI signal transduction. PIP2 produced by PIPKs could either affect nuclear events directly or upon conversion to second messengers, such as inositol triphosphate and diacylglycerol, that can modulate intranuclear Ca2+ levels (Malviya and Rogue, 1998) and PKC activity. In addition to known nuclear substrates of PKC (Matter et al., 1993; Goss et al., 1994; Collas et al., 1997), other attractive targets include transcription factors or SR proteins. As another example, casein kinase Iα, known to be regulated by PIP2 in vitro (Brockman and Anderson, 1991), was recently found to localize to the same nuclear speckles, where it phosphorylates a subset of SR proteins (Gross and Anderson, submitted).
The speckle morphology correlates with transcriptional activity, with speckles becoming small and more diffuse when it is increased (Zeng et al., 1997) and fewer and larger when mRNA transcription is inhibited (Carmo-Fonseca et al., 1992; Misteli et al., 1997). The PIPKs and their product, PIP2, reorganize identically with speckles (Figure 10), both spatially and temporally, suggesting direct interaction of PIPKs with speckle component(s). We are currently examining this possibility. Although factors known to affect nuclear PI turnover, such as insulin-like growth factor 1 in Swiss 3T3 cells, cause translocation of PLCβ and PKC to sites in the nuclear interior (Divecha et al., 1993, 1997; Maraldi et al., 1994), this does not appear to be the case for nuclear PIPKs. We did not observe significant changes in PIPK immunofluorescence in the nuclei of Swiss 3T3 cells treated with insulin-like growth factor 1 or other agonists (our unpublished results). PIPK staining, colocalization with Sm, or the amounts of PIPKIIα in nuclear fractions by Western blotting also did not change appreciably during the cell cycle after NRK cells were released from serum starvation (our unpublished results).
There is good evidence that splicing and transcription are distributed widely throughout the nucleus, even though they appear to be concentrated within and around speckles (Jackson et al., 1993; Singer and Green, 1997). For instance, Neugebauer and Roth (1997) demonstrated that lowering the concentration of splicing factor antibodies used to stain cells resolved the nuclear speckles into more numerous smaller, defined structures, some of which identically colocalize with sites of transcription. A similar result was observed when low concentrations of PIPK antibodies were used. In nonextracted, formaldehyde-fixed cells, nuclei tend to have more diffuse PIPK staining with high antibody concentration, whereas at lower concentrations, speckles became emphasized and eventually were resolved into multiple smaller dots. These smaller dots would thus represent the sites at which the PIPKs are most concentrated.
The PIPKI and PIPKII isoforms have different substrate specificities and regulation (Jenkins et al., 1994; Loijens et al., 1996; Rameh et al., 1997; Zhang et al., 1997; Tolias et al., 1998). The localization of several isoforms to the same foci in nuclei may reflect the diversity of PI signals generated at these sites. Because there is no evidence for the existence of D3-phosphoinositides in nuclei (Divecha et al., 1993), it was important to determine whether the ability of nuclear PIPKs to synthesize these lipids was compromised in favor of other products. As shown in Figure 4, PIPKs from membrane-depleted nuclei still have the potential to generate D3-phosphoinositides in vitro. Because phosphatidylinositol 3-kinase has recently been reported to be present in the nuclear matrix of human osteosarcoma cells (Zini et al., 1996a) or in nuclei of rat liver cells (Lu et al., 1998), it may be necessary to reexamine the phosphoinositides generated in the nuclear PI pathways to determine whether the D3-phosphoinositides are synthesized in nuclei. These lipids could be involved in the modulation of activity of PI3,4P2-dependent protein kinase Akt/PKB that has been recently shown to translocate to nuclei (Meier et al., 1997).
Using Con A and other markers, the nuclear envelope has been recently found to project invaginations that penetrate, and even traverse, the nucleus (Fricker et al., 1997). As seen in Figure 9, speckles containing PIPKs, PIP2, and splicing factors did not colocalize with these invaginations of the nuclear envelope. This implies either that there are membranes at speckles that have yet to be identified, or that speckles are devoid of membrane structures. Polyphosphoinositides have been shown to be tightly associated with nuclei stripped of membranes by detergent (Figure 7 in this study). The absence of membranes at nuclear speckles would necessitate that the kinases that phosphorylate the phosphoinositides be active toward substrates presented in a nonmembranous form, such as bound to proteins. Currently, this model is most consistent with the data presented here and in other reports (Divecha et al., 1993; Lu et al., 1998). Association of PIs with proteins has been reported (Janmey, 1994). Indeed, there is evidence that PLC is capable of using PI4,5P2 bound to the PI transfer protein, a protein reportedly found in nuclei (De Vries et al., 1996; Cockcroft, 1998). Thus, it is plausible that phosphoinositides would remain bound to proteins and could be used by enzymes that generate phosphoinositide messengers. Several PIP2-binding proteins have been shown to be present in nucleus (Iida et al., 1992; Onoda and Yin, 1993; De Vries et al., 1996; Yu et al., 1998), and these could be candidates for assembling proteophosphoinositide complexes. Although PIPKs associate with nuclear speckles that are functionally linked to mRNA metabolism, the role of the phosphoinositides generated at these sites remains to be elucidated.
