Abstract
Coiled coils have a rich history in the field of protein design and engineering. Novel structures, such as the first 7-helix coiled coil, continue to provide surprises and insights. Large-scale data sets quantifying the influence of systematic mutations on coiled-coil stability are a valuable new asset to the area. Scoring methods based on sequence and/or structure can predict interaction preferences in coiled-coil-mediated bZIP transcription factor dimerization. Experimental and computational methods for dealing with the near-degeneracy of many coiled-coil structures appear promising for future design applications.
Introduction
Coiled coils remain beguiling after more than two decades of close scrutiny. Their supercoiled structures are encoded by a seven-residue repeat that can often be detected in sequence data [1,2]. The heptad repeat, denoted [abcdefg]n, typically has hydrophobic residues at a and d, and polar/charged residues at e and g (Figure 1). Many protein engineers have introduced some variant of this pattern into de novo peptide sequences, often with surprising results. Despite an apparent simplicity at the sequence level, a large number of structural variations are observed among coiled coils. Dimers, trimers, tetramers, pentamers and at least one heptamer have been reported, and these can vary in their helix orientations and alignments, as well as in whether they form homo- or hetero-complexes. This poses an interesting specificity problem. How, within the confines of the heptad sequence repeat, are such diverse structures encoded? Design studies, serendipitous discoveries and systematic analyses continue to provide new insights.
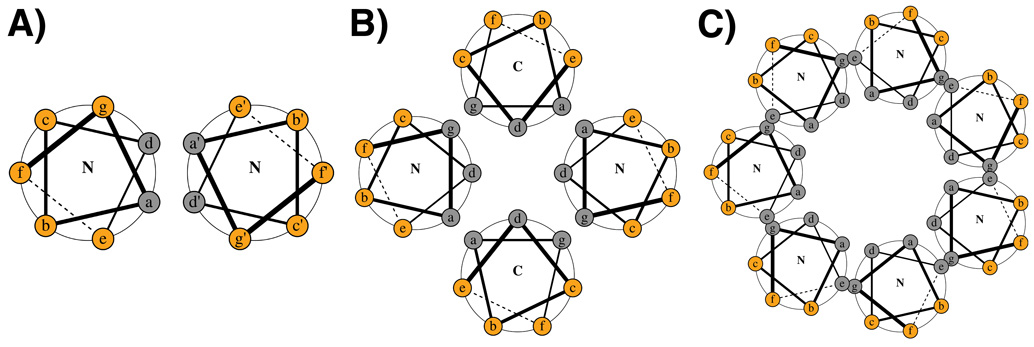
Figure 1.
Helical wheels for coiled coils of varying topology. Heptad positions are shown in small letters, with gray and orange circles indicating predominantly hydrophobic and predominantly polar/charged residues, respectively. A The canonical 3–4 heptad repeat, in which hydrophobic residues are 3 and 4 amino acids apart, is found for many coiled coils including dimers (shown in the figure), trimers and tetramers. Prime notation (e.g. a’) in this figure and throughout the main text is used to indicate a residue on the opposite chain. B An antiparallel tetramer with a 3-3-1 repeat, as in Figure 2B and reference [14]. C A parallel seven-helix coiled coil with a 3-1-2-1 hydrophobic pattern, as in Figure 2D and reference [19]. Note that these hydrophobic patterns do not uniquely specify these structures. Other features, including a-a’ asparagine hydrogen bonding, can be important. Helical wheels were made using DrawCoil 1.0 (http://www.gevorggrigoryan.com/drawcoil/)
Coiled coils offer attractive features to the protein designer and were among the first rationally designed structures [3]. Hydrophobic-polar patterning imposes association of helices, and charge patterning and other features can be used to confer specificity. For example, heterodimerization can be introduced by making one helix basic and another acidic, and a preference for asparagine residues to pair at a-a’ positions can influence oligomerization state and helix orientation [4]. High symmetry simplifies the description of coiled-coil structure, and mathematical methods for describing ideal coiled-coil backbones have been developed [5,6]. Coiled coils present experimental advantages as well. Short peptides of ~30 residues fold to give stable complexes, allowing for facile introduction of both native and non-native amino acids via peptide synthesis. Circular dichroism can report on cooperative folding and association, which are usually reversible.
