Abstract
Land plants evolved a long-distance transport system of water and nutrients composed of the xylem and phloem, both of which are generated from the procambium- and cambium-comprising vascular stem cells. However, little is known about the molecular mechanism of cell communication governing xylem–phloem patterning. Here, we show that a dodecapeptide (HEVHypSGHypNPISN; Hyp, 4-hydroxyproline), TDIF (tracheary element differentiation inhibitory factor), is secreted from the phloem and suppresses the differentiation of vascular stem cells into xylem cells through a leucine-rich repeat receptor-like kinase (LRR-RLK). TDIF binds in vitro specifically to the LRR-RLK, designated TDR (putative TDIF receptor), whose expression is restricted to procambial cells. However, the combined analysis of TDIF with a specific antibody and the expression profiles of the promoters of two genes encoding TDIF revealed that TDIF is synthesized mainly in, and secreted from, the phloem and its neighboring cells. The observation that TDIF is capable of promoting proliferation of procambial cells while suppressing xylem differentiation suggests that this small peptide functions as a phloem-derived, non-cell-autonomous signal that controls stem cell fate in the procambium. Our results indicate that we have discovered a cell communication system governing phloem–xylem cross-talk.
Keywords: CLV3/ESR-related (CLE), leucine-rich repeat receptor-like kinase, phloem, procambium, xylem
Higher eukaryotes have evolved specialized stem cells that can either remain undifferentiated or differentiate into specialized cells. Plant stem cells are found in meristem tissue localized at the tips of roots and shoots (root and shoot apical meristem; RAM and SAM) and in the vascular system (procambium/cambium or vascular meristem) (1). Stem cells in each meristem play a pivotal role in the continuous formation of various tissues during postembryonic development. The fate of stem cells in the SAM and the RAM is controlled by the transcription factors WUSCHEL (WUS) (2) and WUSCHEL-related homeobox 5 (WOX5) (3), respectively. In SAM, the expression of WUS is regulated by the CLAVATA (CLV) system involving CLV3, a member of the CLV3/ESR-related (CLE) family and the CLV1 (LRR-LRK)/CLV2 (a leucine-rich repeat receptor-like protein, LRR-RLP) complex (4–9). In the vascular meristem, a few layers of stem cells proliferate, and their progeny differentiate into apposing xylem and phloem cells (10, 11). Some transcription factors are known to function as transcription switches for xylem cell differentiation (12–14) or to confer phloem cell identity (15). However, little is known about the regulation of the fate of the vascular stem cells (16, 17). We recently isolated the CLE peptide TDIF (tracheary element differentiation inhibitory factor) from a xylogenic Zinnia cell culture (18) and showed that this TDIF can suppress the differentiation of cultured cells into the tracheary element (TE), a component of the xylem in culture (19). In Arabidopsis, two genes, CLE41 and CLE44, encode TDIF. These findings suggest that the fate of vascular stem cells may also be controlled by a CLE receptor system, reminiscent of that of CLAVATA, comprising CLV3, CLV1, and CLV2 (17).
Results
TDIF Promotes Procambial Cell Proliferation While Suppressing Differentiation of These Cells into Xylem Vessel Cells in Planta.
