Abstract
Forces generated by goldfish keratocytes and Swiss 3T3 fibroblasts have been measured with nanonewton precision and submicrometer spatial resolution. Differential interference contrast microscopy was used to visualize deformations produced by traction forces in elastic substrata, and interference reflection microscopy revealed sites of cell-substratum adhesions. Force ranged from a few nanonewtons at submicrometer spots under the lamellipodium to several hundred nanonewtons under the cell body. As cells moved forward, centripetal forces were applied by lamellipodia at sites that remained stationary on the substratum. Force increased and abruptly became lateral at the boundary of the lamellipodium and the cell body. When the cell retracted at its posterior margin, cell-substratum contact area decreased more rapidly than force, so that stress (force divided by area) increased as the cell pulled away. An increase in lateral force was associated with widening of the cell body. These mechanical data suggest an integrated, two-phase mechanism of cell motility: (1) low forces in the lamellipodium are applied in the direction of cortical flow and cause the cell body to be pulled forward; and (2) a component of force at the flanks pulls the rear margins forward toward the advancing cell body, whereas a large lateral component contributes to detachment of adhesions without greatly perturbing forward movement.
INTRODUCTION
Crawling cell locomotion involves protrusion at the leading edge, adhesion to the substratum, and retraction of the trailing edge (Harris, 1990; Lauffenburger and Horwitz, 1996). These movements require cells to generate mechanical forces, both internally, to overcome resistance to movement of the cytoskeletal and membrane systems, and externally, to overcome resistance provided by the aqueous medium, the extracellular matrix, and the cell’s own adhesions to the substratum. Studies of cell motility have frequently used epidermal keratocytes isolated from fish or amphibian skin as a model system owing to their ability to crawl with little change in shape or speed as a result of tight coupling between protrusion at the front and retraction at the rear (Bereiter-Hahn et al., 1981; Eutenuer and Schliwa, 1984; Theriot and Mitchison, 1991; Lee et al., 1993b). Keratocytes are also attractive because their large lamellipodia can locomote independently of the cell body (Eutenuer and Schliwa, 1984) and their ∼0.1-μm thickness makes them ideal for structural study by light and electron microscopy (Bereiter-Hahn et al., 1981; Eutenuer and Schliwa, 1984; Strohmeier and Bereiter-Hahn, 1984; Mittal and Bereiter-Hahn, 1985; Cooper and Schliwa, 1986; Bereiter-Hahn and Voth, 1988; Heath and Holifield, 1991a; Bereiter-Hahn and Luers, 1994; Small et al., 1995; Anderson et al., 1996; Svitkina et al., 1997).
Traction forces during keratocyte locomotion were first studied by Jacobson and colleagues (Lee et al., 1994; Oliver et al., 1995; Dembo et al., 1996) with the use of a modification of the method developed by Harris et al. (1980) in which cell forces applied to silicone rubber substrata produce deformations visible in the light microscope. Lee et al. (1994) were able to fabricate elastic substrata with higher compliance than those used in earlier studies on fibroblasts (Harris et al., 1980), allowing lower forces to be detected. In addition, strain in the elastic substratum was quantified by monitoring the movements of markers incorporated into its surface (Harris, 1984; Lee et al., 1994; Oliver et al., 1995; Dembo et al., 1996) rather than wrinkles, as had been used originally (Harris et al., 1980). Their results showed that lateral forces, directed inward toward the center of the cell, were dominant during forward locomotion (Lee et al., 1994; Oliver et al., 1995). Unexpectedly, lower forces at the front and rear edges appeared to be directed slightly outward, i.e., the bead movements suggested that these cells pushed forward at the leading edge of the lamellipodium and rearward at the trailing edge of the cell body (Dembo et al., 1996).
Recent structural data from keratocytes studied by light and electron microscopy (Small et al., 1995; Anderson et al., 1996; Svitkina et al., 1997) have been used to postulate that locomotive force might be generated by various mechanisms, including actin polymerization via an “elastic brownian ratchet” mechanism (Mogilner and Oster, 1996), myosin I activity (Small et al., 1993), contraction of transverse actin-myosin fibers (Anderson et al., 1996), and contraction of the actin-myosin meshwork along the boundary of the cell body and the lamellipodium (Svitkina et al., 1997). Tests of these proposals require measurements of traction forces with high spatial resolution over a wide range of magnitudes as well as correlation of these forces with close contacts between the cell and the substratum.
In the present study, we used an improved version of the Harris method in which the compliance of the silicone rubber substrata is increased in a controlled manner by illumination with UV light (Burton and Taylor, 1997). Therefore, the sensitivity of the substratum could be adjusted so that weak forces were detectable or strong forces produced small, localized wrinkles that were much smaller than the cell. The silicone used here also improves visualization of close contacts with the use of interference reflection microscopy (IRM). Two types of elastic deformation, wrinkles and substratum displacements, were monitored. The use of wrinkles has several advantages, including high spatial resolution (deformations over as little as ∼1 μm2), linearity in force (Burton and Taylor, 1997), force measurement from a single image (without reference to the unstressed substratum) based on the length of one wrinkle (without the need for iterative model fitting), and direct visualization of sites of force application. Wrinkles and substratum displacement both provide high temporal resolution with moderate deformations in the silicone rubber used here, approaching the unstressed configuration in ≤0.1 s. However, when the wrinkle pattern is dense and complex owing to high substratum compliance relative to cell strength, substratum displacement provides a more direct indicator of the direction of force. In addition, displacements of markers can be used to fit a traction field to integrate forces over an area (Oliver et al., 1995; Peterson, 1996).
Forces at discrete locations were revealed by submicrometer deformations and by larger wrinkles produced in the silicone substratum by the lamellipodium and the cell body. We confirm the earlier findings of Jacobson and colleagues that lateral forces are applied at the flanks of locomoting keratocytes; additionally, we report several new observations on the location, magnitude, and direction of forces applied by all regions of the cell, including lamellipodia, lateral flanks, and the cell body. Our data reveal a mechanism of cell locomotion in which the anterior regions of lamellipodia produce centripetal forces to pull the cell body forward, whereas forces generated along the lamellipodial boundary with the cell body act both to detach trailing flanks from their adhesions and to pull them inward and forward toward the cell body. These retraction forces are mainly perpendicular to the direction of motion in keratocytes and, therefore, perturb smooth forward movement only minimally.
Some of the results described here have been presented in preliminary form (Burton et al., 1996).
MATERIALS AND METHODS
Plating Keratocytes
Keratocytes from goldfish (Carassius auratus) were prepared essentially as described previously (Kolega, 1986). One minor modification was the replacement of Leibovitz’s L-15 medium with RPMI (Roswell Park Memorial Institute) growth medium (no. 31800, Life Technologies, Grand Island, NY) containing 10% fetal calf serum (FCS), because it was found that larger numbers of single keratocytes migrated from epidermal tissue adherent to scales that were placed onto microscope coverslips for observation (Lee et al., 1993b). The source of this improvement is not clear, but it might result from the presence in RPMI medium of glucose rather than galactose, glutathione (reduced), and the amino acids aspartic acid, hydroxyproline, and proline.
When cells were to be plated onto silicone rubber substrata (see below), keratocytes that had migrated from scales onto the glass coverslips were washed with RPMI medium without FCS followed by brief treatment with fish Ringer’s solution ([in mM] 112 NaCl, 2 KCl, 2.4 NaHCO3, 1 Tris, 1.5 MgCl) containing 0.05% trypsin and 0.53 mM EDTA to release cells into the medium. The keratocytes were then gently added to a Petri dish containing a silicone sheet submerged in RPMI medium plus 20% FCS. Within a few minutes, some cells settled and spread onto the silicone sheet. Keratocyte morphology and migration were similar on the silicone sheets and on glass. Serum proteins, along with extracellular matrix proteins secreted by the cells, presumably coated the silicone substrata and allowed the cells to adhere. Unattached cells and any remaining trypsin were washed away after 10–15 min, and the medium was then replaced by a fresh volume of RPMI medium plus 20% FCS. Cells prepared in this way were active for several hours at room temperature.
Preparation of Silicone Sheets
Silicone substrata were prepared from phenylmethyl polysiloxane (Dow Corning, Midland, MI; 710 fluid; viscosity = 500 centistokes). The silicone fluid (9 μl) was held in a square well bounded by four strips of glass (each 1.1 cm × ∼1 mm × ∼100 μm) cut from coverslips and glued (Norland Optical Adhesive, Norland Products, New Brunswick, NJ) onto a circular no. 0 coverslip (40 mm in diameter, thickness ∼ 110 μm) to form a chamber ∼1 cm on each side. The depth of the fluid varied from ∼20 μm in the center of the well to 100 μm at the edges. The surface of the fluid was vulcanized to produce silicone sheets by briefly applying heat with either a Bunsen burner flame, similar to Harris’ original method (Harris, 1984), or a hot tungsten wire. In the first method, coverslips containing the layer of silicone fluid were inverted and placed on a wire screen with a hole cut in the middle that was supported on a wire arm ∼9 cm above a 2.5-cm-diameter Bunsen burner. The wire arm was rotated by an electric motor in a horizontal plane to pass the coverslip through a fully oxygenated flame for a specific duration (∼1 s) at a given height.
Because temperature variation across the flame and its movements resulted in somewhat heterogeneous vulcanization, a more controllable method was sought. A suitable alternative was found to be use of a tungsten wire heated by electric current. This procedure was performed in a vacuum jar normally used for evaporation of carbon or metal onto specimens for electron microscopy, because low air pressure was required to avoid burning the filament. Pressure was reduced to ∼100 millitorr, but a higher vacuum could not be used because the silicone did not vulcanize, presumably because of a requirement for oxygen to cross-link the silicone polymer. The filament was 0.75 mm in diameter and shaped into a single arc or coil at a height 3–10 cm above the coverslip containing the silicone layer; greater heights reduced temperature variation across the silicone. The filament was heated by applying 40 V until it became white hot for ∼0.2 s, after which the voltage was reduced to zero. Current increased transiently to ∼50 A and then decreased to ∼42 A as the wire became white hot; steady current at 40 V was 38 A. Higher currents were not used because they caused the tungsten to be evaporated onto the coverslip. The thickness of the vulcanized layer can be increased by multiple applications of the heating protocol, allowing ∼30 s for the glass and silicone fluid to cool between applications. It is essential that the heating period be very brief, because if it is prolonged the temperature increases in the entire volume of silicone fluid, causing it to flow, round up, and otherwise disrupt the vulcanized surface layer.
The silicone is highly hydrophobic (Harris, 1984, 1988), with the current phenylmethyl polymer appearing to be even more so than the original dimethyl polymer. One important consequence of this is that surface tension between the medium and the rubber sheet is often greater than the strength of the sheet itself. To avoid destroying the sheet when initially adding medium, the coverslips containing silicone had to be rapidly immersed by first floating the coverslip on the surface of a large volume of medium (∼10 ml in a 60-mm Petri dish) and then using blunt forceps to rapidly (∼0.1 s) force the coverslip down into the medium, thus causing the meniscus to flow over the silicone sheet without pausing. Once the coverslips were immersed in the medium, all solution changes, mounting into the experimental chamber, and other maneuvers were carried out without allowing the solution surface to contact the rubber sheet, because this caused “explosive dewetting” and destruction of the sheet and cells (Harris, 1984, 1988).
The poor wetability of the silicone probably also explains the slower cell spreading on the rubber substrata than on glass (Harris, 1984, 1988), presumably as a result of the low affinity between the silicone and the serum proteins or extracellular matrix proteins required for cell-substratum adhesion. When a flame was used to vulcanize the silicone, the extent of cell spreading on the phenylmethyl silicone was similar to that on glass, although it was somewhat slower. The alternative method of hot-wire vulcanization, however, initially produced sheets to which few cells adhered and spread. The difference in the two methods appears to result from the “soot” deposited onto the silicone by the flame (a weakly oxygenated flame would increase this effect [Harris, 1982]), because cells adhered well when carbon was vacuum evaporated onto sheets vulcanized with a hot wire. Carbon evaporation was also observed to speed cell spreading onto flame-vulcanized sheets.
The phenylmethyl polysiloxane used here provided for more highly compliant substrata than the dimethyl polysiloxane used by Harris and colleagues. First, the silicone polymers are shorter, producing a less viscous fluid (500 centistokes compared with 12,000–60,000 centistokes for the dimethyl polysiloxane used previously) and more compliant sheets when vulcanized (Harris, 1988). Second, the phenyl side chains strongly absorb UV light (wavelength ≈ 254 nm), weakening the rubber layer and increasing compliance, as described previously (Burton and Taylor, 1997). The increased sensitivity to traction forces was necessary for measurements on keratocyte lamellipodia.
The compliance of the silicone substrata was calibrated by applying force to the sheets with flexible microneedles and comparing needle bending to wrinkle length, which was nearly linear in the range used (wrinkles of up to ∼100 μm long) (Burton and Taylor, 1997). “Wrinkle stiffness” was thus defined as force per unit of wrinkle length (nanonewton [nN]/μm, where 10 nN = 1 millidyne) (for measurement of wrinkle length, see below under Analysis of Deformations in Silicone Substrata). Microneedles, a few micrometers in diameter and 100–300 μm in length, were pulled by hand from 50-μm optical fiber softened with heat from a nichrome wire coil heated by electric current. Needle stiffness was measured by hanging glass beads of various weights (Sigma Chemical, St. Louis, MO) from the needles. Handles for the beads were fashioned from strings of metal loops cut from electron microscope grids with a razor blade. The handles were glued to the beads with the Norland optical cement mentioned above. Force was applied to sheets via adherent cells fixed with 1% glutaraldehyde (Burton and Taylor, 1997) by either maneuvering the needle against a cell or shifting the stage to move a cell against the needle. The entire length of the flexible needle was visible in the field of view (250 μm). Images were acquired before and after applying each of two to six different loads; substratum compliance was then calculated from wrinkle length and needle bending, as described previously (Burton and Taylor, 1997). Force produced by living cells was estimated from the lengths of single wrinkles or the longest of a group. All calibrated positions were >150 μm from the edges of the sheets, and wrinkle modulus was not observed to be a function of distance from the edge in these regions. All images of living cells were acquired from regions ≥4 mm from the edges of the sheets. Relative force was also judged qualitatively by other types of strain, including wrinkle density (number divided by area), wrinkle depth, as revealed by contrast in the image, and displacement of natural markers in the plane of the substratum (debris, irregular distortions produced by cells, and the wrinkles themselves).