Interest in coiled coils is heightened by their prevalence in biology, and by their potential applications in materials science and synthetic biology [7,8]. Coiled coils are predicted in ~10% of all eukaryotic proteins and are associated with widely ranging functions [9–11]. In materials science and nanotechnology, the rod-like shapes and distinct folded-to-disordered transitions of coiled coils have inspired numerous applications. The utility of charge-pairing rules and other simple principles for controlling helix association are also valuable for engineering nanomaterials with defined structures [12].
Reviews have summarized much of the accumulated knowledge regarding coiled coils [1,4,7,13]. Here, we focus on recent results in three areas. First, coiled coils continue to surprise us with their variability and sensitivity to sequence changes. Second, we describe database analyses, systematic experiments and computational modeling studies that elucidate principles controlling specificity and stability in coiled coils. Finally, experiments and calculations demonstrate progress in identifying synthetic peptide ligands for native sequences that can form coiled coils.
Novel structures, switches and functions
Altering the hydrophobic/polar patterning of the coiled-coil heptad repeat can generate a variety of structures. For example, the Lu group has reported variants of the yeast GCN4 transcription factor coiled coil with hydrophobic substitutions at the e or g positions, generating 3-3-1 hydrophobic repeats. Four peptides with this pattern form homotetramers, although the structures show orientations and axial alignments that differ according to the substitutions made (Figure 2A–B) [14–16]. The GCN4 variants all contain an asparagine residue at one of the hydrophobic positions, and the hydrogen-bonding potential of this group is accommodated in a variety of ways. The resulting topologies reflect the combined influence of the 3-3-1 hydrophobic repeat, the Asn residue that disrupts this repeat, and perhaps other features. Right-handed helices can be encoded by 11-residue 3-4-4 repeats, as in a classic design study by Harbury et al. [17]. The Harbury tetramer contained alloisolucine to satisfy core packing requirements, but more recently the structure of a designed right-handed tetramer with only biological amino acids was solved (Figure 2C) [18].
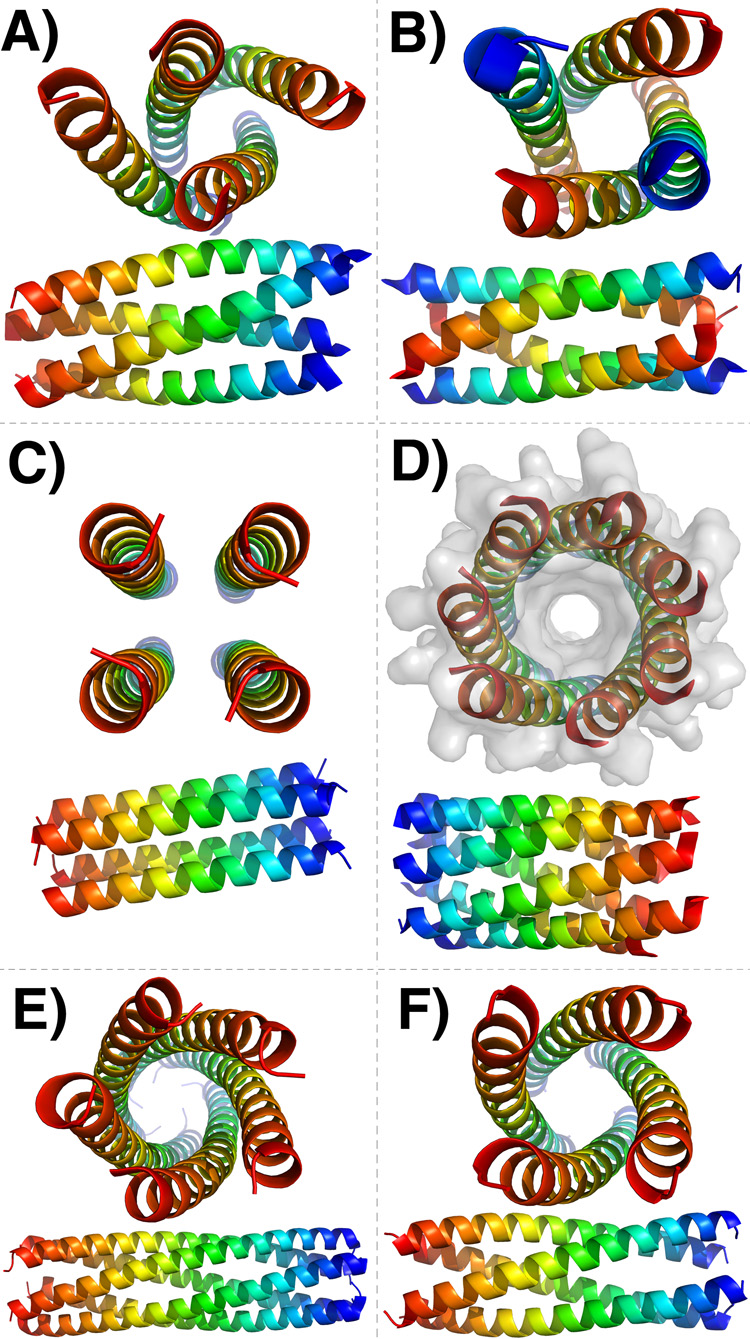
Figure 2.