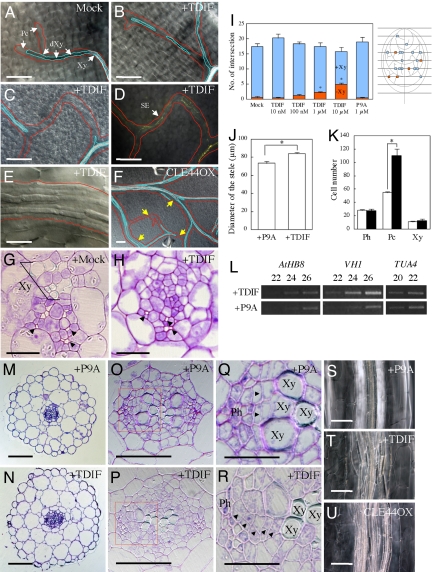
First, to understand how TDIF influences plant development, Arabidopsis seeds were germinated and grown in a liquid medium containing TDIF. TDIF did not significantly affect the overall plant growth. However, TDIF inhibited xylem vessel formation, especially in high-order veins in leaves (Fig. 1 A–D, G, and H), although the phloem differentiation process proceeded normally (Fig. 1 C and D). In contrast, the formation of procambial cells was enhanced in the veins (Fig. 1 E, G, and H). Quantification of discontinuous xylem vessels in the veins indicated that this effect is positively dose dependent on TDIF, whereas a nonfunctional dodecapeptide (P9A), in which proline at the ninth position is replaced by alanine (19), has no effect (Fig. 1I). TDIF application also led to increased stele size in the hypocotyls (Fig. 1 J, N, and P), an effect caused by increased proliferation of the procambial cells between the xylem vessel and the phloem (Fig. 1 K and R). P9A did not promote proliferation of any cells (Fig. 1 J, M, O, and Q). The plane of division in the proliferating procambial cells was formed at random in TDIF-treated hypocotyls (Fig. 1R, arrowheads), whereas procambial cells divided periclinally in P9A-treated hypocotyls (Fig. 1Q, arrowheads). TDIF-induced procambium proliferation often caused secondarily ectopic xylem vessel formation (Fig. 1 S and T). Consistently, TDIF promoted the expression of two procambium-specific marker genes, AtHB8 (A. thaliana homeobox 8; ref. 20) and VH1 (VASCULAR HIGHWAY 1; ref. 21), which correlated with the observed increase in the number of procambial cells (Fig. 1L). These results collectively indicate that TDIF can promote procambial cell proliferation while suppressing the cells' differentiation into xylem vessel cells in planta. Similar phenotypes, including discontinuous formation of xylem vessels (Fig. 1F) and stele enlargement (Fig. 1U), were also observed in transgenic plants overexpressing CLE44. The CLE44-overexpressing plants showed bushy-like appearance (data not shown).
Fig. 1.
Vascular phenotypes caused by excess TDIF. (A–H) TDIF suppresses xylem vessel differentiation in the leaf vein. Pc, procambium; Xy, xylem vessel; dXy, differentiating Xy. (A and G) No peptide application. (B–E) Application of 1 μM TDIF. (H) Application of 10 μM TDIF. (B and C) Discontinuous xylem vessel. (D) Sieve elements (SEs) visualized by aniline blue in the same position as C. (E) Proliferated procambium cells in the region in which TE formation was suppressed. (F) Discontinuous xylem vessel formation in leaves of a CLE44-overexpressing plant; the yellow arrows show the non-TE region in the veins. Veins are outlined in red, and xylem vessels are outlined in blue. (G and H) Sections of the vein; xylem vessels are not often formed in veins of TDIF-treated seedlings. Arrowheads show SEs. (I) Dose-dependent suppression of TE formation in the veins by TDIF. (Right) A schematic illustration of the quantification method. The presence (+Xy) or absence (−Xy) of the xylem vessels was examined at the position where arbitrary transverse lines and veins of a high order crossed. (Left) Based on the method, veins with discontinuous xylem vessel strands were quantified in the presence of TDIF or P9A, an inactive derivative of TDIF. Blue boxes indicate veins with xylem vessels; red boxes, veins without xylem vessels. (J) TDIF enlarges the stele of hypocotyls. (K) The number of procambial cells but not phloem or xylem cells is increased by TDIF application. White boxes indicate P9A application; black boxes, TDIF application. (L) Procambium marker genes (AtHB8 and VH1) were up-regulated by TDIF application. (M–U) TDIF promotes procambial cell proliferation in the hypocotyl. (M, O, Q, and S) Application of 1 μM P9A. (N, P, R, and T) Application of 1 μM TDIF. (M–R) Toluidine blue-stained transverse sections of hypocotyls of 7-day-old Arabidopsis seedlings. (O and P) Magnification of the stele. (Q and R) Magnification of the boxed area in O and P. Arrowheads indicate the division plane of newly divided cells. (S–U) Hypocotyls of 10-day-old seedlings that were treated with P9A and TDIF and that were overexpressing CLE44. TDIF enhanced random division of procambium cells. (Scale bars: A–D, F, M, N, and S–U, 100 μm; E, G, H, Q, and R, 20 μm; O and P, 50 μm.) Error bars indicate SEM, n = 6 (I), 18 (J), and 7 (K). Asterisks indicate significant differences from mock/P9A application by Student's t test (P < 0.01).