To measure forces over a large range, silicone sheets were prepared with stiffnesses between ∼10 and 30 nN/μm. Although the most compliant substrata were deformed by the full range of forces reported here, stiffer substrata were valuable because (1) the strongest forces produced smaller wrinkles that were better isolated from one another (see Figures 1–4), (2) cell-substratum contacts in IRM images were less obscured by wrinkles (compare Figures 2 and 3 with Figures 5 and 6), and (3) forces were nearly isometric, with little displacement in the plane of the substratum (see Figures 2 and 5). Highly compliant substrata were valuable because of their greater force sensitivity, yielding both larger wrinkles and greater displacement of markers in the plane of the substratum. Such movements were used here to confirm the direction of forces inferred from the wrinkle patterns (see Figures 5–7, 9, and 10).
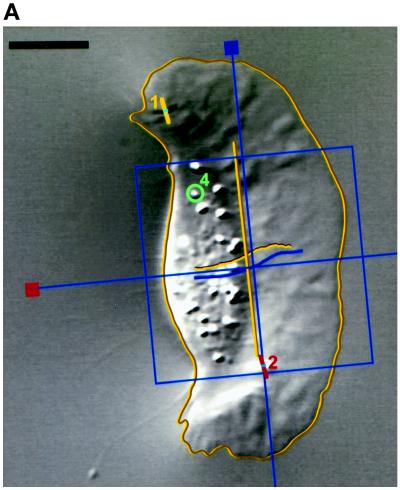
Figure 1.
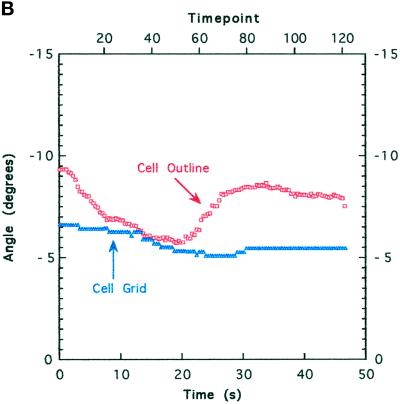
Image analysis. (A) Whole cells and features of interest in time-lapse sequences were tracked to quantitate position and orientation. Features were tracked with the use of numbered graphical objects: short bars with a dot in the middle were positioned at the center of compression wrinkles and oriented in the direction of force (orthogonal to the wrinkle); circles indicate the positions of small features such as organelles or small distortions in the elastic substratum. The positions and orientations of cells were tracked with MacWrinkle software to place a grid on the image or with MacDraw Pro and NIH Image to outline the cell (see MATERIALS AND METHODS). Movements are shown by trajectories drawn on the image. The grid and a long bar show the MacWrinkle and NIH Image estimates of location (near the front of the cell body in this image) and orientation, respectively. These estimates were similar, although the absolute centroids were offset. (B) Cell orientation (grid, triangles; outline, squares) versus time. The upper horizontal axis shows time point (128 images over 47 s). Cell orientation was calculated from the orientation of the grid (MacWrinkle) or from an ellipse fitted to the cell outline (NIH Image). The data in A and B were taken from the same image sequence as the data in Figure 2, A, D, and E. Bar, 10 μm.
Figure 4.
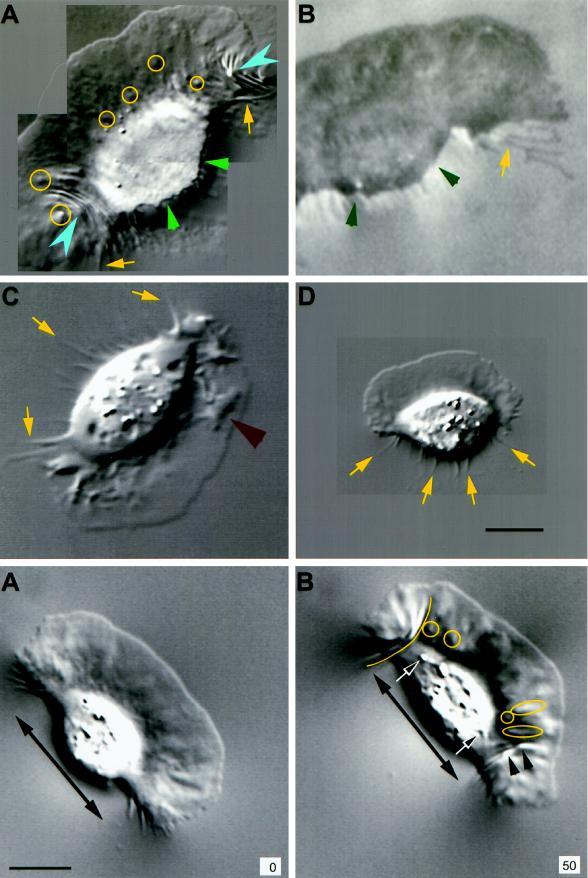
Cell body widening is associated with increased traction force. A cell that initially moved rapidly (A) with a flat lamellipodium and rounded cell body but then slowed (B) as it generated curved wrinkles at its flanks (force = 235 nN for the wrinkle traced at the left flank), force spots under the anterior lamellipodium (circles), and small wrinkles that extended from force spots (ovals and arrowheads). The cell body widened (double-headed arrow is the same length in A and B), and organelles (arrows) moved laterally when force increased (B). Time is shown in seconds. Bar, 10 μm.
Figure 2.
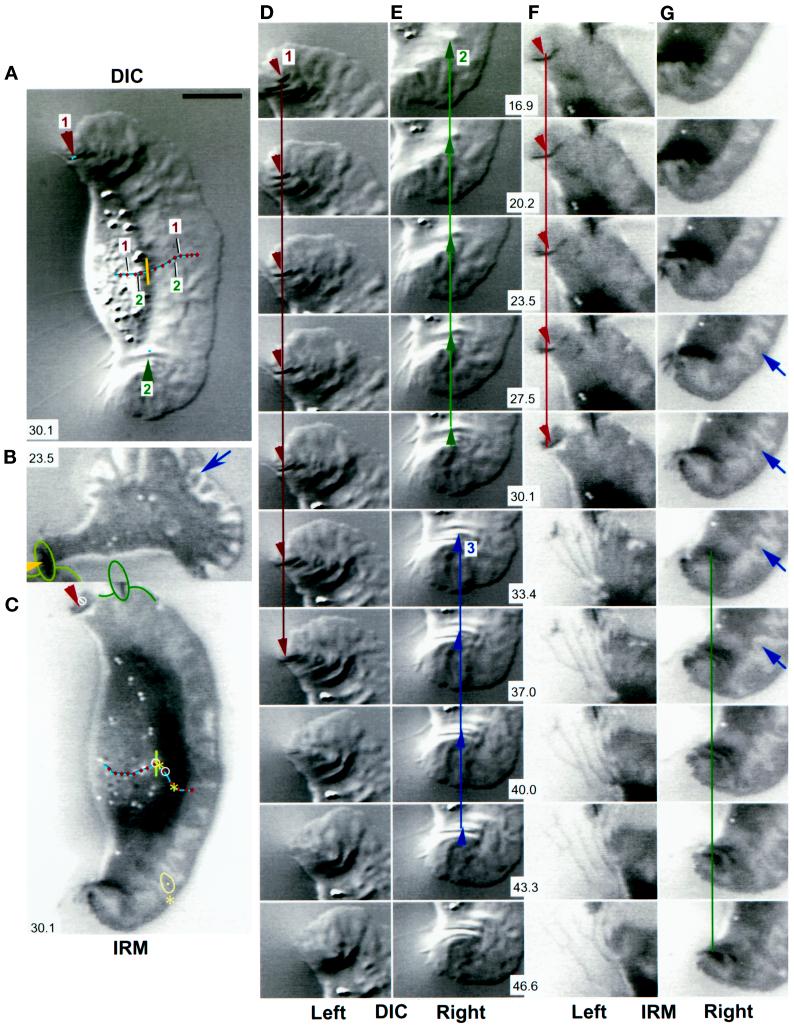
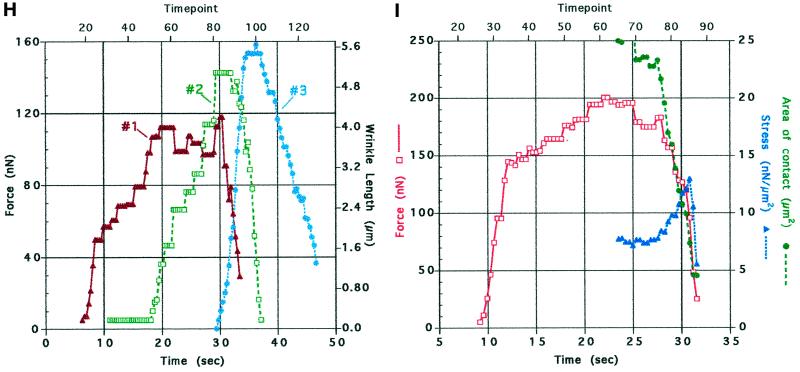
Traction force increases at discrete locations on the substratum. DIC (A, D, and E) and IRM (C, F, and G) images of the same keratocyte locomoting on a stiff substratum. Panels D and E and panels F and G show sequences of the left and right flanks from the full images in A and C, respectively. (B) A cell in which the left flank was stretched into a tail after the cell turned and moved slowly to the right. The cell in B was located just above that in C and F, and the wrinkle at the end of its tail, outlined in B and C, is also visible in F. Each image in A–G is the average of five video frames (0.165 s). The contrast in each image (A–C) or sequence (D–G) was independently adjusted in software to enhance the visibility of the features of interest. Numbers in A–C and E–G (center of each row) refer to time (seconds) elapsed since the cell was at the start of the respective trajectory shown in A and C. The IRM data were acquired 2 min and 10 s after the DIC data. A small vertical bar at the front of the cell body in A and C indicates the location and orientation of the reference used to track the cell’s trajectory (see Figure 1). The trajectories are marked at 3.7-s intervals. Arrowheads indicate the positions and directions of traction forces that produced curved wrinkles. In the DIC images (A, D, and E), force was directed 5–25 degrees forward of the lateral axis of the cell, indicating that the forward component of traction force was 10–40% of the total (see H below). Within measurement accuracy, wrinkles were stationary with respect to the field of view; the small lines on wrinkles 1 and 2 in A show that their displacement was much less than the cell’s movement during the same period (numbered markers on the trajectory in A). The close contact indicated by the arrow at the right front lamellipodium in G also moved only slightly (<0.5 μm, shown by a dot on the contact outlined at the asterisk in C), whereas the cell moved greatly (∼5 μm, shown by two asterisks on the cell trajectory; the average cell velocity before retraction was 20–30 μm/min). Two additional close contacts near the midline of the lamellipodium (at 16.9 and 43.3–46.6 s [data not shown]) also exhibited no measurable displacement. When the close contact at the left flank lifted up and retracted (note the bright region at the left flank in front of the wrinkle at 30.1 s), it changed shape so that it appeared to move (in C, the trajectory within the circle at the left flank shows the movement of the close contact during the period when the cell moved between the circles on its trajectory). In contrast, for the slowly moving tethered cell in B, close contacts made by the lamellipodium (arrow) showed rapid retrograde movement over the substratum (see text). Bar, 18 μm. (H) Traction force versus time for wrinkles 1–3 in A, D, and E. (I) Force, stress, and contact area at the retracting left flank in the IRM images (C, F, and G). Force was calculated from wrinkle length, and stress was calculated by dividing force by the area of close contact behind the wrinkle visible in the IRM image.
Figure 3.
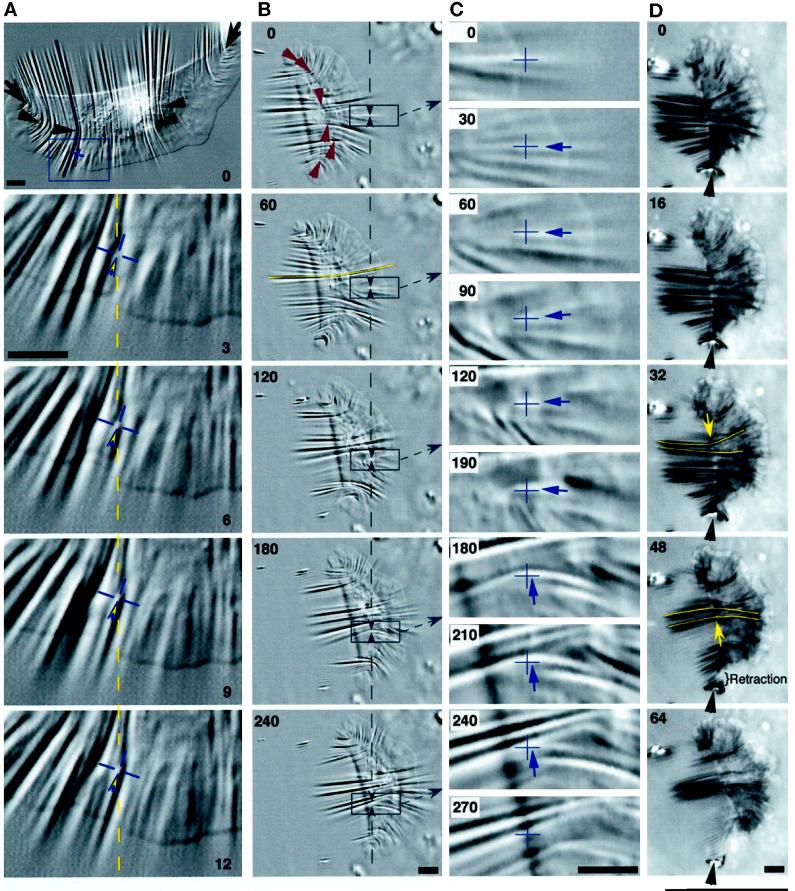
Forward movement is resisted by cell body–substratum interactions. Retraction fibers (yellow arrows) trailing behind the cell suggested that forward movement was resisted by adhesion between the substratum and the cell, including the cell body itself. The retraction fibers in the IRM image in B became bright as they rose upward from the substratum to the cell body. The cell body often had close contacts in IRM images that were as dark as those of the lamellipodium (B; see also Figures 2, 5, and 6), suggesting similar adhesion. DIC (A) and IRM (B) images of the same cell in which the posterior margin was sharply defined where it contacted the substratum (small green arrowheads), suggestive of a “lip” that adhered before releasing upward and forward into the cell body (partially out of focus in A). In A, substratum deformations are shown at high resolution (pixel spacing = 58 nm); yellow circles indicate small distortions (force spots), and blue arrowheads mark wrinkles. (C and D) In the absence of identifiable wrinkles, small force spots were not present, although lamellipodia exhibited ridges and bulges (C, large red arrowhead) or were nearly flat (high-resolution image in D). Bar, 10 μm.