ACKNOWLEDGMENTS
We are grateful to Dr. Kyoko Fukami (University of Tokyo, Toyko, Japan) for generously providing anti-PIP and anti-PIP2 mAbs and to Scott Doughman for discussions and critical reading of the manuscript. We thank Dr. Glenn Prestwich (University of Utah, Salt Lake City, UT) for providing synthetic phosphoinositides. The technical assistance of Gregory J. Parker was appreciated. This work was supported by National Institutes of Health grant GM51968 (to R.A.A.). I.V.B. is a graduate student in the Department of Biomolecular Chemistry, and J.C.L. was a graduate student in the Cellular and Molecular Biology Program.
Abbreviations used:
- Con A
concanavalin A
- DRB
5,6-dichlorobenzimidazole riboside
- ER
endoplasmic reticulum
- PI
phosphatidylinositol
- PI4P
phosphatidylinositol 4-phosphate
- PI3P
phosphatidylinositol 3-phosphate
- PI4
5P2, phosphatidylinositol 4,5-bisphosphate
- PI3
4P2, phosphatidylinositol 3,4-bisphosphate
- PIP
phosphatidylinositol phosphate
- PIP2
phosphatidylinositol bisphosphate
- PIPK
phosphatidylinositol phosphate kinase
- PIPKI
type I phosphatidylinositol phosphate kinase
- PIPKII
type II phosphatidylinositol phosphate kinase
- PKC
protein kinase C
- PLC
phosphoinositide-specific phospholipase C
REFERENCES
- Asano M, Tamiya-Koizumi K, Homma Y, Takenawa T, Nimura Y, Kojima K, Yoshida S. Purification and characterization of nuclear phospholipase C specific for phosphoinositides. J Biol Chem. 1994;269:12360–12366. [PubMed] [Google Scholar]
- Balboa MA, Insel PA. Nuclear phospholipase D in Madin-Darby canine kidney cells. Guanosine 5′-O-(thiotriphosphate)-stimulated activation is mediated by RhoA and is downstream of protein kinase C. J Biol Chem. 1995;270:29843–29847. doi: 10.1074/jbc.270.50.29843. [DOI] [PubMed] [Google Scholar]
- Bazenet CE, Ruano AR, Brockman JL, Anderson RA. The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J Biol Chem. 1990;265:18012–18022. [PubMed] [Google Scholar]
- Boronenkov IV, Anderson RA. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem. 1995;270:2881–2884. doi: 10.1074/jbc.270.7.2881. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Anderson RA. Casein kinase I is regulated by phosphatidylinositol 4,5-bisphosphate in native membranes. J Biol Chem. 1991;266:2508–2512. [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino AM, Parker GJ, Boronenkov IV, Anderson RA, Chao MV. A novel interaction between the juxtamembrane region of the p55 tumor necrosis factor receptor and phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem. 1997;272:5861–5870. doi: 10.1074/jbc.272.9.5861. [DOI] [PubMed] [Google Scholar]
- Cocco L, Gilmour RS, Ognibene A, Letcher AJ, Manzoli FA, Irvine RF. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J. 1987;248:765–770. doi: 10.1042/bj2480765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S. Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. Bioessays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Collas P, Thompson L, Fields AP, Poccia DL, Courvalin JC. Protein kinase C-mediated interphase lamin B phosphorylation and solubilization. J Biol Chem. 1997;272:21274–21280. doi: 10.1074/jbc.272.34.21274. [DOI] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Davis L, Cadrin M, Brown DL, Chaly N. Reversible disassembly of transcription domains in lymphocyte nuclei during inhibition of RNA synthesis by DRB. Biol Cell. 1993;78:163–180. doi: 10.1016/0248-4900(93)90127-z. [DOI] [PubMed] [Google Scholar]
- De Vries KJ, Westerman J, Bastiaens PI, Jovin TM, Wirtz KW, Snoek GT. Fluorescently labeled phosphatidylinositol transfer protein isoforms (alpha and beta), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp Cell Res. 1996;227:33–39. doi: 10.1006/excr.1996.0246. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991;10:3207–3214. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF. Inositides and the nucleus and inositides in the nucleus. Cell. 1993;74:405–407. doi: 10.1016/0092-8674(93)80041-c. [DOI] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Treagus JE, Vann L, Irvine RF, D’Santos C. Nuclear diacylglycerol, the cell cycle, the enzymes and a red herring (or how we came to love phosphatidylcholine) Biochem Soc Trans. 1997;25:571–575. doi: 10.1042/bst0250571. [DOI] [PubMed] [Google Scholar]