Identification of a Putative Receptor (TDR) for TDIF.
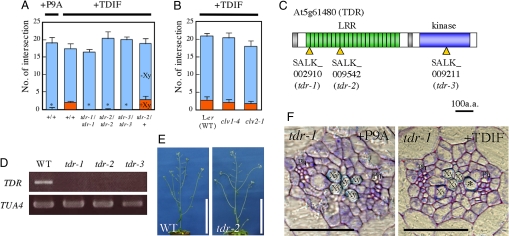
The above observations have motivated us to identify the corresponding receptor by screening mutants that would show insensitivity to TDIF in vascular formation in leaves and hypocotyls. Because the CLV3 receptor CLV1 is an LRR-RLK (1, 5–9), by analogy we expected that a receptor for TDIF might also be an LRR-RLK or an LRR-RLP. In the Arabidopsis genome, more than 200 LRR-RLK and more than 50 LRR-RLP genes have been annotated (22). From 250, we selected 56 genes [supporting information (SI) Table S1] that are predicted to be expressed in the procambium based on the public database AtGenExpress and a gene profiling study in differentiating Zinnia cells (23), and that are homologous to the predicted procambium-associated genes, because the aforementioned results had suggested that procambial cells are the targets of TDIF. We analyzed ≈130 T-DNA insertion lines of the genes and tested their sensitivity to TDIF. Only three T-DNA insertion lines exhibited TDIF-insensitive phenotypes; that is, in the presence of TDIF they did not have veins with discontinuous xylem vessels in the leaves (Fig. 2A). The three lines had a T-DNA insertion at three different positions of the same gene, At5g61480 (Fig. 2C). Therefore, we designated this gene TDR (putative TDIF receptor gene) and the three alleles as tdr-1, tdr-2, and tdr-3. The TDR gene encodes an LRR-RLK belonging to the subclass XI, which includes CLV1 (22) (Fig. S1). Genotyping revealed that plants homozygous for any tdr allele exhibit insensitivity to TDIF (Fig. 2A). The T-DNA insertions in both the tdr-1 and tdr-2 alleles are in the same exon near the 5′ end of the gene, and the TDR transcript is below the detectable level (Fig. 2D), suggesting that these are null alleles. In contrast to the tdr mutants, clv1–4 and clv2–1 are unresponsive to CLV3 in the shoots (24) but remain sensitive to TDIF (Fig. 2B). This result suggests that CLV1 and CLV2 are not involved in the TDIF signaling pathway. No T-DNA insertion line from the other 25 members of the LRR-RLK subclass XI, including two closely related homologues of TDR, was insensitive to TDIF. These results suggest that TDR is involved specifically in the TDIF signaling pathway. Recently, Fisher and Turner (25) reported that pxy-1 (phloem intercalated with xylem-1), which may act as an antimorphic allele, induces smaller procambium and phloem adjacent to the xylem in flower stalks. Because this mutation occurred at At5g61480, the TDR locus was the same as that already published as PXY.
Fig. 2.
Identification of a putative TDIF receptor. (A) Each of three homozygous alleles in an LRR-RLK gene (tdr) resulted in xylem vessel formation that was insensitive to exogenous TDIF. (B) Xylem vessel formation in clv1 and clv2 was TDIF sensitive. Error bars indicate SEM (n = 6). Asterisks indicate that the ratios of the number of non-TE positions to all examined positions were significantly different compared with wild type (WT) treated with 1 μM TDIF by Student's t test (P < 0.01). (C) Predicted TDR protein structure and positions of T-DNA insertions. (D) Levels of TDR mRNA in tdr-1, tdr-2, and tdr-3 measured with RT-PCR. (E) A 5-week-old tdr plant. (F) A transverse section of the hypocotyl of a 7-day-old tdr seedling grown in the presence of P9A or TDIF peptide. Note that phloem cells (Ph) are located close to xylem vessel cells (Xy) and, in an extreme case, are adjacent to a xylem vessel cell (asterisk). (Scale bars: E, 10 cm; F, 50 μm.)