Figure 5.
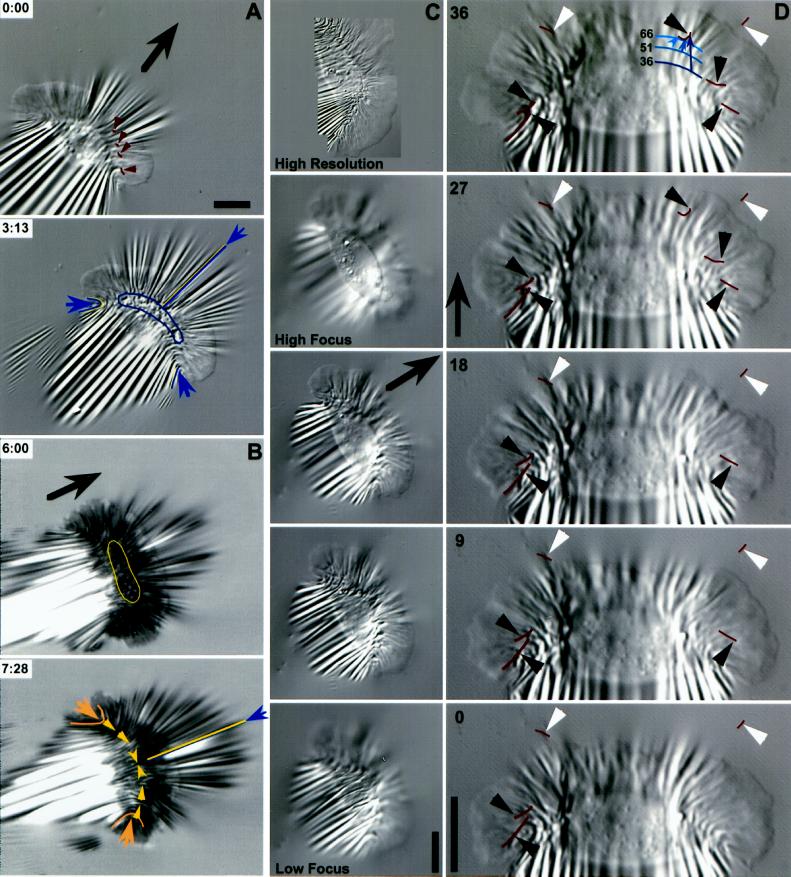
Traction forces are applied across the entire cell. (A) A large keratocyte that generated high forces on a stiff substratum (wrinkle traced at cell’s right corresponds to 680 nN). Wrinkles were produced at the flanks (arrows) and across the entire cell body (arrowheads). Most wrinkles were located along the boundary of the lamellipodium and the cell body, and the pattern was symmetrical about the midline of the cell. The box at 0 s is magnified in the lower four panels (3–12 s). The cross hair marks the position of the origin of a tension wrinkle, with the direction of force indicated by the arrowhead. The dashed line and the cross hair are at the same position in the field of view in each image, showing very little displacement of the origin of the wrinkle. Numbers indicate seconds. Bars, 5 μm. (B–D) A cell observed in DIC (B and C) and IRM (D). (B) Single arrowheads at 0 s indicate the location and orientation of traction forces producing curved compression wrinkles. The traced wrinkle at the left side of the midline at 60 s corresponds to 600 nN, whereas the force at the right flank was 70 nN. Boxed areas are magnified in C, and intermediate time points are shown. Paired arrowheads in B bracket a constant location in the field of view where a wrinkle was tracked as the cell passed over, shown by cross hairs in C. The arrows in C indicate the direction of force, which rotated by ∼90 degrees as the front of the cell body passed over (see text). (D) Sequence demonstrating the continued presence of central wrinkles when the right flank retracted (arrowheads). The direction of force near the midline reversed as the center of the cell shifted to the left (arrows at 32 and 48 s). Bars, 5 μm.
Figure 6.
Deformation of highly compliant substrata. (A and B) DIC and IRM images of one keratocyte on a highly compliant substratum. Outlined areas under the cell body show where the silicone rubber was crinkled, obscuring wrinkles that were otherwise present (compare times 6 min and 7 min 28 s in B). Wrinkles were out of focus under the cell body in A. Arrowheads in A (time 0:00) point to wrinkles and distortions that exhibited small inward lateral displacements (durations, 17–26 s). Arrowheads in B (time 7:28) indicate the positions and directions of forces that produced compression wrinkles centered near the boundary of the cell body and the lamellipodium. Force was symmetrical about the midline, where wrinkle curvature reversed (pair of angled arrowheads). Small arrows in A and B point to wrinkles used to estimate traction force (left, middle, and right): 120, 600, and 130 nN in A and 150, 700, and 146 nN in B. (C) A different cell showing a through-focus series (lower four panels). The cell body depressed the substratum so that wrinkles under the cell body were out of focus when wrinkles under the lamellipodium were in focus. The ventral wrinkles originated near the front of the cell body (bottom and top panels; the top panel shows a high-resolution three-part montage taken at a later time). (D) The same cell 45 s after the images in C, showing a time-lapse sequence (seconds) in which displacements of wrinkles (black arrowheads) and natural markers in the substratum (white arrowheads) were tracked. Arrowheads point to the position of the marker in each image, and lines show the trajectories of the markers. A portion of the boundary of the cell body and the lamellipodium is indicated by a curved line at 36, 51, and 66 s. At each time point, an arrow connects the boundary to the position of a small wrinkle that moved rearward before turning inward as the cell body approached (see trajectory at 27 and 36 s). Large arrows indicate the direction of cell movement. Bars, 10 μm.
Figure 7.
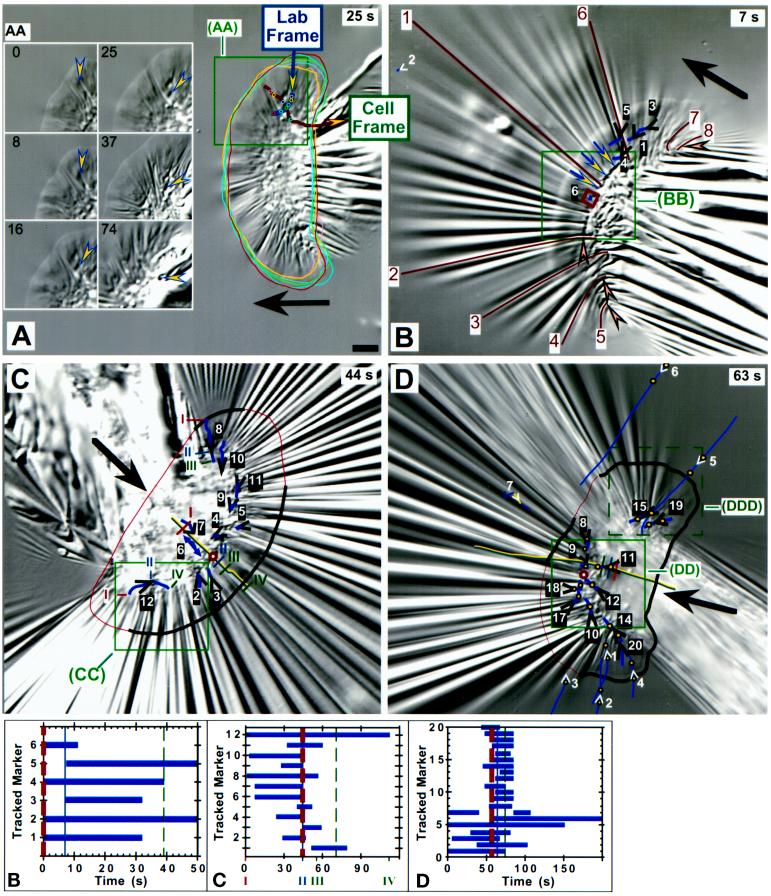
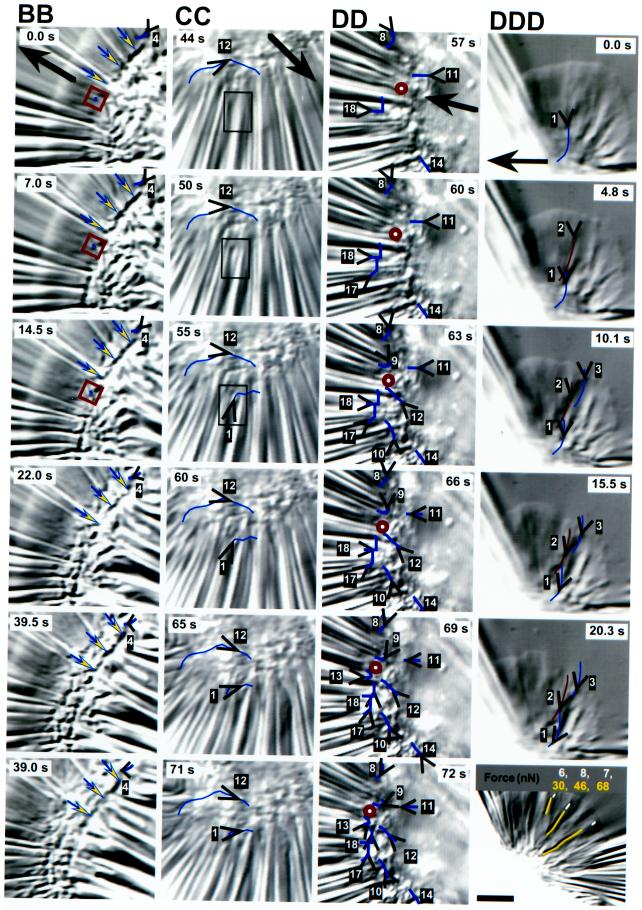
Displacements of markers in very highly compliant substrata. (A) The box on the right side of the whole cell indicates the region shown in time lapse in AA, in which an arrowhead tracks the instantaneous position and direction of one force spot in the elastic substratum from near the right front edge of the lamellipodium to behind the cell body. Movement of the force spot with respect to the field of view (corrected for drift) is shown by the trajectory that passes down and then forward (Lab Frame) within the boxed area in A. A second trajectory showing movement with respect to the cell (Cell Frame) was produced by taking cell outlines from each time point and overlaying them onto the image in A (those from the six images in AA are shown). Dots on each trajectory refer to the position of the force spot at each time point shown in AA; the two trajectories cross at the time of the image in A (25 s). Force was initially lateral and inward under the flank and became forward directed as the cell body passed over. (B–D) Boxes on images of whole cells indicate regions magnified in the time-lapse sequences of BB–DDD. Open arrowheads indicate markers used to monitor substratum displacement shown by trajectories on the images. Each marker is numbered for reference to graphs B–D and to magnified images in BB–DD; the respective arrowhead gives the location and direction at the time of the image shown. As shown in graphs B–D, natural markers in the substratum (white arrowheads) could be tracked for longer periods than force spots produced by the cells (black arrowheads). In the graphs, thick and thin dashed lines bracket the time intervals of the image sequences shown in BB–DD, whereas the solid vertical lines refer to time points of the whole-cell images of B–D. Large arrows indicate the direction of cell movement. (B) The triplet of arrows indicates a long deformation that remained visible as the cell body moved over. The small square is positioned on a force spot (6) that showed very little displacement near the midline (compare 1 and 3–5). Marker 2 (far upper left in B) was used to correct for drift. Angled arrowheads in B point to curved compression wrinkles located along the boundary of the lamellipodium and the cell body. Lines on wrinkles 1–8 were used to calculate traction forces increasing from 60 and 65 nN at the flanks (5 and 8) to 600 nN at the midline (1) (force = 85, 200, 340, 345, and 480 nN for wrinkles 7, 4, 3, 6, and 2, respectively). (C) The cell trajectory is shown along the midline, marked at positions I–IV for reference to movements of force spots 8 and 12 (time points indicated on the x axis of graph C); position II corresponds to the image in C. The trajectory of force spot 6 is given by a double-headed arrow because it moved directly rearward under the lamellipodium and then reversed direction and moved forward as the cell body passed over. The cell outline is indicated by a thick line where it was visible and a thin line where it was estimated from images at other time points and focal planes. The circle on the midline at the boundary of the cell body and the lamellipodium indicates the point of force symmetry estimated from marker displacements (see text). (CC) The boxed area shows the appearance of force spot 1 (not present in C [44 s]), which initially moved with a large rearward component but then tracked laterally along the front of the cell body. (D) Symbols and lines are as in C. Cell movement during the sequence shown in DD is indicated on the cell trajectory in D by three vertical bars, the times of which are given by the three vertical lines in graph D. Paired dots on trajectories refer to marker positions at the beginning and end of the sequence shown in DD; only one dot is shown if the marker appeared or disappeared during that period. The larger circle at the front of the cell body to the left side of the cell trajectory shows the estimated point of force symmetry. Marker 7 was out of focus in the image shown in D (63 s) but in focus at earlier and later times (see graph D). (DDD) Displacements of tension wrinkles at the right flank of the cell in D (dashed box) but imaged 6 min and 15 s later when the video camera had been adjusted to provide maximum image quality there. The bottom panel shows a high-resolution image taken after the time-lapse sequence (2 min and 30 s). Force magnitudes in the lamellipodium were calculated from three wrinkles and three force spots (indicated by tracings on the image). Bars, 5 μm.
Figure 9.