- Divecha N, Truong O, Hsuan JJ, Hinchliffe KA, Irvine RF. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem J. 1995;309:715–719. doi: 10.1042/bj3090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J Cell Biol. 1984;98:358–363. doi: 10.1083/jcb.98.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fukami K, Matsuoka K, Nakanishi O, Yamakawa A, Kawai S, Takenawa T. Antibody to phosphatidylinositol 4,5-bisphosphate inhibits oncogene-induced mitogenesis. Proc Natl Acad Sci USA. 1988;85:9057–9061. doi: 10.1073/pnas.85.23.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami K, Takenawa T. Quantitative changes in polyphosphoinositides 1,2-diacylglycerol and inositol 1,4,5-trisphosphate by platelet-derived growth factor and prostaglandin F2 alpha. J Biol Chem. 1989;264:14985–14989. [PubMed] [Google Scholar]
- Goss VL, Hocevar BA, Thompson LJ, Stratton CA, Burns DJ, Fields AP. Identification of nuclear beta II protein kinase C as a mitotic lamin kinase. J Biol Chem. 1994;269:19074–19080. [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Hardin JA, Lerner MR, Lerner EA, Steitz JA. New directions in antinuclear antibody research: the Sm, RNP, Ro, and La antigens are found on small-RNA protein particles. Am J Kidney Dis. 1982;2:98–100. [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Matsumoto S, Yahara I. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct. 1992;17:39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J Biol Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- Itoh T, Takeshi I, Takenawa T. A novel phosphatidylinositol-5-phosphate 4-kinase (phosphatidylinositol-phosphate kinase IIγ) is phosphorylated in the endoplasmic reticulum in response to mitogenic signals. J Biol Chem. 1998;273:20292–20299. doi: 10.1074/jbc.273.32.20292. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- Kuriki H, Tamiya-Koizumi K, Asano M, Yoshida S, Kojima K, Nimura Y. Existence of phosphoinositide-specific phospholipase C in rat liver nuclei and its change during liver regeneration. J Biochem. 1992;111:283–286. doi: 10.1093/oxfordjournals.jbchem.a123750. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Liu N, Fukami K, Yu H, Takenawa T. A new phospholipase C delta 4 is induced at S-phase of the cell cycle and appears in the nucleus. J Biol Chem. 1996;271:355–360. doi: 10.1074/jbc.271.1.355. [DOI] [PubMed] [Google Scholar]
- Loijens JC, Anderson RA. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem. 1996;271:32937–32943. doi: 10.1074/jbc.271.51.32937. [DOI] [PubMed] [Google Scholar]
- Loijens JC, Boronenkov IV, Parker GJ, Anderson RA. The phosphatidylinositol 4-phosphate 5-kinase family. Adv Enzyme Regul. 1996;36:115–140. doi: 10.1016/0065-2571(95)00005-4. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Hsu AL, Wang DS, Yan HY, Yin HL, Chen CS. Phosphoinositide 3-kinase in rat liver nuclei. Biochemistry. 1998;37:5738–5745. doi: 10.1021/bi972551g. [DOI] [PubMed] [Google Scholar]
- Malviya AN, Rogue PJ. “Tell me where is calcium bred”: clarifying the roles of nuclear calcium. Cell. 1998;92:17–23. doi: 10.1016/s0092-8674(00)80895-8. [DOI] [PubMed] [Google Scholar]
- Maraldi NM, Cocco L, Capitani S, Mazzotti G, Barnabei O, Manzoli FA. Lipid-dependent nuclear signalling: morphological and functional features. Adv Enzyme Regul. 1994;34:129–143. doi: 10.1016/0065-2571(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Maraldi NM, Zini N, Ognibene A, Martelli AM, Barbieri M, Mazzotti G, Manzoli FA. Immunocytochemical detection of the intranuclear variations of phosphatidylinositol 4,5-bisphosphate amount associated with changes of activity and amount of phospholipase C beta 1 in cells exposed to mitogenic or differentiating agonists. Biol Cell. 1995;83:201–210. doi: 10.1016/0248-4900(96)81309-8. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Gilmour RS, Bertagnolo V, Neri LM, Manzoli L, Cocco L. Nuclear localization and signalling activity of phosphoinositidase C beta in Swiss 3T3 cells. Nature. 1992;358:242–245. doi: 10.1038/358242a0. [DOI] [PubMed] [Google Scholar]
- Matter N, Ritz MF, Freyermuth S, Rogue P, Malviya AN. Stimulation of nuclear protein kinase C leads to phosphorylation of nuclear inositol 1,4,5-trisphosphate receptor and accelerated calcium release by inositol 1,4,5-trisphosphate from isolated rat liver nuclei. J Biol Chem. 1993;268:732–736. [PubMed] [Google Scholar]
- Matuoka K, Fukami K, Nakanishi O, Kawai S, Takenawa T. Mitogenesis in response to PDGF and bombesin abolished by microinjection of antibody to PIP2. Science. 1988;239:640–643. doi: 10.1126/science.2829356. [DOI] [PubMed] [Google Scholar]