The overall morphology of the tdr mutant plants appeared normal (Fig. 2E). However, the number of procambial cells in tdr-1 hypocotyls (31.3 ± 2.8, n = 7) was significantly lower than that in wild-type hypocotyls (55.1 ± 3.0, n = 7), although there were no significant differences in the numbers of xylem (10.9 ± 0.4 vs. 11.1 ± 1.1) and phloem (27.4 ± 1.0 vs. 28.1 ± 1.4) cells between tdr and wild-type hypocotyls. In such hypocotyls, xylem vessel cells were sometimes formed adjacent to the phloem (Fig. 2F, asterisk). TDIF did not enhance procambial proliferation in tdr hypocotyls (Fig. 2F).
TDIF Binds Specifically to TDR.
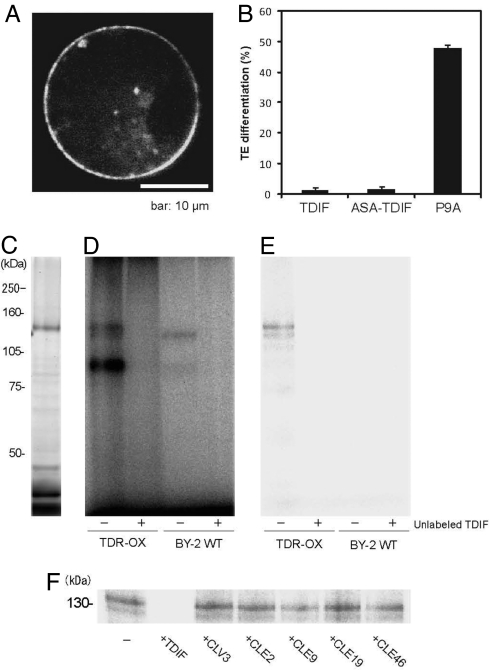
To test the idea that TDR is the receptor of the TDIF ligand, we first examined the subcellular localization of the TDR protein. The chimeric gene p35S::TDR-GFP was transiently expressed by transfection into Arabidopsis protoplasts. Fluorescence was detected primarily on the plasma membrane, but some signals were also observed in intracellular vesicles (Fig. 3A). Next, we examined the direct interaction between TDIF and TDR according to Ogawa et al. (9). For photoaffinity labeling, [(4-azidosalicyl)Lys2]TDIF (ASA-TDIF) was synthesized and purified. ASA-TDIF is as efficient as the unmodified TDIF in inhibiting TE differentiation in Zinnia culture (Fig. 3B). We expressed in tobacco BY2 cells a TDR gene encoding a variant in which the cytoplasmic kinase domain was replaced by the HaloTag (TDR-ΔKD-HT; Promega). The variant can be detected as a 130-kDa protein from the plasma membrane fraction (Fig. 3C). Incubation of the membrane fractions with a photoactivatable 125I-labeled ASA-TDIF followed by cross-linking by UV irradiation resulted in labeling of the 130-kDa and 120-kDa bands and a broad 90-kDa band (Fig. 3D). Immunoprecipitation with an anti-HaloTag antibody confirmed that the 130-kDa protein is the TDR-ΔKD-HT (Fig. 3E). The 130-kDa band disappeared after the membrane fraction was preincubated with excess TDIF (Fig. 3D), but not significantly after preincubation with CLE46, CLV3, CLE2, CLE9, CLE19, or P9A peptides, which are structurally related to TDIF (Fig. 3F and Fig. S2). These results indicate that TDIF binds TDR in a highly specific manner.
Fig. 3.