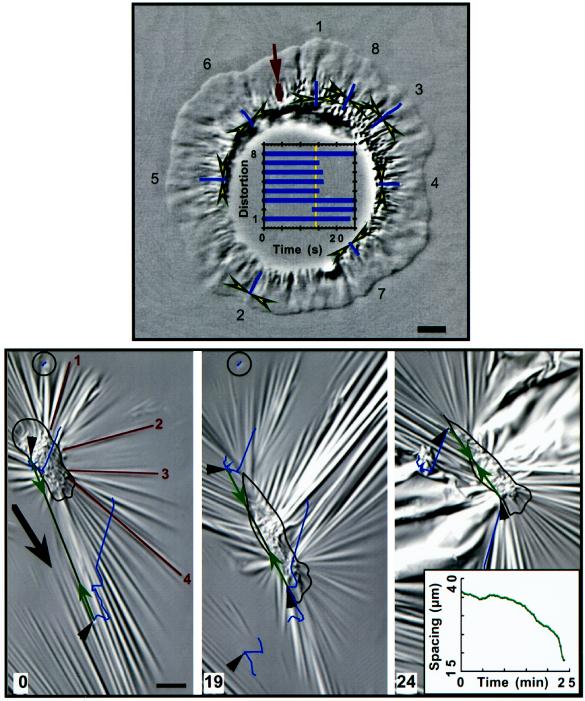
Fried-egg–shaped cell. Paired arrowheads bracket substratum deformations that exhibited centripetal movement under the lamellipodium. The distortions are numbered in the image, and their movements were tracked for the periods shown in the graph (the vertical line refers to the time of the image shown). The clustered wrinkles near the cell body were produced by forces of several 10s of nanonewtons. The cell body was digitally removed from the image (unsharp masking) to facilitate increased contrast in the lamellipodium. Bar on wrinkle at arrow, 40 nN; horizontal bar, 5 μm.
Figure 10.
The fibroblast lamellum pulls rearward and the tail pulls forward. These images were focused on the substratum to show crinkles located underneath the cell body and wrinkles extending out from the cell; the outline of the cell is taken from images acquired 15 s later at a higher focal plane. Note that the time is in minutes. Forces estimated for wrinkles 1–4 at time 0 were 265, 500, 480, and 800 nN, respectively. The substratum was compressed by the cell when it formed a tail and contracted, as shown by two substratum markers in front of and behind the cell (arrowheads connected by a double-arrow line; separation quantitated in graph inset [24 min]). A second marker located farther in front of the cell was also pulled rearward between 12.5 and 19 min. Contraction of this cell and others outside of the field of view caused large shifts in the substratum, and individual marker trajectories reflect all of these movements. To better visualize the relative displacements of markers, the images at 0 and 19 min were shifted so that a reference marker behind the cell (circled) was positioned at the same location in the image. The reference marker had moved out of the field of view in the last image shown (24 min), and no correction was applied; hence, the displacement of all features in the image contains a large upward-right component. Bar, 20 μm.
Data Acquisition
A ×100, 1.3 numerical aperture Plan-Neofluar oil objective was used with video-enhanced Nomarski differential interference contrast (DIC) microscopy to visualize cell morphology and structure as well as strain in the silicone substratum; IRM was used to visualize close contacts. In both modes of microscopy, long-wavelength illumination (>600 nm) was used to reduce intrinsic absorbance by cellular proteins. The silicone used for the elastic substratum is transparent to visible wavelengths and has a relatively high index of refraction (1.536), close to that of glass, which is conferred by the phenyl side chains (Burton and Taylor, 1997). This improves contrast in IRM images as a result of the increased index differential between the substratum and the medium. DIC and IRM images were acquired on a multimode microscope based on a Zeiss (Thornwood, NY) Axiovert microscope that had been automated to allow changes in optical configuration (optical path, DIC analyzer, color and neutral density filters, camera and lamp shutters, stage position, and focus) within a period of a few seconds under software control (STC-View, the multimode microscopy imaging system developed for the Science and Technology Center at Carnegie Mellon University), with simultaneous control of image acquisition and storage into computer memory (Macintosh Quadra 950; Apple, Cupertino, CA) and onto disk (Giuliano et al., 1990; Farkas et al., 1993). Images were usually acquired by a video camera (C-2400 Newvicon, Hamamatsu, Bridgewater, NJ) and stored directly into computer memory or onto videotape. Images stored in memory were sampled for periods of 30–380 s at rates of 0.3–5 s−1. Data stored on videotape was sampled at video rates (30 s−1) and digitized with frame averaging after the experiment as needed. In some cases, single images were magnified by a ×4 coupling lens placed in front of a cooled charge-coupled device camera (Photometrics, Tucson, AZ).
Image Processing
Digital images were either 512 pixels × 474 pixels × 8 bits (video camera) or 512 pixels × 382 pixels × 12 bits (charge-coupled device camera). In most cases, images were divided by a featureless background (“flat-field”) image to reduce the effects of inhomogeneous illumination and unwanted objects in the optical path. In all cases, linear intensity scaling was applied to enhance contrast in the areas of interest. In some cases, out-of-focus scattering from objects near the cell caused significant intensity variations over areas of the image that were large compared with the sizes of the features of interest. Such diffuse intensity signals were removed by subtracting a smoothed version of the image from the original to leave sharper features of interest (“unsharp masking”). Some images were enlarged in software by floating-point linear interpolation between pixels in the original image. Batch processing of large sets of time-lapse images was carried out by the “movie-processing” application within STC-View, including resident routines for contrast enhancement, flat-field division, and enlarging as well as calls to subroutines for additional procedures written by the authors (e.g., unsharp masking). Time-lapse sequences were usually viewed with STC-View to display movies at video rates (30 frames/s) on the computer monitor and also on an external video monitor, but NIH Image (written by Wayne Rasband at the U.S. National Institutes of Health [Bethesda, MD] and available in the public domain over the Internet at zippy.nimh.nih.gov) was also used to display images at 100 frames/s on a Power Macintosh. NIH Image was used for cropping and contrast enhancement of images to be used in the figures. Photoshop (Adobe Systems, Mountain View, CA) was used for placement of groups of images into figures, and MacDraw Pro (Claris, Santa Clara, CA) was used to add text and graphical annotation to figures.
Analysis of Deformations in Silicone Substrata
The positions, lengths, orientations, and movements of cells and other features of interest in digitized time-lapse image sequences (e.g., markers and distortions in the elastic substrata; see Figure 1) were quantitated with either NIH Image or MacWrinkle software (developed within the National Science Foundation Science and Technology Center at Carnegie Mellon University). MacWrinkle provides graphical objects (lines, circles, and grids) that are manually positioned in images on features of interest and adjusted as needed in time-lapse sequences. Positions and orientations of objects are expressed relative to the field of view, as well as relative to the cell, and can be viewed in time lapse with either coordinate system. All quantities, including velocities (speed and direction) calculated from changes in position, are saved in MacWrinkle movie files and in spreadsheet format on disk for separate graphical analysis.
MacWrinkle was used to calculate displacements of markers in the substratum to indicate the direction of traction force and relative force increments (see Figures 5–7, 9, and 10). Absolute force magnitudes were not calculated from displacements (see the description of force calibration based on wrinkles above). The pattern of strain in the elastic substratum was often complex when compliance was high; thus, criteria were established for choosing a distortion or wrinkle to be tracked: (1) it had to be in focus throughout the analysis; (2) it had to be a discrete entity distinguishable from neighboring distortions; and (3) its size and shape had to remain relatively constant during one sample period, because the size and shape of individual distortions and the pattern of strain as a whole often changed greatly over several time points. Groups of adjacent distortions moved together, providing a guide to the movement of any one distortion.
NIH Image was used to measure wrinkle length to estimate force as described above (Preparation of Silicone Sheets). Measurements of wrinkle length required the ends of wrinkles to be identified, and for those wrinkles that gradually faded into the background, the end was defined by the signal-to-noise ratio at the position where intensity variation across the wrinkle was twice the SD of pixel intensity in the adjacent background.
Errors in the measurement of displacement can be introduced by stage drift. Drift was accounted for in some cases by making measurements on natural markers in the substratum distant from the cell, and a correction was made when the movements of these markers were significant compared with those of the features of interest. In cases in which drift could not be discerned explicitly (e.g., when there were displacements in the substratum caused by other cells outside of the field of view), relative displacements between markers were used to ascertain changes in strain. As a measure of the uncertainty in estimated position, a comparison was made between analyses carried out on the same data set by three independent investigators, each repeated 10 times. The error in the estimated position of a point distortion was within one pixel (0.18 μm).
Tracking Movements of Cells
MacWrinkle software was usually used to estimate cell centroid by positioning a grid of adjustable size and orienting it on the cell (Figure 1). One purpose of this measurement was to check for gross effects of substratum compliance on cell speed (Pelham and Wang, 1997; Thomas, 1998). The grid was positioned with one arm along the midline of the cell in the direction of motion and the other arm along the lateral axis of the cell at a consistent position under the cell body (e.g., the approximate center or front margin). The uncertainty in calculated cell speed was estimated to be ∼2% of the mean by repeating measurements on the same sequence three times. To reduce overestimates of cell speed arising from the components of movement perpendicular to the average direction (as a result of measurement error and real lateral shifts), a simple measure of distance was taken as the net movement over a period of time during which the cell moved in a nearly straight line. The accuracy of the MacWrinkle measurements was estimated by comparing them with the same quantities obtained with the use of a combination of MacDraw Pro and NIH Image. The cell was outlined in MacDraw Pro, and the outline was adjusted for each image in a time-lapse sequence. The outlines were then exported to NIH Image, and calculations were made of area centroid and orientation (based on an ellipse fitted to the outline). Movements calculated in this way were similar with either method, although the cell centroids were offset (Figure 1A; this is the same cell that is shown in Figure 2, A, D, and E). The orientation of the cell was different by as much as 3 degrees (Figure 1B), owing in part to changes in the shape of the cell during small turns. These differences do not alter the conclusions of the study.
Interpretation of Wrinkle Patterns
Wrinkles in silicone sheets are of two general types (Harris, 1984; Peterson, 1996; Burton and Taylor, 1997): (1) curved “compression” wrinkles that originate from force that is applied at the center on the concave side and is directed toward the convex side (see Figures 1–5); and (2) straight “tension” wrinkles that originate from force applied at the end that is narrowest and deepest (higher-image contrast) and is directed along the wrinkle away from its center (see Figure 5A). As force increases and wrinkles lengthen, both ends of a compression wrinkle extend away from the center, whereas most of the increase in length of a tension wrinkle results from movement of the end opposite to the site of force application.
Several points should be noted when inferring forces from deformations in the rubber substrata. The first is that the appearance of isolated wrinkles under a cell localizes force application to those sites. Experiments with needles have shown that wrinkles originate at the sites where traction force is applied (Burton and Taylor, 1997), in contrast to displacements in the elastic substratum that can be influenced by distant forces. On the other hand, wrinkles can be detected only where local stress (force divided by area) is great enough to visibly bend the rubber; the absence of wrinkles elsewhere does not imply an absence of force. For example, even though the cell in Figure 2B was moving forward, there was only one wrinkle at the end of the tail resulting from a localized force that resisted forward movement. Finally, forces applied at wrinkles need not have been generated within the cell at that position but may have originated elsewhere and been transmitted through the cytoskeleton to cell-substratum adhesions located there.
Centripetal force in the lamellipodium frequently produced tension wrinkles that extended out from the cell (see Figures 5, A–C, 7DDD, and 10) in the same direction as compression wrinkles from lateral forces (see Figure 5, A–C). Therefore, such wrinkles probably had contributions from (1) lateral forces along the boundary of the lamellipodium and the cell body, and (2) lower centripetal forces in the lamellipodium directed perpendicular to that boundary (see RESULTS). The presence of closely adjacent orthogonal forces was shown directly by displacements of force spots. Distinguishing the relative contributions of these two sets of forces to patterns of wrinkles would require detailed analysis with the use of methods such as those of Peterson (1996).
RESULTS
Forces Are Applied at Stationary Adhesions
Figure 2 shows DIC and IRM time-lapse sequences of a keratocyte on a silicone sheet in which deformations revealed the locations, directions, and magnitudes of traction forces. This freely locomoting keratocyte possessed a classic fan shape and moved rapidly at nearly constant velocity (15–25 μm/min). Localized traction forces in the lamellipodium were revealed by small irregularly shaped distortions (Figures 2, 3A, and 4B) that grew in size while remaining nearly stationary with respect to the substratum as the cell moved over (Figure 2, A and C–G). Traction forces are necessarily applied at cell-substratum adhesions, and, consistent with wrinkle positions, close contacts observed in IRM images also moved little (e.g., 0.4 μm in 5.5 s; Figure 2, C and G). As reported previously (Lee et al., 1994; Oliver et al., 1995), force typically increased to a maximum at the rear margin of the lamellipodium, reaching 100–200 nN for the individual wrinkles indicated in Figure 2, D–F, H, and I (substratum stiffness = 30–35 nN/μm [force divided by unit of wrinkle length; see MATERIALS AND METHODS]). The magnitude of these lateral forces is similar to the magnitudes described in previous reports (Oliver et al., 1995; Dembo et al., 1996).
Traction Forces Pull Retracting Margins Forward and Inward
Curved compression wrinkles at the lateral margins (“flanks”) of the lamellipodium (Anderson et al., 1996) indicated that forces were directed mainly laterally toward the center of the cell, as first shown by Lee et al. (1994). However, wrinkles at the flanks also contained a forward-directed component that increased posteriorly (Figures 2, 3A, and 4B). This component of “lateral” force tends to pull the posterior regions of the flanks forward, consistent with movements observed when a flank retracts (Figure 2F, note paths of retraction fibers in IRM image sequence; see also Lee et al., 1993b).
Traction force may contribute to loss of cell-substratum adhesion. To address this question, we monitored force and contact area in IRM images and calculated changes in stress (force divided by area) at the site of retraction. Figure 2I shows such a calculation for the IRM images shown in Figure 2F, where force was applied at the close contact under the left flank. The area of close contact decreased faster than force during retraction, so that stress increased from 80 to 130 nN/μm2 before abruptly decreasing as the cell pulled away. This result shows that retraction results in part from increasing force applied to adhesions.
The sudden retraction of one flank caused tension to be reduced across the cell so that force decreased at the opposing flank. In the example shown in Figure 2, F and G, the IRM images show that as the left flank lifted from the substratum (Figure 2F, 30.1 s), the wrinkle under the right flank nearly disappeared (Figure 2G). Such asymmetrical retraction caused cells to turn briefly (this cell veered to the right by 55 degrees [see cell trajectory in Figure 2C] and rotated clockwise by ∼6 degrees).
Cell-Substratum Adhesions Resist Forward Movement
IRM images showed that close contacts between the cell and the substratum were often similar under the lamellipodium and the cell body, on both silicone rubber substrata (see Figures 2B, 3B, and 5D) and on glass (data not shown). The close contacts observed under the cell body suggested that it adhered to the substratum and would resist forward movement. Consistent with this interpretation, the ventral surface often appeared to be dragged backward, as shown by a thin “lip” at the rear of the cell in high-resolution images (Figure 3A), a sharp posterior edge in IRM (Figure 3B), and short retraction fibers trailing behind the cell body (Figure 3, A–D). These observations suggest that cells must overcome resistance to movement provided by the cell body; additional evidence for this kind of load is reported below (see Figures 6 and 7).
