Abstract
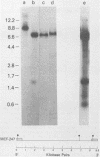
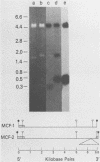
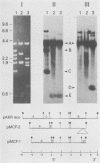
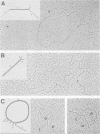
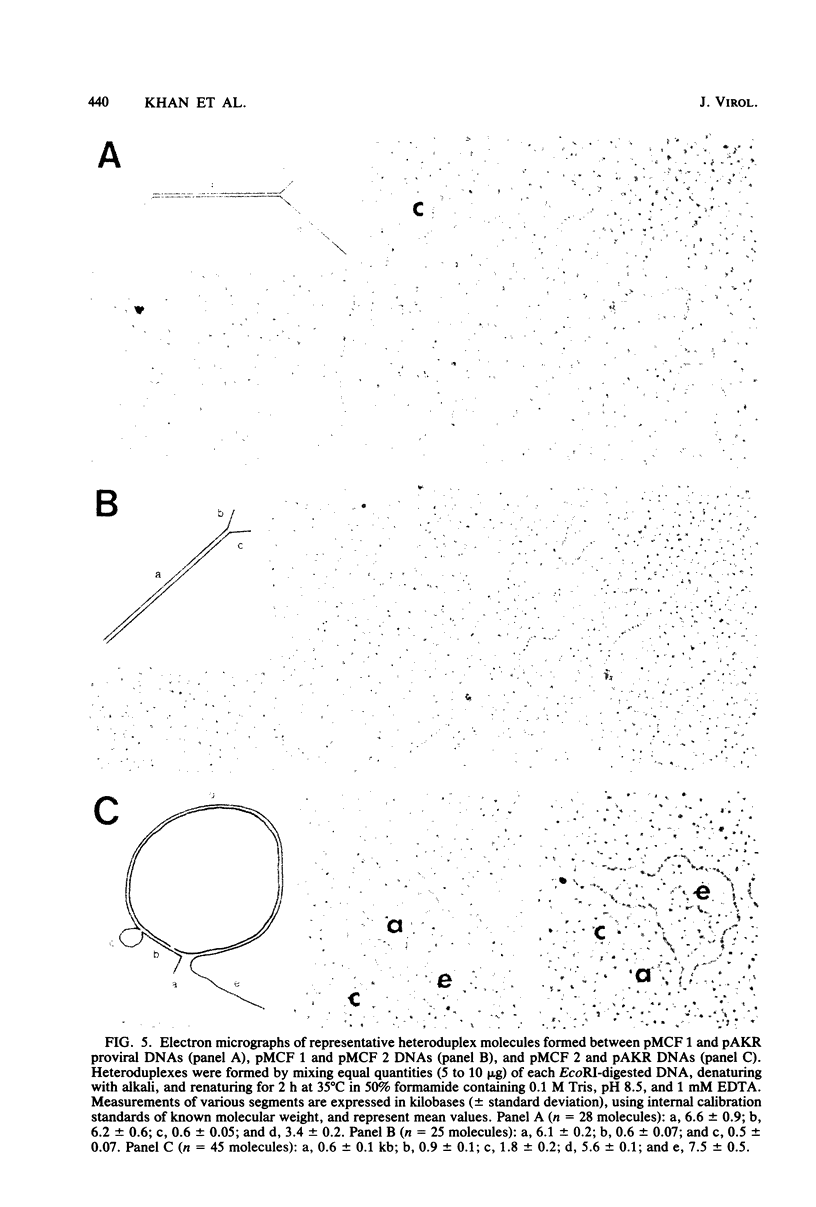
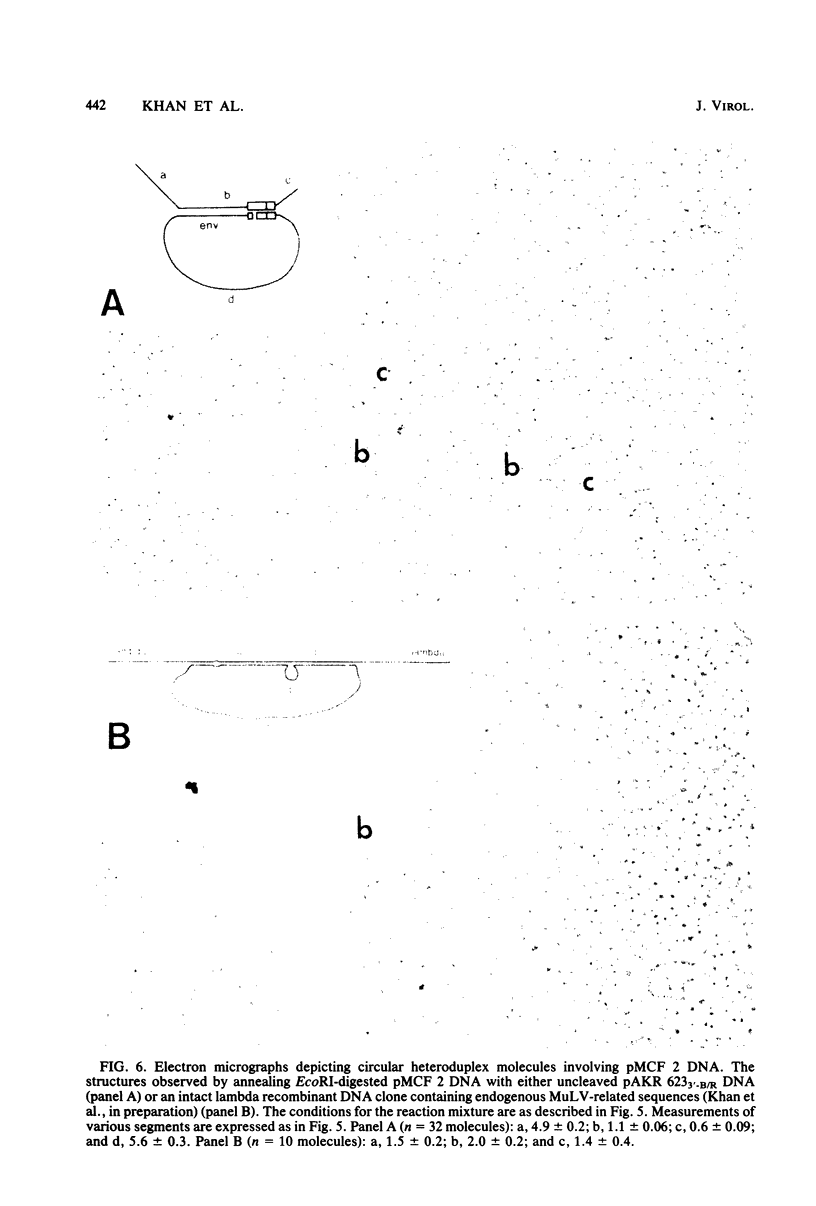
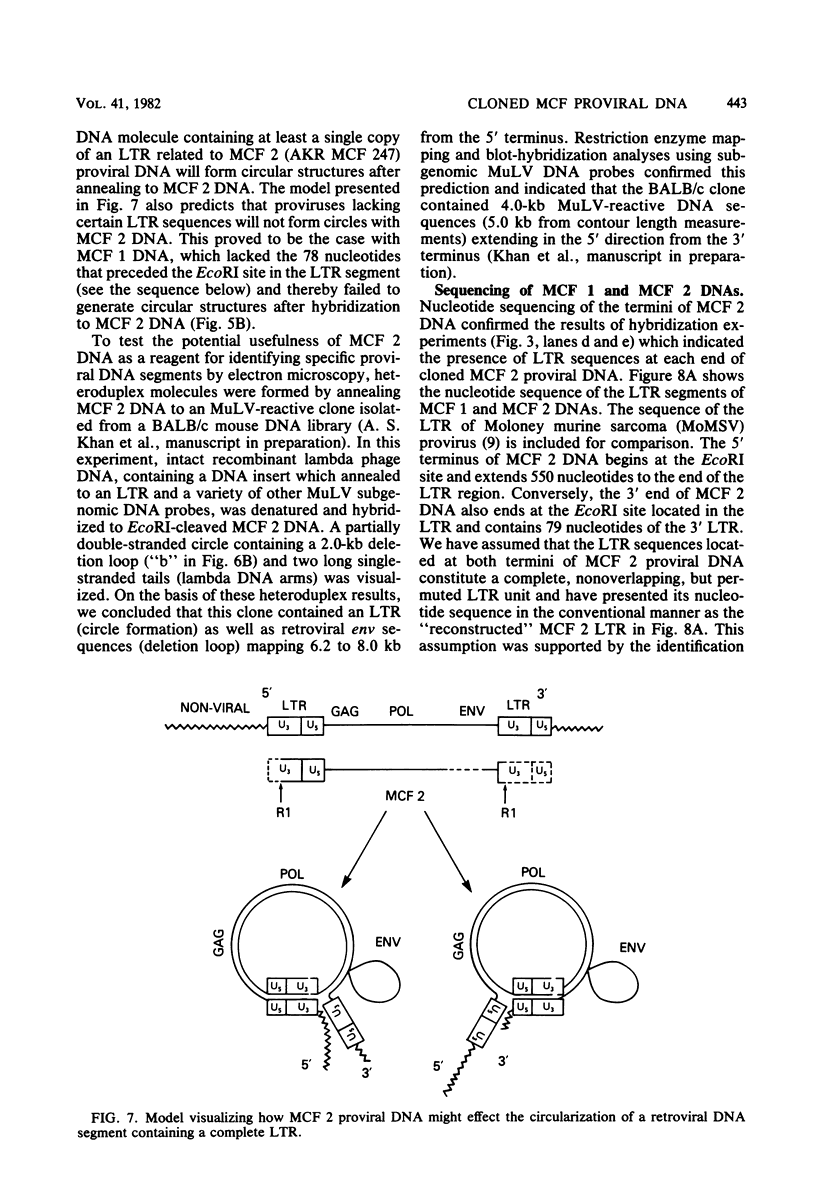
Two proviruses were cloned from EcoRI-digested DNA extracted from mink cells chronically infected with AKR mink cell focus-forming (MCF) 247 murine leukemia virus (MuLV), using a lambda phage host vector system. One cloned MuLV DNA fragment (designated MCF 1) contained sequences extending 6.8 kilobases from an EcoRI restriction site in the 5' long terminal repeat (LTR) to an EcoRI site located in the envelope (env) region and was indistinguishable by restriction endonuclease mapping for 5.1 kilobases (except for the EcoRI site in the LTR) from the 5' end of AKR ecotropic proviral DNA. The DNA segment extending from 5.1 to 6.8 kilobases contained several restriction sites that were not present in the AKR ecotropic provirus. A 0.5-kilobase DNA segment located at the 3' end of MCF 1 DNA contained sequences which hybridized to a xenotropic env-specific DNA probe but not to labeled ecotropic env-specific DNA. This dual character of MCF 1 proviral DNA was also confirmed by analyzing heteroduplex molecules by electron microscopy. The second cloned proviral DNA (designated MCF 2) was a 6.9-kilobase EcoRI DNA fragment which contained LTR sequences at each end and a 2.0-kilobase deletion encompassing most of the env region. The MCF 2 proviral DNA proved to be a useful reagent for detecting LTRs electron microscopically due to the presence of nonoverlapping, terminally located LTR sequences which effected its circularization with DNAs containing homologous LTR sequences. Nucleotide sequence analysis demonstrated the presence of a 104-base-pair direct repeat in the LTR of MCF 2 DNA. In contrast, only a single copy of the reiterated component of the direct repeat was present in MCF 1 DNA.