- Meier R, Alessi DR, Cron P, Andjelkovic M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase B beta. J Biol Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Umeda M, Horikoshi T, Yanagisawa K, Yoshioka T, Inoue K. Production and characterization of monoclonal antibodies that bind to phosphatidylinositol 4,5-bisphosphate. Mol Immunol. 1988;25:1025–1031. doi: 10.1016/0161-5890(88)90010-7. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes & Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- Onoda K, Yin HL. gCap39 is phosphorylated. Stimulation by okadaic acid and preferential association with nuclei. J Biol Chem. 1993;268:4106–4112. [PubMed] [Google Scholar]
- Payrastre B, Nievers M, Boonstra J, Breton M, Verkleij AJ, Van Bergen en Henegouwen PM. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J Biol Chem. 1992;267:5078–5084. [PubMed] [Google Scholar]
- Pombo A, Cook PR. The localization of sites containing nascent RNA and splicing factors. Exp Cell Res. 1996;229:201–203. doi: 10.1006/excr.1996.0360. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate (see comments) Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol, 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH, Green MR. Compartmentalization of eukaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Smith CD, Wells WW. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J Biol Chem. 1983;258:9368–9373. [PubMed] [Google Scholar]
- Spector DL. Nuclear organization and gene expression. Exp Cell Res. 1996;229:189–197. doi: 10.1006/excr.1996.0358. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Sun B, Murray NR, Fields AP. A role for nuclear phosphatidylinositol-specific phospholipase C in the G2/M phase transition. J Biol Chem. 1997;272:26313–26324. doi: 10.1074/jbc.272.42.26313. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Rameh LE, Ishihara H, Shibasaki Y, Chen J, Prestwich GD, Cantley LC, Carpenter CL. Type I phosphatidylinositol-4-phosphate 5-kinases synthesize the novel lipids phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5-phosphate. J Biol Chem. 1998;273:18040–18046. doi: 10.1074/jbc.273.29.18040. [DOI] [PubMed] [Google Scholar]
- Vann LR, Wooding FB, Irvine RF, Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem J. 1997;327:569–576. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan KM, Nickerson JA, Krockmalnic G, Penman S. The B1C8 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. Proc Natl Acad Sci USA. 1994;91:594–598. doi: 10.1073/pnas.91.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Majerus PW. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–7850. [PubMed] [Google Scholar]
- York JD, Saffitz JE, Majerus PW. Inositol polyphosphate 1-phosphatase is present in the nucleus and inhibits DNA synthesis. J Biol Chem. 1994;269:19992–19999. [PubMed] [Google Scholar]
- Yu H, Fukami K, Watanabe Y, Ozaki C, Takenawa T. Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur J Biochem. 1998;251:281–287. doi: 10.1046/j.1432-1327.1998.2510281.x. [DOI] [PubMed] [Google Scholar]
- Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Loijens JC, Boronenkov IV, Parker GJ, Norris FA, Chen J, Thum O, Prestwich GD, Majerus PW, Anderson RA. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J Biol Chem. 1997;272:17756–17761. doi: 10.1074/jbc.272.28.17756. [DOI] [PubMed] [Google Scholar]
- Zini N, Martelli AM, Cocco L, Manzoli FA, Maraldi NM. Phosphoinositidase C isoforms are specifically localized in the nuclear matrix and cytoskeleton of Swiss 3T3 cells. Exp Cell Res. 1993;208:257–269. doi: 10.1006/excr.1993.1245. [DOI] [PubMed] [Google Scholar]
- Zini N, Ognibene A, Bavelloni A, Santi S, Sabatelli P, Baldini N, Scotlandi K, Serra M, Maraldi NM. Cytoplasmic and nuclear localization sites of phosphatidylinositol 3-kinase in human osteosarcoma sensitive and multidrug-resistant Saos-2 cells. Histochem Cell Biol. 1996a;106:457–464. doi: 10.1007/BF02473307. [DOI] [PubMed] [Google Scholar]
- Zini N, Sabatelli P, Faenza I, Ognibene A, Maraldi NM. Interleukin-1 alpha induces variations of the intranuclear amount of phosphatidylinositol 4,5-bisphosphate and phospholipase C beta 1 in human osteosarcoma Saos-2 cells. Histochem J. 1996b;28:495–504. doi: 10.1007/BF02331409. [DOI] [PubMed] [Google Scholar]