Similarly, keratocytes occasionally possessed an elongated “tail” that tethered the cell, as judged by forward-directed force applied at its end (Figure 2B) and by slowed movement (see Figure 10). These cells often had lamellipodia that adhered less well to the substratum, as judged in IRM by brighter areas and by centripetal movement of gray close contacts. For example, the close contact shown in Figure 2B (arrow) moved rearward over the substratum at 23 μm/min, whereas the cell moved forward at only ∼2 μm/min. Lamellipodia that appeared to “slip” in this way did not produce detectable traction force.
An Increase in Traction Force Is Associated with Widening of the Cell Body
Actin-myosin stress fibers are oriented across the cell body perpendicular to the direction of motion (Euteneuer and Schliwa, 1984; Anderson et al., 1996; Svitkina et al., 1997; Park and Taylor, unpublished observations), and it has been suggested that the characteristic spindle shape of keratocytes is maintained by lateral tension generated in these fibers (Cooper and Schliwa, 1986; Anderson et al., 1996). We tested this relationship by observing locomoting cells in which the width of the cell body fluctuated with time (Figure 4), as has been observed in keratocytes (Svitkina et al., 1997). When force was low or undetectable, cell bodies were more circular in projection (Figure 4A), but when force increased so that wrinkles and smaller distortions formed under the flanks, cell bodies widened (Figure 4B). When the cell body widened, it narrowed in the direction of motion and its thickness decreased (as judged by adjusting focus), and organelles were observed to move outward toward the flanks. In addition, stress fibers were visible in DIC only when the cell body was spindle shaped and the flanks generated relatively high lateral forces. A contractile mechanism to explain these observations is proposed below (see DISCUSSION).
Keratocytes Can Produce High Lateral Forces
When cells were stronger than those shown Figures 2–4, wrinkles and distortions were larger and appeared under more areas of the cell (Figure 5). At the flanks, curved wrinkles showed that forces of a few 10s to ∼200 nN were directed inward and slightly forward, similar to those described above. However, more centrally located wrinkles were progressively less curved and faced less forward, becoming straight at the midline (see Figure 6B). These wrinkles were often longer than those at the flanks, indicating higher forces (680 nN for the example shown in Figure 5A). The curved wrinkles in Figure 5 show that the orientation of lateral forces generally conformed to the shape of the cell body, with forces near the front being directed along an arc, whereas force near the rear of the cell body was more purely lateral. The strong cell shown in Figure 5A is the largest keratocyte we observed, but there was no obvious correlation between force and cell size, nor were correlations observed between force and the anatomical position of the scale from which cells were derived or the time spent by cells in culture. The magnitude of these forces is considered further below (see DISCUSSION).
It is unlikely that the more centrally located wrinkles arose from forces applied solely at the flanks and propagated through the rubber sheet. IRM images showed that close contacts were present where wrinkles were produced (Figure 5D; IRM not shown for Figure 5A), and when one entire flank suddenly retracted, the central wrinkles did not disappear but reversed direction as the center of the cell shifted laterally (Figure 5D; see also Figure 8 below), suggesting that force was applied at that location.
Figure 8.
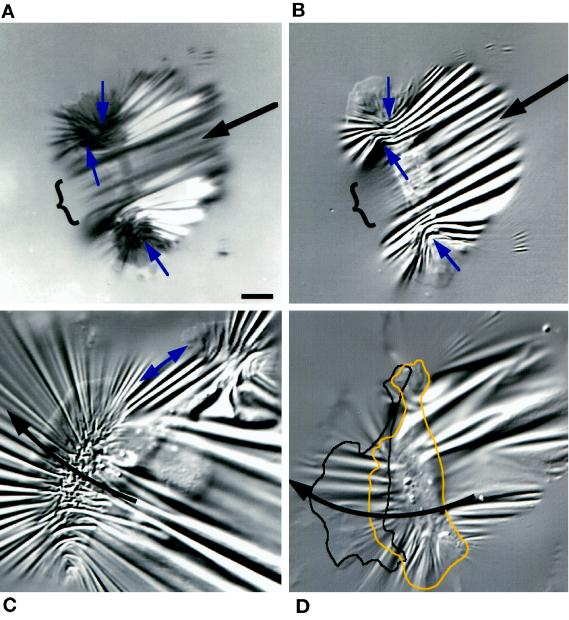
Bipolar cells. (A and B) IRM and DIC images taken 10 s apart. Small arrows indicate the direction of lateral forces compressing the substratum at each lamellipodium. The central region ({) produced few wrinkles in front of the cell. Large arrows show the direction of motion. (C) A pair of lamellipodia stretched the substratum between them (double-headed arrow). The larger lamellipodium (left) migrated faster than the smaller one, causing the cell to turn right (curved arrow). (D) Another cell that was being turned by a dominant lamellipodium, outlined in the image shown and at its position 50 s later. Bar, 10 μm.
On more compliant substrata (20 nN/μm), the substratum was compressed laterally into a dense pattern of curved wrinkles centered near the front of the cell body (Figure 6B) that extended from well behind the cell to well in front, where it formed a radial array of wrinkles (Figure 6, A and B). As with the cells shown in Figure 5, force near the midline was usually higher (600–700 nN) than that at the flanks (120–150 nN), where it remained near the levels observed for cells on stiffer substrata (Figures 2–4).
Lamellipodia Apply Centripetal Forces at Localized Sites
Forces applied by the lamellipodium over small areas in front of the cell body were revealed by punctate distortions (“force spots”) in the substratum that became visible at the same time as wrinkles at the flanks (Figures 3A, 4B, and 5A). Such force spots in these moderately stiff substrata were stationary in the field of view within measurement accuracy. Force spots visible in DIC were always located in regions of close contact in IRM (Figure 3B; see also Figure 5A [IRM not shown]), but a correlation with focal contact–like dark spots in IRM was not established because (1) the spots were fleeting, allowing little time to switch between DIC and IRM, and (2) the deformation itself generated contrast in IRM.
In strong cells, such as those shown in Figure 5, nascent wrinkles often originated at force spots (Figure 5, A–C). Wrinkles produced by lamellipodia in front of the cell body had characteristics of tension wrinkles, in which force applied at the narrow end stretched the substratum along the wrinkle (see MATERIALS AND METHODS). In Figure 5A (times 6–12 s), force was applied at the stationary end of the wrinkle nearest the cell body, and the distal end extended outward as force increased. In Figure 5C, the straight wrinkle under the lamellipodium at times 30–90 s was produced by force directed rearward and perpendicular to the front of the cell body. However, as the front of the cell body moved over (times 120–190 s), the wrinkle became curved and centered on the same location (times 180–240 s), indicating that force had become directed inward and parallel to the front of the cell body. The site of force application remained nearly stationary except for a slight inward displacement in the direction of force. This apparent 90-degree rotation of force at the boundary of the lamellipodium and the cell body was also apparent from displacements observed in more compliant substrata described below (Figures 6D and 7, C and D).
The Cell Body Depresses Highly Compliant Substrata
Wrinkles often appeared to originate at the front edge of the cell body on more compliant substrata. This was partly caused by the cell body pressing the substratum down and out of focus (Figure 6, A, C, and D) and by wrinkles that were disrupted by “crinkling” of the substrata (Figures 6, A and B, and 7). These images suggest that wrinkles radiating from the front of the cell body on highly compliant substrata (Figures 6–8) were produced by lateral forces applied there.
Although the silicone rubber substratum undoubtedly provided resistance to forward movement when the cell body pressed down into it, the velocity of movement was little reduced (17 ± 10 to 13 ± 4 μm/min [mean ± SD, n = 4–5], respectively, for the stiffest to the most compliant substrata used). Forward-directed force would stretch the sheet behind the cell body and probably contributed to the long, straight wrinkles that extended rearward (Figure 6C, lower focus). In contrast, no evidence was found for rearward-directed forces at the posterior margin of the cell body that would indicate that cells “push” themselves forward.
Lamellipodial Force Is Centripetal and Pulls the Cell Body Forward
This and the next section describe detailed analyses of patterns of forces inferred from both wrinkles and displacements of markers in highly compliant substrata. Both the entire cell and magnified views are shown, including time-lapse sequences. The entire pattern of forces can be appreciated only by viewing the entire cell, whereas individual markers used to track displacements are often difficult to discern at low magnification. A schematic summary of substratum deformations across the entire cell and the forces inferred from them is provided in Figure 11, which can serve as a reference for the image sequences and analyses shown in Figures 6 and 7.
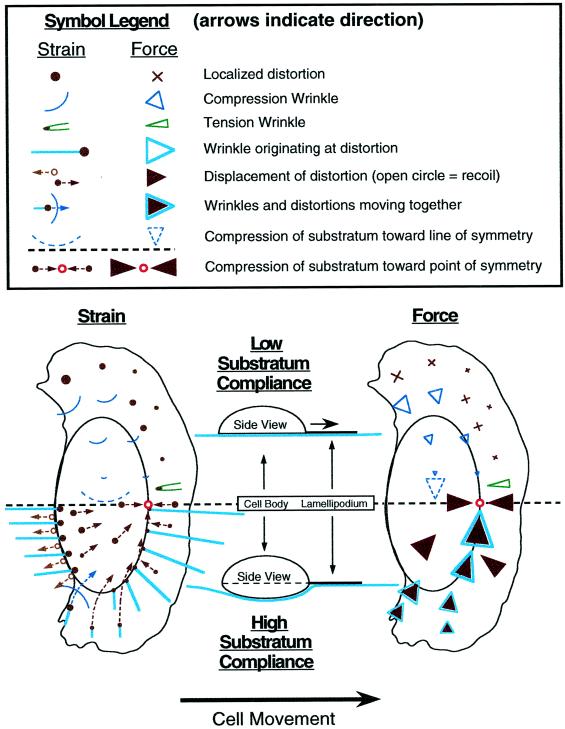
Figure 11.
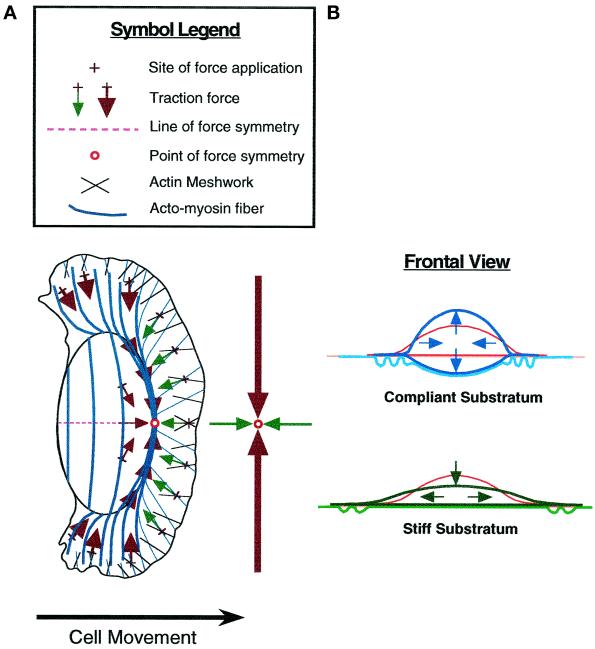
Summary of keratocyte traction forces inferred from substratum deformation. The drawings show average patterns of deformation in elastic substrata (left) and traction forces (right) produced by locomoting keratocytes. The upper part of the diagram (left side of the cell) indicates observations on stiff substrata that are not greatly deformed by the cell, remaining nearly flat under the cell body (see diagrammatic “Side View”), whereas the lower part of the diagram (right side of the cell) refers to highly compliant substrata that are greatly deformed by the cell and are depressed by the cell body. Symbols referring to strain and traction force are defined in the legend. The trajectories of force spots (filled circles) and wrinkles (curved lines) given by broken arrows refer to instantaneous displacement in the reference frame of the cell; individual markers generally do not follow these trajectories over time because of cell movement.
Displacements of markers in the plane of highly compliant substrata provided an important check on the directions of forces inferred from wrinkles. In agreement with the previous results of Jacobson and colleagues, who embedded beads in silicone rubber substrata (Lee et al., 1994; Oliver et al., 1995), lateral forces caused wrinkles at the flanks to be pulled inward toward the cell body (Figure 6, A and D), followed by recoil of the substratum as the cell passed by. Additionally, however, we observed that there were also rearward components in displacements under the flanks and in front of the cell (Figure 6D), consistent with the directions of forces inferred from tension wrinkles (Figure 5, A–C). Displacements under the lamellipodium in front of the cell body were approximately perpendicular to their boundary (Figure 7, C, D, and DDD), so that near the midline force was almost directly rearward. This pattern was observed with either distortions produced by the lamellipodium (Figure 7C, distortion 6) or natural markers in front of the cell (Figure 7D, marker 7), and is expected from cortical flow in other cell types (Bray and White, 1988; Fisher et al., 1988). In contrast to the results of Dembo et al. (1996), we observed no forward-directed displacements in front of or under the lamellipodium that might indicate pushing forces that retard forward movement (see DISCUSSION).
Increasing substratum compliance also provided direct evidence of localized force application nearly everywhere under the lamellipodium, even near the edge (Figure 7, A and DDD). In some cases, one distortion could be tracked from its appearance under the lamellipodium all the way to the rear of the cell (Figure 7A, inset), showing that force could be applied at a single adhesion site while the entire cell passed over.
Differences in substratum displacements at two locations indicate relative changes in force. Rearward displacements in front of the cell body in Figure 6 were 5–10 times smaller than lateral displacements at the flanks during the same period, indicating that the increase in retrograde force was smaller than the increase in lateral force. However, for the most highly compliant substrata, changes in pulling forces in front of the cell body were relatively stronger, being less than the lateral forces at the flanks by only a factor of 2 (Figure 7D; dots on trajectories indicate displacements over the 15-s period shown in Figure 7DD). The absolute value of lateral force at the midline of the cells shown in Figure 7 was very high, ranging from several hundred nanonewtons (600 nN in Figure 7B) to about 1 μN (Figure 7, C and D).