Direct interaction between TDR and TDIF. (A) Subcellular localization of TDR. (B) Suppression of TE differentiation in Zinnia cell culture. TDIF, ASA-TDIF, and the nonfunctional peptide P9A are shown. (C) Visualization of TDR-ΔKD-HT in transgenic tobacco BY-2 cells overexpressing TDR-ΔKD-HT. (D) Photoaffinity labeling of TDR-ΔKD-HT by 125I-ASA-TDIF in the absence (−) or presence (+) of excess unlabeled TDIF in microsomal fractions from TDR-ΔKD-HT-overexpressing cells (TDR-OX) or nontransformed tobacco BY2 cells (BY2-WT). (E) Immunoprecipitation with an anti-HaloTag antibody for the photoaffinity-labeled samples used in D. (F) Photoaffinity labeling of TDR-ΔKD-HT by 125I-ASA-TDIF in the absence (−) or presence of excess unlabeled TDIF, CLV3, CLE2, CLE9, CLE19, or CLE46 peptide.
Spatial Relationship Between TDIF and TDR.
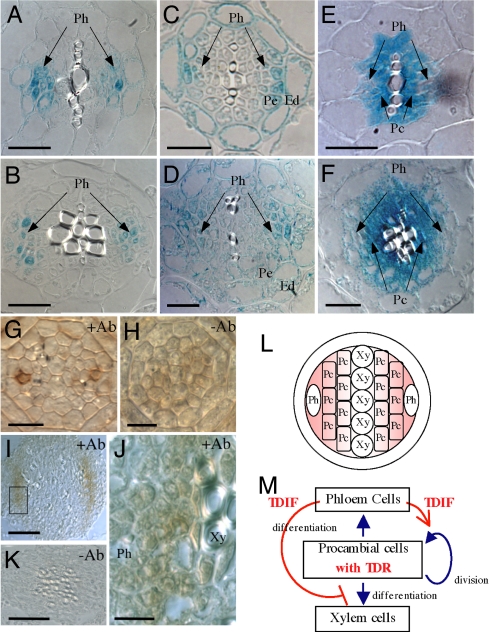
To determine the tissue-specific expression of TDR and TDIF, we expressed them as promoter–β-glucuronidase (GUS) reporter gene fusions (pTDR::GUS, pCLE41::GUS, and pCLE44::GUS) (Fig. 4 A–F and Fig. S3). pCLE41::GUS activity was observed along the vascular strands in all organs examined, such as cotyledons, leaves, and roots, but not in SAM (Fig. S3 A, D, G, and J). Its expression was restricted to the phloem and the neighboring pericycle cells in the roots and hypocotyls (Fig. 4 A and B). pCLE44::GUS was expressed more widely along the vascular strands than was pCLE41::GUS (Fig. S3 B, E, H, and K). In the roots and hypocotyls, pCLE44::GUS expression occurred in endodermal cells as well as cells in the phloem and the adjacent pericycle (Fig. 4 C and D). pTDR::GUS was also expressed along the vascular bundles in all organs of seedlings (Fig. S3 C, F, I, and L). Cross-sections showed procambial cell-specific expression of TDR in the roots and hypocotyls (Fig. 4 E and F and Fig. S3M). To localize the TDIF peptide in situ, we raised an antibody against TDIF. The affinity-purified antibody specifically recognized TDIF (CLE41/CLE44 peptide) and weakly recognized the CLE42 peptide, which has an amino acid replacement and can suppress xylem formation (Fig. S4). In the root tip, the antibody to TDIF stained the apoplast surrounding two phloem precursor cells in a specific manner (Fig. 4 G and H). In the hypocotyl, the antibody stained the region between the phloem cells and developing xylem cells (Fig. 4 I–K). These results support the idea that TDIF is secreted from the phloem cells and is distributed in the procambial region.
Fig. 4.