Full text
PDF

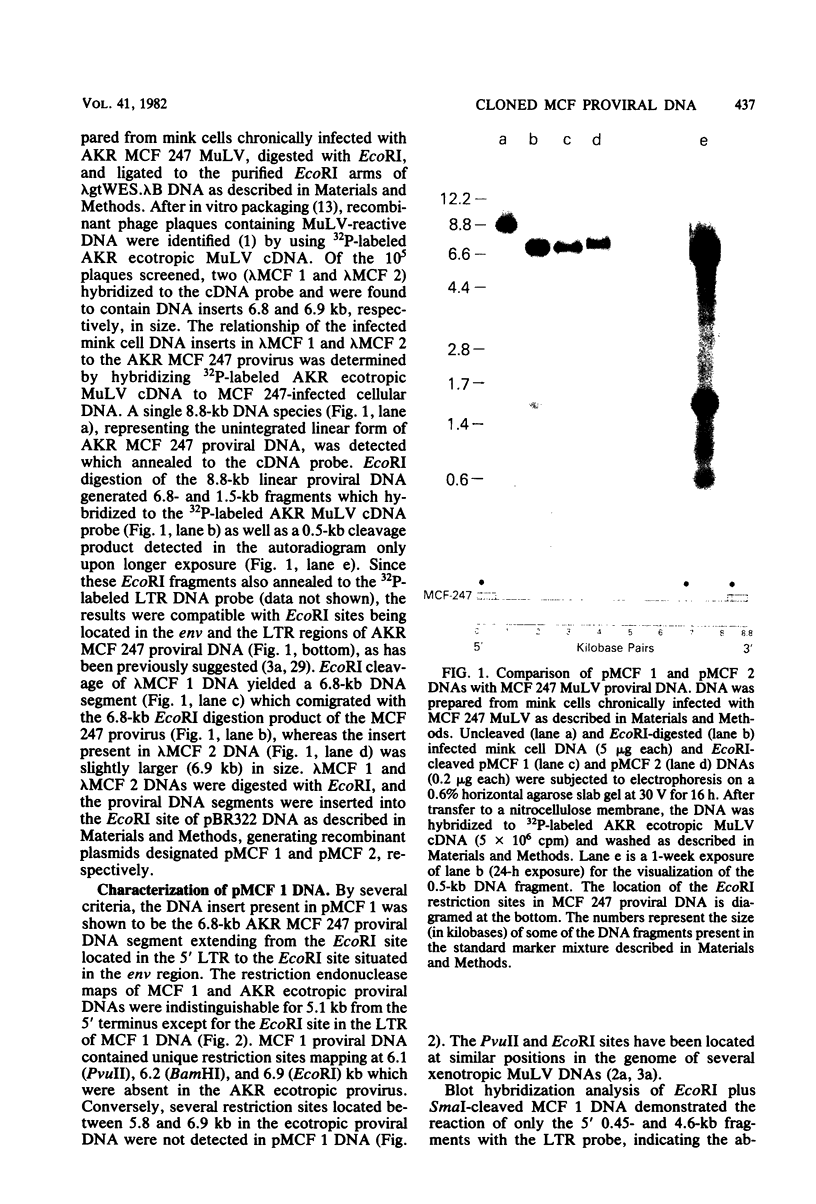






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berns A. J., Lai M. H., Bosselman R. A., McKennett M. A., Bacheler L. T., Fan H., Maandag E. C., van der Putten H. V., Verma I. M. Molecular cloning of unintegrated and a portion of integrated moloney murine leukemia viral DNA in bacteriophage lambda. J Virol. 1980 Oct;36(1):254–263. doi: 10.1128/jvi.36.1.254-263.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler C. E., Hoggan M. D., Chan H. W., Sears J. F., Khan A. S., Moore J. L., Hartley J. W., Rowe W. P., Martin M. A. Cloning and characterization of an envelope-specific probe from xenotropic murine leukemia proviral DNA. J Virol. 1982 Jan;41(1):228–236. doi: 10.1128/jvi.41.1.228-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. W., Bryan T., Moore J. L., Staal S. P., Rowe W. P., Martin M. A. Identification of ecotropic proviral sequences in inbred mouse strains with a cloned subgenomic DNA fragment. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5779–5783. doi: 10.1073/pnas.77.10.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lander M. R., Gupta S., Rands E., Lowy D. R. Origin of mink cytopathic focus-forming (MCF) viruses:comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981 Sep;113(2):465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Verma I. M., Shih T. Y., Scolnick E. M., Davidson N. Heteroduplex analysis of the sequence relations between the RNAs of mink cell focus-inducing and murine leukemia viruses. J Virol. 1978 Oct;28(1):352–360. doi: 10.1128/jvi.28.1.352-360.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloyd M. W., Hartley J. W., Rowe W. P. Cell-surface antigens associated with recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1979 Mar 1;149(3):702–712. doi: 10.1084/jem.149.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloyd M. W., Hartley J. W., Rowe W. P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980 Mar 1;151(3):542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. H., Gautsch J. W., Jensen F. C., Lerner R. A., Hartley J. W., Rowe W. P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger P. J., Ihle J. N., de Noronha F., Bolognesi D. P. Oncogenic and immunogenic potential of cloned HIX virus in mice and cats. Med Microbiol Immunol. 1977;164(1-3):119–129. doi: 10.1007/BF02121308. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager G. L., Chang E. H., Chan H. W., Garon C. F., Israel M. A., Martin M. A., Scolnick E. M., Lowy D. R. Molecular cloning of the Harvey sarcoma virus closed circular DNA intermediates: initial structural and biological characterization. J Virol. 1979 Sep;31(3):795–809. doi: 10.1128/jvi.31.3.795-809.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. C., Lieber M. M., Todaro G. J. Mink cell line Mv 1 Lu (CCL 64). Focus formation and the generation of "nonproducer" transformed cell lines with murine and feline sarcoma viruses. Virology. 1974 Jul;60(1):282–287. doi: 10.1016/0042-6822(74)90386-9. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Karshin W. L., Arcement L. J., Naso R. B., Arlinghaus R. B. Common precursor for Rauscher leukemia virus gp69/71, p15(E), and p12(E). J Virol. 1977 Sep;23(3):787–798. doi: 10.1128/jvi.23.3.787-798.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K., Ikeda H., Hartley J. W., Stockert E., Rowe W. P., Old L. J. Changes in expression of murine leukemia virus antigens and production of xenotropic virus in the late preleukemic period in AKR mice. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4680–4684. doi: 10.1073/pnas.73.12.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy D. R., Rands E., Chattopadhyay S. K., Garon C. F., Hager G. L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc Natl Acad Sci U S A. 1980 Jan;77(1):614–618. doi: 10.1073/pnas.77.1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung M. L., Hering C., Hartley J. W., Rowe W. P., Hopkins N. Analysis of the genomes of mink cell focus-inducing murine type-C viruses: a progress report. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1269–1274. doi: 10.1101/sqb.1980.044.01.138. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
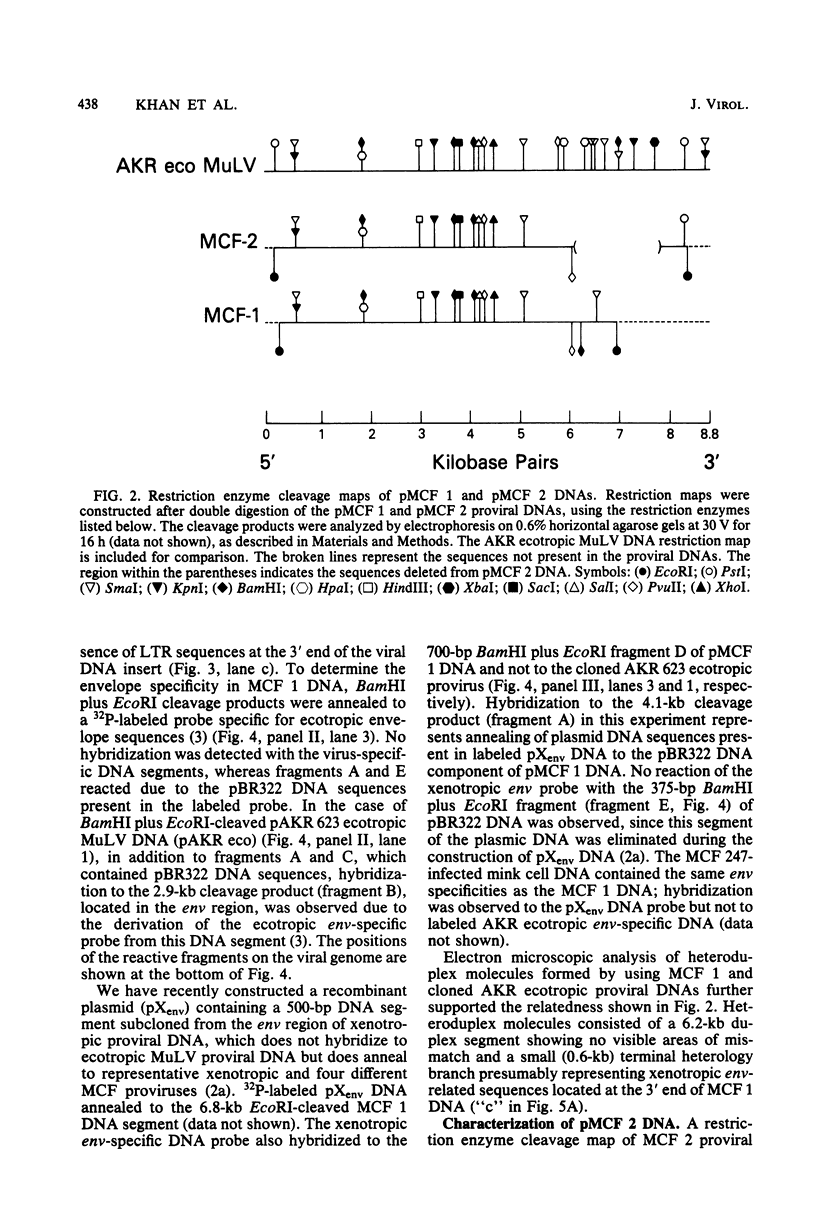
- Martin M. A., Bryan T., McCutchan T. F., Chan H. W. Detection and cloning of murine leukemia virus-related sequences from African green monkey liver DNA. J Virol. 1981 Sep;39(3):835–844. doi: 10.1128/jvi.39.3.835-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Nowinski R. C., Hays E. F. Oncogenicity of AKR endogenous leukemia viruses. J Virol. 1978 Jul;27(1):13–18. doi: 10.1128/jvi.27.1.13-18.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. V., Stockert E., Obata Y., DeLeo A. B., Old L. J. Murine-leukemia-virus-related cell-surface antigens as serological markers of AKR ecotropic, xenotropic, and dualtropic viruses. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1255–1264. doi: 10.1101/sqb.1980.044.01.136. [DOI] [PubMed] [Google Scholar]