Lamellipodial Force Abruptly Increases and Becomes Lateral at the Cell Body
As the cell body approached a small wrinkle under the lamellipodium shown in Figure 6D (right of the midline at times 27 and 36 s), the wrinkle was displaced rearward but then began to move laterally and inward, similar to the rotation of force inferred from the wrinkle in Figure 5, B and C. This turning was more dramatic on the most highly compliant substrata (Figure 7, A, CC, DD, and DDD): force spots moved rearward under the lamellipodium, turned inward at the transition with the cell body, and then moved forward behind the advancing boundary. For markers near the midline, the change was more purely a reversal of direction from rearward to forward (Figure 7C, distortion 6). These forward displacements demonstrate that the cell body applies forward-directed forces on the substratum, consistent with observations of cell morphology (Figure 3). For the cell as a whole, instantaneous displacements were generally directed toward a “point” of force symmetry near the front of the cell body at the midline (indicated by an open square or circle in Figure 7, B–D, BB, and DD; see also Figure 11). When cells moved directly forward, this point of force symmetry was located on the geometrical midline of the cell (Figure 7, B and C), but in the example shown in Figure 7D, in which the cell was turning left, it deviated to the left of the midline.
Paired Lamellipodia Independently Produce Force and Movement in Bipolar Cells
A useful model system to distinguish between forces at the lamellipodium and at the cell body is a bipolar cell with two “lateral” lamellipodia (Bereiter-Hahn et al., 1981; Cooper and Schliwa, 1986; Anderson et al., 1996) that locomotes similarly to cells with a single continuous lamellipodium (Anderson et al., 1996). Figure 8 shows bipolar cells on substrata comparable to those shown in Figure 7, but unlike in cells with a single continuous lamellipodium, there were no wrinkles in the central region. This behavior was always observed and suggests that a lamellipodium is necessary for force production along the front of the cell body. This apparent coupling between the lamellipodia and lateral forces is considered in light of the structural data and model of Svitkina et al. (1997) (see DISCUSSION).
Consistent with the evidence shown in Figure 5D, this result also suggests that wrinkles in central regions (Figure 7) are not produced by forces transmitted through the substratum from the cell’s flanks.
Long wrinkles, however, did extend behind the cell body in the bipolar cell shown in Figure 8, A and B, similar to those observed in cells with complete single lamellipodia (Figures 5–7). As discussed above (Figure 6C), this effect is expected if the cell body stretches the substratum as it is pulled forward. Note that the cell body was bowed backward at its ventral surface where it adhered to the substratum (IRM image; Figure 8A) but was unbowed above the substratum (DIC image; Figure 8B). Such wrinkles would not arise if the cell body pushed itself forward.
Lamellipodia of bipolar cells were similar to those of monopolar cells in generating lateral forces, as shown by a radial arrays of wrinkles (Figure 8C), compression wrinkles at the respective flanks (Figure 8, B and C, arrows at right-hand lamellipodium), and/or tension wrinkles between the lamellipodia (Figure 8C, double-headed arrow). A dominant lamellipodium caused that side to move faster, causing the cell to turn (Figure 8, C and D).
Forces of Cortical Flow in the Lamellipodia of Stationary Cells
Keratocytes that possess a “fried-egg” shape (Bereiter-Hahn et al., 1981) have a rounded cell body encircled by a continuous lamellipodium (Figure 9). Such cells provided the opportunity to study forces produced by lamellipodia in the absence of movement along, or against, the substratum. These lamellipodia exhibited continuous centripetal flow with little change in size or shape, similar to stationary fibroblasts (Fisher et al., 1988) or neuronal growth cones from sea hares (Forscher and Smith, 1988). In Figure 9, the cell body depressed and severely crinkled the substratum (data not shown) but did not produce the large radial array of wrinkles that were present when moving cells pressed forward into similar substrata. The lamellipodium, however, did produce force spots and wrinkles that were like those under locomoting cells in several respects: (1) in the reference frame of the cell, force spots and wrinkles in the lamellipodium moved centripetally, ultimately combining with crinkles under the cell body; (2) force spots and wrinkles in a given area moved together, suggesting that they were located at fixed locations on a moving substratum; (3) force spots could appear very near the edge of the lamellipodium; and (4) the size of a given distortion increased with time and displacement from the edge of the lamellipodium, demonstrating increasing force. However, unlike freely locomoting cells, the average force was constant around the entire cell, with displacements being absolutely perpendicular to the boundary of the lamellipodium and the cell body.
Reduced Speed in Cells Tethered by a Tail
To extend our results to cell motility in general, we compared traction forces of locomotion in keratocytes and fibroblasts with and without tails. Keratocytes occasionally form an elongated tail when one side of the lamellipodium does not retract while the cell continues to move (Figures 2B and 8, C and D). Wrinkles located at the end of the tail in both cell types showed that force was directed forward (Figures 2B and 10). The fibroblast shown in Figure 10 was initially like a keratocyte in having no tail, moving relatively rapidly (∼5 μm/min), and possessing a large leading lamellum. During the initial period of rapid locomotion, a generally radial pattern of wrinkles was produced by forces of 300–800 nN, no higher than for keratocytes. The fibroblast eventually formed an elongated tail, at which time force increased greatly and movement slowed (Figure 10, 19 min). The substratum was compressed when the leading edge continued to move forward against the adherent tail (Figure 10, line with arrowhead pair). Finally, the cell contracted and massively distorted the substratum (Figure 10, 24 min). Substratum markers under the front of the cell and in the substratum ahead of the cell moved rearward (Figure 10, arrowheads), showing that as with keratocytes (Figures 6D and 7D) the fibroblast applied forces in front of the cell body that acted to propel it forward.
DISCUSSION
Summary of Traction Forces in Locomoting Keratocytes
Figure 11 schematically shows substratum deformations observed for locomoting cells (left) and the distribution of traction forces inferred from these patterns of strain (right) for cells on both stiff substrata (left half of cell) and highly compliant substrata (right half of cell). The most general conclusion is that traction force in keratocytes is centripetal, as Harris (1984) previously observed in fibroblasts, adding that there was no apparent evidence of pushing by a fibroblast against the rubber substratum. Thoumine and Ott (1996) observed radially oriented wrinkles produced by rounded fibroblasts, probably tension wrinkles resulting from centripetal contraction. In keratocytes, the centripetal nature of traction force is most apparent in circular cells (Figure 9).
High-resolution measurements, however, have revealed a wealth of detail critical to understanding the mechanisms of motility. We confirm the previous observations of Jacobson and colleagues (Lee et al., 1994; Oliver et al., 1995) that lateral traction forces of ∼30–150 nN are applied at the flanks of keratocytes and that force increases posteriorly. In addition, we report several new observations that provide a much more comprehensive description of traction forces: (1) forces are applied by lamellipodia over small (∼1 μm2) areas (force spots); (2) forces in lamellipodia are in the retrograde direction, acting to pull the cell body forward; (3) force at the posterior margins of the flanks has a forward component that adds to movement; (4) sites of force application are stationary with respect to the substratum; (5) a single site of force application can be maintained as the entire cell passes over; (6) force abruptly changes direction at the boundary of the lamellipodium and the cell body; (7) high forces are often localized near the boundary of the lamellipodium and the cell body; (8) force at the midline can be much higher than at the flanks; (9) axes of force symmetry cross at the front of the cell body to define a point of force symmetry; (10) the point of force symmetry shifts in the direction toward which cells turn; (11) stress increases even as force decreases at retracting cell-substratum contacts visualized by IRM; (12) lateral force at the flanks correlates with the width of the cell body in individual cells; and (13) keratocytes can generate a wide range of forces, even equal to those of fibroblasts. Observations 1, 4, 5, and 11 take advantage of the high spatial resolution of wrinkles and would be difficult or impossible to make with substratum displacements.
Traction Forces Parallel to the Direction of Motion Act to Pull the Cell Forward
Observations 2 and 3 above suggest that the force needed for forward movement is produced in two regions, first by rearward-directed forces in the lamellipodium that pull the cell body forward and second by forward-directed forces in the posterior flanks that pull them toward the cell body.
Tension wrinkles under the lamellipodium in front of the cell body suggested rearward-directed force (Figure 5, A–C) (observation 2 above). Such retrograde lamellipodial force was more directly demonstrated by displacement of force spots under the lamellipodium (Figure 7) and substratum markers in front of cells (Figures 7D and 10) on more compliant substrata. This result is clearly expected from the direction of cortical flow in the lamellipodia of many types of cells (Bray and White, 1988; Fisher et al., 1988; Forscher and Smith, 1988). In contrast, Dembo et al. (1996) interpreted the displacements of beads on the substratum to suggest that keratocytes push forward at the leading edge, thus resisting their own motion. The reason for the difference in the results of the two studies is not clear, especially because our interpretation is based on both substratum displacements and wrinkles. In the study by Dembo et al. (1996), it was shown that compression of the substratum between a pair of needles caused the substratum to be displaced perpendicular to, and away from, the axis of compression. This property could have contributed to outward displacements parallel to keratocyte movement when the flanks “pinched” the substratum, although the authors’ methods of analysis would be expected to account for this behavior. We note that retrograde forces often were not detectable in the present work (Figure 2) but became more apparent when lateral force increased near the midline (Figure 5).
In the posterior region of the cell, curved wrinkles at the flanks showed that lateral force contained a forward component (observation 3 above). In addition, displacements of force spots under most of the cell body contained both lateral and longitudinal components, gradually changing from entirely forward at the midline to entirely inward along the front of the cell body. Although there was no lateral displacement of force spots at the midline, indicating zero net lateral force, wrinkles symmetrical about the midline showed the substratum to be under compression (Figures 5 and 6, A and B). The forward-directed forces are consistent with retraction of the cell’s posterior margins forward into the cell body, thus contributing to overall forward movement. These forward-directed forces were most apparent when the cell body pressed into the substratum as the cell moved forward. It is not clear to what extent this may have occurred in previous studies of locomoting keratocytes.
High-Traction Forces in Keratocytes
Our measurements have revealed an extremely large range of traction forces, varying from the limit of sensitivity (a few nanonewtons) at localized spots to hundreds of nanonewtons at large compression wrinkles near the boundary of the cell body and the lamellipodium (Figures 5A and 7, C and D), substantially greater than previously reported (Lee et al., 1994; Oliver et al., 1995; Dembo et al., 1996). For cells on the stiffest substrata used here, forces were often detectable mainly at the flanks, and in those cases they were directed laterally and slightly forward, with magnitudes in the range of a few 10s of nanonewtons to ∼200 nN (Figures 2, 3A, and 4), generally consistent with values reported previously (∼30–140 nN; Lee et al., 1994; Oliver et al., 1995; Dembo et al., 1996). However, on the same substrata, many cells produced no detectable force (i.e., < ∼20 nN) (Figure 3, B–D) and others produced >500 nN. These cells were easily identified during the experiment by the lengths of wrinkles, a task that would have been much more difficult had we relied on observations of beads in real time.
It is important to realize that the low forces we report (Figure 7DDD) refer to small wrinkles in localized areas and not to force integrated over the cell. The highest forces were applied near the center of the cell, in contrast to previous work on keratocytes in which the highest forces were located at the flanks.
Other than the positive relationship between cell body width and lateral force (see Relationships between Lateral Force and Cell Body Width below), there was no obvious correlation between force and cell size, shape, and velocity. Measured traction force depends on many factors, including cell-substratum adhesions, the proportion of myosin(s) and actin assembled into filaments and cytoskeletal fibers, and the proportion of the force-generating machinery active at any one time (see also Additional Processes Are Required for Motility below).
Five observations together suggest an explanation for some of these varied patterns of traction forces: (1) the cell body appeared to be dragged along stiff substrata (Figure 3), which probably provided some “frictional” resistance to cell movement (see also Anderson et al., 1996); (2) lateral force applied at the flanks was not noticeably dependent on substratum compliance; however, (3) medial force was often much higher for cells moving on highly compliant substrata; (4) the cell body usually depressed highly compliant substrata; and (5) high lateral forces were not present in stationary cells (Figure 9). On the basis of these observations, we suggest that one factor that may elicit high force production in keratocytes is resistance to forward movement provided by the interaction between the substratum and the cell body.
We suggest that increased force in response to a load is to be expected in the short term on the basis of actin-myosin interactions known from studies of muscle (Hill, 1938) and in the longer term from the assembly of stress fibers in nonmuscle cells in response to applied stress (Kolega, 1986). These arguments do not exclude other possibilities, such as cell shape change induced by compliant substrata or changes in surface characteristics caused by UV irradiation of silicone rubber. However, even on stiff substrata that had been irradiated little and on which the cell body depressed the substratum only slightly (wrinkles were in focus everywhere under the cells), some cells produced high forces (Figure 5). Therefore, the ability to generate high forces appears to be an inherent property of some keratocytes, which undoubtedly would confer an advantage to cells having to pull themselves through tissue matrices in vivo.
Cell-Substratum Contacts
Consistent with the widespread application of traction forces, all areas of the keratocyte appear able to adhere to the substratum, as judged by IRM (Bereiter-Hahn et al., 1981; Kolega et al., 1982; Strohmeier and Bereiter-Hahn, 1984; Mittal and Bereiter-Hahn, 1985; Bereiter-Hahn and Voth, 1988; Lee et al., 1993b; Bereiter-Hahn and Luers, 1994; Lee and Jacobson, 1997; de Beus and Jacobson, 1998; the present work) and the distribution of integrins and other adhesion proteins (Lee and Jacobson, 1997; de Beus and Jacobson, 1998). In the present work, close contacts were often homogeneous under both the cell body and the lamellipodium in rapidly locomoting cells, although with much variability (Figures 2, B and D, 3B, 5D, and 6B). In contrast, close contacts under the lamellipodium were often lost when the lamellipodium appeared to slip and the cell slowed (Figures 2B and 5D).
These IRM data are consistent with sites of force application remaining stationary even while the entire cell passes over. This observation predicts that adhesion complexes would remain stationary during the same period. Stationary sites of force application are also consistent with “rolling” of the cell body, at least at its adherent ventral surface (Bereiter-Hahn et al., 1981; Anderson et al., 1996).
Possible Contributions of Myosin II and F-Actin to Traction Forces
The distribution of myosin II (muscle-type myosin that forms bipolar filaments, referred to simply as myosin below) in keratocyte lamellipodia has been studied in fixed and living cells at the light and electron microscope levels (Bereiter-Hahn et al., 1981; Anderson et al., 1996; Svitkina et al., 1997; see also Comparison with Other Types of Cells below) and shows a strong correspondence to traction forces in lamellipodia studied here: (1) discrete force spots and myosin spots are present in the lamellipodium; and (2) the size and density of spots of force and myosin increase from the periphery to the cell body.