Vascular cell-specific localization of TDIF and TDR. (A–F) Distinctive vascular cell-specific expression of the CLE41, CLE44, and TDR genes in roots (A, C, and E) and hypocotyls (B, D, and F). (A and B) pCLE41::GUS. (C and D) pCLE44::GUS. (E and F) pTDR::GUS. Ph, phloem; Pc, procambium; Pe, pericycle; En, endodermis. (G–K) Immunohistochemical localization of the TDIF peptide in the root tip (G and H) and hypocotyls (I–K). (J) Magnification of the box in I. Note that the signal is detected around the phloem cells. (L and M) Positional (L) and functional (M) models of the TDIF/TDR signaling system, which regulates the fate of procambial cells in a non-cell-autonomous manner. (Scale bars: A–F, 20 μm; G and H, 10 μm; I and K, 50 μm; J, 10 μm.)
Discussion
A Non-Cell-Autonomous Model of Cell Fate Determination Between the Differentiation and Maintenance of Vascular Stem Cells.
Based on these data, we propose a non-cell-autonomous model of cell fate determination between the differentiation and maintenance of vascular stem cells (Fig. 4 L and M). Phloem and their neighboring cells produce and secrete TDIF. Procambial cells, which are vascular stem cells, perceive the TDIF signal through the TDR. The signal promotes maintenance of procambial cells through proliferation while inhibiting their differentiation into xylem cells. Our finding is the first indication of cell–cell communication being critical between xylem and phloem cells to form organized vascular tissues. ALTERED PHLOEM DEVELOPMENT (APL) gene, which encodes an MYB transcription factor, represses xylem development and promotes asymmetric cell division at the phloem side (15, 26). Therefore, the TDIF/TDR system might affect APL and suppress xylem differentiation. However, the ability of TDIF to activate proliferation of the procambium is not explained simply by APL. We therefore sought to identify the key transcription factor controlling the switching between cell proliferation and differentiation of vascular stem cells.
In this study, we discovered the TDIF/TDR system, which involves a member of the CLE peptide ligand family and its receptor belonging to the subclass XI of LRR-RLKs. This system plays a determining role in the fate of vascular stem cells in a non-cell-autonomous manner. The CLV3/CLV1 system, which governs stem cell fates in the SAM, is also composed of a CLE and a member of the LRR-RLK subclass XI. Therefore, different sets of CLE and the subclass XI of LRR-RLKs may determine the fate of different types of stem cells through a non-cell-autonomous mechanism. A phylogenetic tree drawn based on sequences of the LRR domain, to which CLE ligands would bind, reveals a clade comprising TDR/PXY, CLV1, BAM1 (barely any meristem 1), BAM2, BAM3, PXL1 (PXY-like 1), and PXL2, among 26 members of the subclass XI (Fig. S1). Mutation in BAM genes and PXL genes causes abnormality of the SAM or vascular tissues or both (25, 27). Therefore, five genes other than TDR/PXY and CLV1 in the same clade might also encode specific receptors for distinctive CLE peptides, which play determining roles in the fate of some stem cells in the meristem.
Materials and Methods
Observation of Vasculature.
Seeds of Arabidopsis thaliana mutants and the wild type were germinated in half-MS liquid medium and grown with or without chemically synthesized TDIF (HEVHypSGHypNPISN) or P9A (HEVHypSGHypNAISN) at 1 or 10 μM at 22°C under continuous light with shaking. Leaves were fixed in a 1:3 mixture of acetic acid/ethanol and mounted in a mixture of chloral hydrate/glycerol/water (8:1:2). Visualization of the sieve elements was performed as described previously (28). For sectioning, leaves, hypocotyls, and roots were fixed in 4% paraformaldehyde and embedded in Technovit 7100 (Heraeus Kulzer), and 3-μm- or 6-μm-thick sections were cut. The sections were stained with 0.02–0.05% toluidine blue O and observed under a light microscope (BX51; Olympus).
Subcellular Localization.
The TDR coding sequence was cloned into the pSHOI vector, a derivative of pHTS13 (29), to produce the construct of the translational fusion of TDR–GFP expressed under the cauliflower mosaic virus 35S promoter. Transient expression of the GFP fusion gene was performed as described previously (29). GFP fluorescence was detected with a confocal laser microscope (BX60; Olympus) equipped with a confocal scanner (Model CSU10; Yokogawa Electric).