- Pedersen F. S., Buchhagen D. L., Chen C. Y., Hays E. F., Haseltine W. A. Characterization of virus produced by a lymphoma induced by inoculation of AKR MCF-247 virus. J Virol. 1980 Jul;35(1):211–218. doi: 10.1128/jvi.35.1.211-218.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Rands E., Lowy D. R., Lander M. R., Chattopadhyay S. K. Restriction endonuclease mapping of ecotropic murine leukemia viral DNAs: size and sequence heterogeneity of the long terminal repeat. Virology. 1981 Jan 30;108(2):445–452. doi: 10.1016/0042-6822(81)90451-7. [DOI] [PubMed] [Google Scholar]
- Rassart E., Jolicoeur P. Restriction endonuclease mapping of unintegrated viral DNA of B- and N-tropic BALB/c murine leukemia virus. J Virol. 1980 Sep;35(3):812–823. doi: 10.1128/jvi.35.3.812-823.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Canaani E., Robbins K. C., Tronick S. R., Zain S., Aaronson S. A. Nucleotide sequence analysis of the transforming region and large terminal redundancies of Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5234–5238. doi: 10.1073/pnas.77.9.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J., Faller D. V., Hopkins N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc Natl Acad Sci U S A. 1978 Jan;75(1):495–499. doi: 10.1073/pnas.75.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Cloyd M. W., Hartley J. W. Status of the association of mink cell focus-forming viruses with leukemogenesis. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1265–1268. doi: 10.1101/sqb.1980.044.01.137. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Green N., Liu F. T., Niman H. L., Lerner R. A. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980 Oct 30;287(5785):801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxler D. H., Lowy D., Howk R., Young H., Scolnick E. M. Friend strain of spleen focus-forming virus is a recombinant between ecotropic murine type C virus and the env gene region of xenotropic type C virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4671–4675. doi: 10.1073/pnas.74.10.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxler D. H., Yuan E., Linemeyer D., Ruscetti S., Scolnick E. M. Helper-independent mink cell focus-inducing strains of Friend murine type-C virus: potential relationship to the origin of replication-defective spleen focus-forming virus. J Exp Med. 1978 Sep 1;148(3):639–653. doi: 10.1084/jem.148.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]