Recent measurements of myosin distribution in living keratocytes and electron microscopy of fixed cells (Svitkina et al., 1997) have greatly strengthened the correspondence between myosin spots and our measurements of force spots: (2a) force spots and myosin spots both grow with time; (2b) force spots and myosin spots are stationary with respect to the substratum in locomoting cells; (2c) the density of force spots and myosin spots increases at the lamellipodium/cell body boundary; (2d) myosin spots become aligned with transverse fibers and move inward in the direction of large lateral displacements of force spots along the lamellipodium/cell body boundary; and (2e) under the cell body, force spots and myosin spots are displaced forward. These similarities strongly suggest that myosin is the primary generator of traction forces.
Actin polymerization is clearly initiated at the leading edge of migrating cells (Wang, 1985; Fisher et al., 1988; Forscher and Smith, 1988; Small et al., 1995) and could exert traction as well as protrusive force if it were to push against both the leading edge and the substratum (Mogilner and Oster, 1996; Abraham et al., 1999); indeed, keratocyte lamellipodia have been reported to push latex beads in their path (Bereiter-Hahn et al., 1981). However, we think that extension of the leading edge is unlikely to contribute significantly to traction forces for the following reasons. (1) Traction force is often much higher than predicted for leading edge protrusion based on actin polymerization (Mogilner and Oster, 1996; Abraham et al., 1999). (2) The gradient of F-actin density often observed in keratocyte lamellipodia is opposite to that of traction force and myosin (Strohmeier and Bereiter-Hahn, 1984; Bereiter-Hahn and Luers, 1994; Anderson et al., 1996; Svitkina et al., 1997). (3) Traction force in the lamellipodium is in the same direction as retrograde flow (Fisher et al., 1988; Forscher and Smith, 1988; our Figure 9), which is thought to require myosin activity but not actin polymerization; cytochalasin inhibition of actin polymerization does not inhibit the retrograde flow of lamellipodia in sea hare neuronal growth cones (Forscher and Smith, 1988) or the forward movement of the cell body in trout keratocytes (Anderson et al., 1996), whereas inhibition of myosin does inhibit retrograde flow (Lin et al., 1996). It is expected that such treatments would affect lamellipodial traction forces similarly. (4) We did not detect pushing forces at the leading edges of lamellipodia, even when the leading edge appeared to adhere to the substratum (as judged by IRM; see Lee and Jacobson [1997]). (5) Similarly, other actin-based structures, including filopodia, exert traction force toward the cell body after protruding and adhering to the substratum (Burton and Taylor, unpublished observations). (6) Retraction of the flanks into the cell body cannot be caused by actin polymerization.
Proposed Mechanical Mechanism of Cell Crawling
In this section, we propose an integrated, two-phase mechanism of motility based on patterns of traction forces, followed in the next section by a comparison with keratocyte cytoskeletal structure.
We observed that traction forces can be grouped into two phases that form a continuum on the basis of location, orientation, and magnitude. In the first phase, low forces (a few nanonewtons) under the lamellipodium are applied at force spots that grow and become centripetal, thus acting to pull the cell body forward. In the second phase, the lateral forces previously reported by Lee et al. (1994) and confirmed here arise directly from centripetal forces in the lamellipodium, serving (1) to release the flanks from the substratum (Lee et al., 1994; Oliver et al., 1995) and (2) to pull them forward into the advancing cell body.
Adhesions are broken when stress is higher than adhesive strength. We have shown that keratocytes can increase stress at an adhesion site during retraction even while force decreases. In addition, we have observed that force at a wrinkle usually increases monotonically until the cell retracts at the posterior margin (consistent with the results described by Lee et al., 1994), again strongly supporting our assertion that cells pull harder on their adhesions until they come loose. Our measurements do not exclude the possibility that a cell also weakens its adhesions by chemical (e.g., proteolytic) mechanisms; indeed, there is evidence for this in other types of cells.
Lateral force appears to be ideally positioned and oriented to contribute to retraction without perturbing the smooth gliding motion exemplified by keratocytes. The major component of lateral force is oriented perpendicular to the direction of motion, thus neither promoting nor retarding movement while breaking cell-substratum adhesions as the cell body passes. In this way, keratocytes can minimize the jerky motion characteristic of fibroblasts (Abercrombie et al., 1970), in which the tail applies high force that resists forward movement (Harris et al., 1981; Galbraith and Sheetz, 1997; Pelham and Wang, 1997; our Figure 10).
Comparison with Cytoskeletal Structure
Figure 12A compares the distribution of traction forces and the organization of actin filaments and fibers known from light and electron microscopy of keratocytes (Eutenuer and Schliwa, 1984; Strohmeier and Bereiter-Hahn, 1984; Cooper and Schliwa, 1986; Bereiter-Hahn and Luers, 1994; Small et al., 1995; Anderson et al., 1996; Svitkina et al., 1997).
Figure 12.
Mechanical and structural model of keratocyte locomotion. (A) Schematic diagram of a proposed two-phase mechanism of keratocyte locomotion. In the first phase, rearward-directed forces generated by the actin and myosin filament meshwork in the anterior lamellipodium pull the front of the cell body forward. In the second phase, higher lateral forces generated by actin-myosin fibers oriented across the cell body cause adhesions to detach, pulling the flanks and posterior cell body inward and forward. The midline represents a line of morphological, structural, and lateral force symmetry. A second line of symmetry is defined by smaller forces about the boundary of the lamellipodium and the cell body. These two lines cross to define a “point” of force symmetry. The large arrows in front of the cell refer to stronger lateral forces generated by transverse fibers, and the small arrows refer to weaker forces generated by network contraction, the net direction of which is parallel to the direction of motion. (B) Frontal view showing alternative consequences of transverse fiber contraction. (Top) Contraction of a cage of fibers tends to cause the cell body to round up on a compliant substratum that is easily pulled in by the flanks and depressed by the cell body; the cell body can also round up if it loses adhesion to the substratum. (Bottom) Internal shortening of curved transverse fibers causes them to straighten if they remain attached to stiff substrata at their ends. The fibers press inward on the cell body (down in the drawing), squeezing cytoplasm out toward the flanks.
We suggest that the first phase of the mechanism proposed here corresponds to graded assembly of myosin into spots in the lamellipodium (Strohmeier and Bereiter-Hahn, 1984; Anderson et al., 1996; Svitkina et al., 1997; see also DeBiasio et al. [1988] and Bailey et al. [1993] for myosin in fibroblast lamellipodia). The force spots and gradient of force in the lamellipodium are consistent with these structural data, but direct comparisons in individual cells are needed.
Svitkina et al. (1997) observed compression of groups of myosin spots and alignment of actin filaments into transverse bundles at the boundary of the lamellipodium and the cell body, which is strikingly consistent with our observation that compliant substrata are compressed across the same boundary. They proposed a “dynamic network contraction” model to account for these changes in cytoskeletal structure. Our force measurements are clearly consistent with their suggestion that contraction of the actin meshwork into transversely aligned fibers generates force to pull the cell body forward, equivalent to phase 1 of our mechanism.
It is highly likely that the lateral forces observed by Jacobson and colleagues (Lee et al., 1994; Oliver et al., 1995; Dembo et al., 1996) are produced by transverse actin-myosin fibers that terminate at the flanks (Strohmeier and Bereiter-Hahn, 1984; Anderson et al., 1996; Svitkina et al., 1997). Myosin is probably responsible for generating forces to detach adhesions, as suggested by experiments in Dictyostelium amoebae in which myosin was removed (Jay et al., 1995).
The suggested role of transverse fiber contraction in pulling the posterior flanks forward is in contrast to two recent proposals that were based on structural measurements (Anderson et al., 1996; Svitkina et al., 1997). Anderson et al. (1996) suggested that the fibers might generate rearward-directed forces at their termini, but our force measurements suggest that force is directed slightly forward in that region of freely locomoting cells. This forward-directed component of force pulls the posterior flanks forward, rather than the cell body backward, in contrast to the suggestion of Svitkina et al. (1997).
A lamellipodium appears to be necessary for lateral force production, because force was absent in regions lacking this structure (Figure 8). This observation is consistent with the suggestion of Svitkina et al. (1997) that transverse fibers responsible for lateral forces arise directly from the actin meshwork in the lamellipodium. They suggested that alignment of filaments increases in response to increased myosin activity in regions where actin filament density is also reduced. As they noted, this type of mechanism, whereby movement in response to contractile force is increased by reducing cytoskeletal stiffness, has been outlined in the solation-contraction coupling hypothesis (Taylor and Fechheimer, 1982). This process has been demonstrated in in vitro models (Taylor et al., 1973; Janson and Taylor, 1993).
Effects of Lateral Forces on Cell Body Width
Lateral forces have been suggested to arise from transverse fibers (Lee et al., 1994), which include fibers that arc up and over the cell body of keratocytes (Bereiter-Hahn et al., 1981; Anderson et al., 1996) and fibroblasts (Heath and Holifield, 1993; Kolega and Taylor, 1993; Giuliano and Taylor, 1994). The cell body tends to round up when a flank slips from the substratum, as expected for contraction of such a “cage” of fibers (Anderson et al., 1996), because they can shorten without changing the volume of the cell body (i.e., the surface/volume ratio decreases). If the substratum is highly compliant, the cell body can “round down” when adhesions are maintained (Figure 12B, top), suggesting a mechanism whereby highly compliant substrata are depressed by the cell body.
However, transverse fibers have also been suggested to maintain the characteristic spindle shape of keratocytes (Cooper and Schliwa, 1986; Anderson et al., 1996). Consistent with this suggestion, the cell body widened when lateral force increased, and vice versa (Figure 4), but the mechanism is not immediately obvious because actin-myosin fibers shorten when they contract (Kolega et al., 1991; Katoh et al., 1998). We suggest that curved fibers can shorten by straightening if their ends remain fixed to a stiff substratum (Figure 12B). A compressive force is then applied perpendicular to the fibers so that cytoplasm is “squeezed” toward the flanks, consistent with the observed decrease in the thickness of the cell body when it widened. Organelles also moved toward the flanks as force increased, reminiscent of cytoplasmic streaming in the contracting tails of amoebae (Taylor and Fechheimer, 1982). Cell body widening is not easily explained by increased actin polymerization, because evidence has been provided that protrusion is minimal at the flanks (Lee et al., 1993b). The suggested mechanism illustrates how axial sliding between actin and myosin filaments (Huxley and Hansen, 1954; Huxley and Niedergerke, 1954) can be converted into movement perpendicular to filament axes.
Additional Processes Are Required for Motility
Generation of traction is only part of the overall mechanism of motility because crawling movement requires a complete sequence of protrusion at the leading edge, adhesion to the substratum, application of traction force, and finally a reversal of the first two steps, i.e., detachment and retraction (Harris, 1990; DiMilla et al., 1991; Jay et al., 1995; Lauffenburger and Horwitz, 1996). The smooth motion of keratocytes implies a precise coupling between protrusion and phases 1 and 2 of traction force generation, the mechanism(s) of which will be a fruitful area for further investigation.
Actin and myosin monomers must be recycled forward from more posterior regions for filament assembly to occur at the leading edge (Bereiter-Hahn et al., 1981; Kolega et al., 1991; Janson and Taylor, 1993; Post et al., 1995; Anderson et al., 1996; DeBiasio et al., 1996). In keratocytes, fibers were observed by Svitkina et al. (1997) to disassemble under the cell body, analogous to earlier observations of stress fiber dissolution in the cell bodies of fibroblasts (Giuliano and Taylor, 1990; Kolega et al., 1991) and reduced assembly of actin and myosin in the cell body relative to the lamellum (Kolega and Taylor, 1993; Giuliano and Taylor, 1994).
Spatial gradients of contractility in keratocyte lamellipodia, lowest at the leading edge and highest at the retracting flanks (Lee et al., 1994), may be determined in part by gradients of myosin regulation. Brust-Mascher and Webb (1998) recently reported calcium waves in keratocyte lamellipodia that correlate with cell movements. In migrating fibroblasts, free Ca2+ concentration, calmodulin activation (Hahn et al., 1992), and phosphorylation of the regulatory light chain of myosin (Post et al., 1995) are on average higher in the cell body and tail than in the lamellum and lamellipodium. Although these gradients of regulatory properties have not been reported within lamellipodia per se, they do suggest a functional analogy between increased contraction in fibroblast tails and keratocyte flanks. In addition, inspection of images of calcium, calcium-calmodulin, and myosin light-chain phosphorylation in the lamellae of migrating fibroblasts shows that their distribution is not uniform; rather, they are localized into domains or “spots” that may represent sites of myosin activation. A direct correlation of myosin localization, regulation, and traction force will be required to test this possibility.
Comparison with Other Types of Cells
The mechanism proposed here contains elements similar to those of previous models of crawling motility. For example, Dunn (1980) proposed that cell movement results from a continuous contraction of the cytoskeleton after assembly at the leading edge; similarly, Bray and White (1988) suggested that the cortical flow observed in many types of cells (Fisher et al., 1988; Forscher and Smith, 1988) is fundamental to forward locomotion. Kolega and Taylor (1993) subsequently proposed an integration of the cortical flow (Bray and White, 1998) and solation-contraction coupling (Taylor et al., 1980; Taylor and Fechheimer, 1982) hypotheses to explain locomotion in terms of gradients of contractility.
We expect lamellipodia of most cell types to be capable of generating the kinds of traction forces observed here because their cytoskeletons appear to be similar. As in keratocytes, a network of actin filaments is present in fibroblasts (Small, 1981; Fisher et al., 1988; Abraham et al., 1999), macrophages (Rinnerthaler et al., 1991), and neuronal growth cones (Lewis and Bridgman, 1992; Lin et al., 1996). Likewise, transverse fibers are often located at the rear margin of fibroblast lamellipodia (DeBiasio et al., 1988; Fisher et al., 1988; Heath and Holifield, 1993) and may be related to the dense band of actin present in the plasma-gel sheet behind the leading edge of amoebae (Taylor et al., 1980). Myosin has been observed in lamellipodia of several cell types: fibroblasts (DeBiasio et al., 1988; Conrad et al., 1989; McKenna et al., 1989; Bailey et al., 1993 [myosin spots]; Verkhovsky et al., 1995), aortic endothelial cells (Kolega, 1998 [myosin in punctate aggregates]), Xenopus tissue cells (Kelley et al., 1996), rat neuronal growth cones (Bridgman and Dailey, 1989; Rochlin et al., 1995), and keratocytes (as discussed above).