Immunohistochemistry.
A rabbit antibody against TDIF/CLE41/CLE44 was generated against a synthetic peptide of TDIF (HEVHypSGHypNPISN) and purified with affinity chromatography. For immunohistochemical analysis, tissue samples were prepared from 5-day-old Arabidopsis seedlings as described previously (30). Immunohistochemical staining was performed using a Vectastain elite ABC kit according to the manufacturer's instructions (Vector Laboratories).
RT-PCR.
Total RNA was extracted from 2-week-old seedlings of wild-type and tdr mutant alleles for the detection of TDR expression, and from the leaves of 10-day-old wild-type seedlings for semiquantitative RT-PCR.
Preparation of Photoactivatable 125I-ASA-TDIF.
ASA-TDIF was synthesized and purified by the method shown in Ogawa et al. (9). ASA-TDIF was radioiodinated, and it was purified by reverse-phase HPLC to yield analytically pure 125I-ASA-TDIF with specific radioactivity of 200 Ci/mmol.
Bioassay for ASA-TDIF.
In vitro Zinnia cell culture was performed by using the method described by Motose et al. (31). Mesophyll cells were cultured in D medium with 100 pM ASA-TDIF, 100 pM TDIF, or 100 pM P9A peptide (mock). The TE differentiation rates were counted 80 h after the start of cell culture.
Expression of TDR-ΔKD-HT.
To overexpress TDR-ΔKD-HT in tobacco BY-2 cells, we used PCR to amplify the 2.3-kb genomic fragment of TDR, which corresponds to the Met-1 to Gly-726 region, and the 0.8-kb cDNA fragment of HaloTag (Promega), which contains the entire ORF of HaloTag. These two fragments were cloned in translational fusion by three-component ligation in an XbaI/SacI-digested binary vector, pBI121. The recombinant plasmid was introduced into Agrobacterium tumefaciens strain C58C1 (pMP90) by electroporation. BY-2 tobacco cells were cocultivated with the transformed Agrobacterium for 2 days. The transformed BY-2 cells were selected on MS agar medium containing 200 mg/liter kanamycin and 500 mg/liter carbenicillin for several weeks until transformed colonies were formed. Selected cell lines were transferred into MS liquid medium containing 100 mg/liter kanamycin to initiate the suspension culture. Six-day-old BY-2 cells (150 g fresh weight) were homogenized in a Waring blender (20,000 rpm for 5 minutes) in 200 ml of extraction buffer containing 25 mM Tris·HCl (pH 7.0), 10 mM MgCl2, 2 mM DTT, 2 μM leupeptin, 2 mM phenylmethanesulfonyl fluoride, and 250 mM sucrose at 4°C. The slurry was filtered and centrifuged at 10,000 × g for 15 minutes at 4°C. The supernatant was then centrifuged at 100,000 × g for 30 minutes at 4°C to give a pellet of the microsomal fractions. For the fluorescent visualization of the expressed TDR-ΔKD-HT, a 100-μg aliquot of microsomal fraction was suspended in 20 μl of suspension buffer (20 mM Hepes-KOH, pH 7.0) containing 5 μM HaloTag TMR reagent (Promega), followed by incubation at 37°C for 30 min. The sample was then separated by SDS/PAGE on a 7.5% acrylamide gel and analyzed in a Typhoon 9400 imager (GE Healthcare) with a 523-nm excitation filter and a 580-nm emission filter.
Photoaffinity Labeling.