Keratocytes and fibroblasts may represent two extremes of a continuum of morphologies: cultured fibroblasts usually possess a tail and keratocytes usually do not. But a keratocyte flank can adhere so strongly that it is stretched out (Lee et al., 1993a; Small et al., 1993; our Figure 2), and a fibroblast can briefly take on a keratocyte shape that has no tail (Kolega et al., 1982; Heath and Holifield, 1991b; Kolega and Taylor, 1993; our Figure 10). In both cases, force is applied in regions where actin-myosin fibers terminate at the rear margin of the cell, i.e., in the tail and/or flanks. Our observations of centripetal forces in the leading lamellae and tail of fibroblasts (Figure 10) agree with those of Galbraith and Sheetz (1997), who used microcantelevers to show rearward-directed forces in front of the nucleus and forward-directed forces in the tail of fibroblasts. The tail of a fibroblast, therefore, can slow forward movement by pulling against the leading edge (Harris et al., 1981). Keratocytes, in contrast, apply force largely perpendicular to the direction of motion (Lee et al., 1994), which we suggest addresses the problem of pulling adhesions loose without perturbing smooth forward locomotion.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge many insightful conversations with Dr. Gregory Fisher on cell motility in general and lamellipodial retrograde flow in particular. We thank Dr. John Kolega for assistance with isolating and culturing fish keratocytes. This work was supported by grants from the National Institutes of Health (AR-32461) and the National Science Foundation (Science and Technology Center grant MCB-8920118).
Abbreviations used:
- DIC
Nomarski differential interference microscopy
- IRM
interference reflection microscopy
Footnotes
REFERENCES
- Abercrombie M, Heaysman JEM, Pegrum SM. The locomotion of fibroblasts in culture. I. Movements of the leading edge. Exp Cell Res. 1970;59:393–398. doi: 10.1016/0014-4827(70)90646-4. [DOI] [PubMed] [Google Scholar]
- Abraham VC, Krishnamurthi V, Taylor DL, Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K, Wang Y-L, Small JV. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J Cell Biol. 1996;134:1209–1218. doi: 10.1083/jcb.134.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366:44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Luers H. The role of elasticity in the motile behavior of cells. In: Akkas N, editor. Biomechanics of Active Movement and Division of Cells, NATO ASI Series. H84. Berlin: Springer-Verlag; 1994. pp. 181–230. [Google Scholar]
- Bereiter-Hahn J, Strohmeier R, Kunzenbacher I, Beck K, Voth M. Locomotion of Xenopus epidermis cells in primary culture. J Cell Sci. 1981;52:289–311. doi: 10.1242/jcs.52.1.289. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Ionic control of locomotion and shape of epithelial cells. II. Role of monovalent cations. Cell Motil Cytoskeleton. 1988;10:528–536. doi: 10.1002/cm.970100409. [DOI] [PubMed] [Google Scholar]
- Bray D, White JG. Cortical flow in animal cells. Science. 1988;239:883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- Bridgman PC, Dailey ME. The organization of myosin and actin in rapid frozen nerve growth cones. J Cell Biol. 1989;108:95–109. doi: 10.1083/jcb.108.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Webb WW. Calcium waves induced by large voltage pulses in fish keratocytes. Biophys J. 1998;75:1669–1678. doi: 10.1016/S0006-3495(98)77609-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton K, Park JH, Taylor DL. Lamellar traction forces are centripetal and oriented in a continuous and symmetrical distribution about the direction of movement in crawling cells. Mol Biol Cell. 1996;7(suppl):391a. (abstract). [Google Scholar]
- Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997;385:450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- Conrad PA, Nederlof MA, Herman IM, Taylor DL. Correlated distribution of actin, myosin, and microtubules at the leading edge of migrating Swiss 3T3 fibroblasts. Cell Motil Cytoskeleton. 1989;14:527–543. doi: 10.1002/cm.970140410. [DOI] [PubMed] [Google Scholar]
- Cooper M, Schliwa M. Motility of cultured fish epidermal cells in the presence and absence of DC electric fields. J Cell Biol. 1986;102:1384–1399. doi: 10.1083/jcb.102.4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beus E, Jacobson K. Integrin involvement in keratocyte locomotion. Cell Motil Cytoskeleton. 1998;41:126–137. doi: 10.1002/(SICI)1097-0169(1998)41:2<126::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- DeBiasio RL, LaRocca GM, Post PL, Taylor DL. Myosin II transport, organization, and phosphorylation: evidence for cortical flow/solation-contraction coupling during cytokinesis and cell locomotion. Mol Biol Cell. 1996;7:1259–1282. doi: 10.1091/mbc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBiasio RL, Wang LL, Fisher GW, Taylor DL. The dynamic distribution of fluorescent analogs of actin and myosin in protrusions at the leading edge of migrating Swiss 3T3 fibroblasts. J Cell Biol. 1988;107:2631–2645. doi: 10.1083/jcb.107.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70:2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GA. Mechanisms of fibroblast locomotion. In: Curtis ASG, Pitts JD, editors. Cell Adhesion and Motility, 3rd Symposium B.S.C.B. Cambridge, UK: Cambridge University Press; 1980. pp. 409–423. [Google Scholar]
- Eutenuer U, Schliwa M. Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature. 1984;310:58–61. doi: 10.1038/310058a0. [DOI] [PubMed] [Google Scholar]
- Farkas DL, Baxter G, DeBiasio RL, Gough A, Nederlof MA, Pane D, Pane J, Patek DR, Ryan KW, Taylor DL. Multimode light microscopy and the dynamics of molecules, cells, and tissues. Annu Rev Physiol. 1993;55:785–817. doi: 10.1146/annurev.ph.55.030193.004033. [DOI] [PubMed] [Google Scholar]
- Fisher GW, Conrad PA, DeBiasio RL, Taylor DL. Centripetal transport of cytoplasm, actin, and the cell surface in lamellipodia of fibroblasts. Cell Motil Cytoskeleton. 1988;11:235–247. doi: 10.1002/cm.970110403. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA. 1997;94:9114–9118. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano KA, Nederlof MA, DeBiasio R, Lanni F, Waggoner AS, Taylor DL. Multimode light microscopy. In: Herman B, Jacobson K, editors. Optical Microscopy for Biology. New York: Wiley-Liss; 1990. pp. 543–557. [Google Scholar]
- Giuliano KA, Taylor DL. Formation, transport, contraction and disassembly of stress fibers in fibroblasts. Cell Motil Cytoskelelton. 1990;16:14–21. doi: 10.1002/cm.970160104. [DOI] [PubMed] [Google Scholar]
- Giuliano KA, Taylor DL. Fluorescent actin analogs with a high affinity for profilin in vitro exhibit an enhanced gradient of assembly in living cells. J Cell Biol. 1994;124:971–983. doi: 10.1083/jcb.124.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K, DeBiasio R, Taylor DL. Patterns of elevated free calcium and calmodulin activation in living cells. Nature. 1992;359:736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- Harris AK. Traction, and its relations to contraction in tissue cell locomotion. In: Bellairs R, Curtis A, Dunn G, editors. Cell Behavior. Cambridge, UK: Cambridge University Press; 1982. pp. 109–134. [Google Scholar]
- Harris AK. Tissue culture cells on deformable substrata: biomechanical implications. J Biomech Eng. 1984;106:19–24. doi: 10.1115/1.3138449. [DOI] [PubMed] [Google Scholar]
- Harris AK. Fibroblasts and myofibroblasts. Methods Enzymol. 1988;163:623–642. doi: 10.1016/0076-6879(88)63053-9. [DOI] [PubMed] [Google Scholar]
- Harris AK. Protrusive activity of the cell surface and the movements of tissue cells. In: Akkas N, editor. Biomechanics of Active Movement and Deformation of Cells, NATO ASI Series. H42. Berlin: Springer-Verlag; 1990. pp. 249–294. [Google Scholar]
- Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Heath JP, Holifield B. Actin alone in lamellipodia. Nature. 1991a;352:107–108. doi: 10.1038/352107a0. [DOI] [PubMed] [Google Scholar]
- Heath JP, Holifield B. Cell locomotion: new research tests old ideas on membrane and cytoskeletal flow. Cell Motil Cytoskeleton. 1991b;18:245–257. doi: 10.1002/cm.970180402. [DOI] [PubMed] [Google Scholar]
- Heath JP, Holifield B. On the mechanism of cortical actin flow and its role in cytoskeletal organization of fibroblasts. In: Jones G, Wigley C, Warn R, editors. Cell Behavior: Adhesion and Motility, Symposia of the Society for Experimental Biology 47. Cambridge, UK: The Company of Biologists; 1993. pp. 35–56. [PubMed] [Google Scholar]
- Hill AV. The heat of shortening and the dynamic constants of muscle. Proc R Soc Lond B Biol Sci. 1938;126:136–195. doi: 10.1098/rspb.1949.0019. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. Interference microscopy of living muscle fibers. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- Janson LW, Taylor DL. In vitro models of tail contraction and cytoplasmic streaming in amoeboid cells. J Cell Biol. 1993;123:345–356. doi: 10.1083/jcb.123.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay PY, Pham PA, Wong SA, Elson EL. A mechanical function of myosin II in cell motility. J Cell Sci. 1995;108:387–393. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kano Y, Masuda M, Onishi H, Fujiwara K. Isolation and contraction of the stress fiber. Mol Biol Cell. 1998;9:1919–1938. doi: 10.1091/mbc.9.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CA, Sellers JR, Gard D, Bui D, Adelstein RS, Baines IC. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Effects of mechanical tension on protrusive activity and microfilament and intermediate filament organization in an epidermal epithelium moving in culture. J Cell Biol. 1986;102:1400–1411. doi: 10.1083/jcb.102.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Cytoplasmic dynamics of myosin IIA and IIB: spatial ‘sorting’ of isoforms in locomoting cells. J Cell Sci. 1998;111:2085–2095. doi: 10.1242/jcs.111.15.2085. [DOI] [PubMed] [Google Scholar]
- Kolega J, Janson LW, Taylor DL. The role of solation-contraction coupling in regulating stress fiber dynamics in nonmuscle cells. J Cell Biol. 1991;114:993–1003. doi: 10.1083/jcb.114.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J, Shure MS, Chen W-T, Young ND. Rapid cellular translocation is related to close contacts formed between various cultured cells and their substrata. J Cell Sci. 1982;54:23–34. doi: 10.1242/jcs.54.1.23. [DOI] [PubMed] [Google Scholar]
- Kolega J, Taylor DL. Gradients in the concentration and assembly of myosin II in living fibroblasts during locomotion and fiber transport. Mol Biol Cell. 1993;4:819–836. doi: 10.1091/mbc.4.8.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lee J, Ishihara A, Jacobson K. The fish epidermal keratocyte as a model system for the study of cell locomotion. In: Jones G, Wigley C, Warn R, editors. Cell Behavior: Adhesion and Motility, Symposia of the Society for Experimental Biology 47. Cambridge, UK: The Company of Biologists; 1993a. pp. 73–89. [PubMed] [Google Scholar]
- Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993b;362:167–171. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- Lee J, Jacobson K. The composition and dynamics of cell-substratum adhesions in locomoting fish keratocytes. J Cell Sci. 1997;110:2833–2844. doi: 10.1242/jcs.110.22.2833. [DOI] [PubMed] [Google Scholar]
- Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. Traction forces generated by locomoting keratocytes. J Cell Biol. 1994;127:1957–1964. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- McKenna NM, Wang Y-L, Konkel ME. Formation and movement of myosin-containing structures in living fibroblasts. J Cell Biol. 1989;109:1163–1172. doi: 10.1083/jcb.109.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal AK, Bereiter-Hahn J. Ionic control of locomotion and shape of epithelial cells. I. Role of calcium influx. Cell Motil. 1985;5:123–136. doi: 10.1002/cm.970050205. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T, Dembo M, Jacobson K. Traction forces in locomoting cells. Cell Motil Cytoskeleton. 1995;331:225–240. doi: 10.1002/cm.970310306. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MA. Theory of deformable substrates for cell motility studies. Biophys J. 1996;71:657–669. doi: 10.1016/S0006-3495(96)79266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post PL, DeBiasio RL, Taylor DL. A fluorescent protein biosensor of myosin II regulatory light chain phosphorylation reports a gradient of phosphorylated myosin II in migrating cells. Mol Biol Cell. 1995;6:1755–1768. doi: 10.1091/mbc.6.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler G, Herzog M, Klappacher M, Kunka H, Small JV. Leading edge movement and ultrastructure in mouse macrophages. J Struct Biol. 1991;106:1–16. doi: 10.1016/1047-8477(91)90058-5. [DOI] [PubMed] [Google Scholar]
- Rochlin MW, Itoh K, Adelstein RS, Bridgman PC. Localization of myosin II A and B isoforms in cultured neurons. J Cell Sci. 1995;108:3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- Small JV. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J Cell Biol. 1981;91:695–705. doi: 10.1083/jcb.91.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Herzog M, Anderson K. Actin filament organization in the fish keratocyte lamellipodium. J Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Rohlfs A, Herzog M. Actin and cell movement. In: Jones G, Wigley C, Warn R, editors. Cell Behavior: Adhesion and Motility, Symposia of the Society for Experimental Biology 47. Cambridge, UK: The Company of Biologists; 1993. pp. 57–71. [PubMed] [Google Scholar]
- Strohmeier R, Bereiter-Hahn J. Control of cell shape and locomotion by external calcium. Exp Cell Res. 1984;154:412–420. doi: 10.1016/0014-4827(84)90165-4. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade K, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL, Condeelis JS, Moore PL, Allen RD. The contractile basis of amoeboid movement. I. The chemical control of motility in isolated cytoplasm. J Cell Biol. 1973;59:378–394. doi: 10.1083/jcb.59.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL, Fechheimer M. Cytoplasmic structure and contractility: the solation-contraction coupling hypothesis. Philos Trans R Soc Lond B Biol Sci. 1982;299:185–197. doi: 10.1098/rstb.1982.0125. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Wang Y-L, Heiple JM. Contractile basis of ameboid movement. VII. The distribution of fluorescently labeled actin in living amebas. J Cell Biol. 1980;86:590–598. doi: 10.1083/jcb.86.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Thomas T. The Motility and Morphology of Glioblastoma Cells on Polymeric Compliant Substrata. Thesis. Pittsburgh, PA: Carnegie Mellon University; 1998. [Google Scholar]
- Thoumine O, Ott A. Influence of adhesion and cytoskeletal integrity on fibroblast traction. Cell Motil Cytoskeleton. 1996;35:269–280. doi: 10.1002/(SICI)1097-0169(1996)35:3<269::AID-CM8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.