Photoaffinity labeling was performed basically according to Ogawa et al. (9). Aliquots of microsomal proteins (250 μg) were suspended in 250 μl of binding buffer (50 mM Mes-KOH, pH 5.5, with 100 mM sucrose) containing 10 nM 125I-ASA-TDIF in the presence or absence of 3 μM P9A or10 μM other competitor peptides and then incubated for 15 min on ice. The bound and free 125I-ASA-TDIF were separated by layering the reaction mixture onto 900 μl of washing buffer (50 mM Mes-KOH, pH 5.5, with 500 mM sucrose) and centrifuging for 5 min at 100,000 × g at 4°C. The supernatant was discarded, and the pellet was irradiated on ice for 10 min with a UV lamp (model ENF-260C/J, 365 nm; Spectronics) at a distance of 1 cm. The cross-linked membrane proteins (250 μg) were solubilized with 100 μl of solubilization buffer (20 mM Hepes-KOH, pH 7.5, 150 mM KCl, and 1.0% Triton X-100). The lysate was kept on ice for 30 min and centrifuged for 30 min at 100,000 × g at 4°C to remove insoluble material. The supernatant was then incubated for 1 h with 5-μl aliquots of anti-HaloTag antibody (Promega) at 4°C. The resulting immune complexes were immunoprecipitated with 50-μl aliquots of protein A–Sepharose (GE Healthcare) for 1 h at 4°C, and the immunoprecipitated proteins were separated by SDS/PAGE on a 7.5% acrylamide gel. The dried gels were exposed to the bioimaging plate (MS 2025; Fujifilm) for 2 days at room temperature, and the plates were analyzed by using an imaging plate reader and bioimaging analyzer (BAS 5000; Fujifilm).
Supplementary Material
Acknowledgments.
We thank J. Leung for critical reading of this manuscript; H. Nakayama, T. Ueda, and A. Nakano for technical support; and T. Demura for the gift of Arabidopsis seeds harboring pTDR::GUS. This work was supported in part by Grants-in-Aid from the Ministry of Education, Sports, Culture, Science, and Technology, Japan (19060009 to H.F. and 19060010 to Y.M.), the Japan Society for the Promotion of Science (17207004 to H.F. and 18687003 to Y.M.), and the Program of Basic Research Activities for Innovative Biosciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808444105/DCSupplemental.
References
- 1.Simon R, Stahl Y. Botany. Plant cells CLEave their way to differentiation. Science. 2006;313:773–774. doi: 10.1126/science.1131833. [DOI] [PubMed] [Google Scholar]
- 2.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 4.Kondo T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 5.Diévart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- 6.Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiers M, Ku KL, Liu CM. CLE peptide ligands and their roles in establishing meristems. Curr Opin Plant Biol. 2007;10:39–43. doi: 10.1016/j.pbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Jun JH, Fiume E, Fletcher JC. The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci. 2008;65:743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 10.Roberts LW, Gahan PB, Aloni R. Vascular Differentiation and Plant Growth Regulators. Berlin: Springer; 1988. [Google Scholar]
- 11.Esau K. Anatomy of Seed Plants. 2nd Ed. New York: Wiley; 1977. [Google Scholar]
- 12.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda H. Signals that control plant vascular cell differentiation. Nat Rev Mol Cell Biol. 2004;5:379–391. doi: 10.1038/nrm1364. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda H, Hirakawa Y, Sawa S. Peptide signaling in vascular development. Curr Opin Plant Biol. 2007;10:477–482. doi: 10.1016/j.pbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda H, Komamine A. Establishment of an experimental system for the study of tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 1980;65:57–60. doi: 10.1104/pp.65.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito Y, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 20.Baima S, et al. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- 21.Clay NK, Nelson T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell. 2002;14:2707–2722. doi: 10.1105/tpc.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demura T, et al. Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA. 2002;99:15794–15799. doi: 10.1073/pnas.232590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 25.Fisher K, Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- 26.Carlsbecker A, Helariutta Y. Phloem and xylem specification: Pieces of the puzzle emerge. Curr Opin Plant Biol. 2005;8:512–517. doi: 10.1016/j.pbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 27.DeYoung BJ, et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 28.Carland FM, et al. Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell. 1999;11:2123–2137. doi: 10.1105/tpc.11.11.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mähönen AP, et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motose H, Sugiyama M, Fukuda H. An arabinogalactan protein(s) is a key component of a fraction that mediates local intercellular communication involved in tracheary element differentiation of zinnia mesophyll cells. Plant Cell Physiol. 2001;42:129–137. doi: 10.1093/pcp/pce014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.