New and interesting coiled-coil structures. Each coiled coil is shown axially and from the side. Color indicates helix orientation: blue – N-terminus, red – C-terminus. A, B and D Show variants of a GCN4-derived coiled coil with altered hydrophobic-polar patterning. A When all e positions are mutated to Val, departing from the canonical 3–4 repeat and creating a 3-3-1 hydrophobic repeat, the resulting sequence gives a parallel tetramer [15]. B When all g positions are substituted with either Val or Ala, also producing a 3-3-1 repeat, the result is an anti-parallel tetramer [14]. C A right-handed parallel coiled-coil tetramer [18]. D A parallel heptamer is formed when both e and g positions of GCN4 are substituted with Ala [19]. E–F A coiled coil with phenylalanine at all a and d positions folds as a parallel pentamer (E), while if just one of these phenylalanines is substituted by methionine the resulting structure is a tetramer (F) [20]
A fascinating new topology, the first 7-helix coiled coil, is formed when both e and g positions of the GCN4 leucine zipper are replaced with non-polar alanine (Figure 2D) [19]. The structure has parallel helices, a large superhelical radius and a continuous channel through the middle of the coil. Interestingly, the rotational symmetry typical of parallel coiled coils is broken, and a 1-residue axial shift per helix gives a screw axis instead. The seven-helix coiled coil accommodates the a-position asparagine residue through a continuous pattern of Asn-Asn hydrogen bonding that is disrupted only at one helix-helix interface. The buried Asn residues may be as critical as the hydrophobic patterning for determining the structure.
Many coiled-coil structures lie close together in sequence and/or energy space, and this has led to the discovery and design of peptides that switch topology given small changes in sequence or conditions. Liu et al. report a parallel five-helix coiled coil with Phe at all a and d positions. When a single Phe is mutated to Met, a tetramer is formed instead (Figure 2E and F) [20]. Yadav et al. similarly identified a tetrameric GCN4 variant in which a single amino-acid change alters topology [21]. Substitution of glutamate with cysteine at an e position switched a parallel tetramer to an anti-parallel one. Strikingly, substitution with serine led to a sequence that crystallized both as parallel and anti-parallel tetramers. Molecular dynamics simulations reproduced the parallel preference of the parent peptide and the anti-parallel preference of the cysteine mutant.
It is not yet possible to systematically predict the structures that form, or the changes in structure that occur, in cases such as those discussed above. Predicting structure is complicated by an enormous diversity of possible topologies. At least five different packing geometries have been reported for antiparallel tetramers alone [16], and continuous axial shifts, as seen in the parallel heptamer, present a new parameter to be considered. Coiled-coil structures are not determined by simple rules for hydrophobic packing. For example, whereas Lu and colleagues found a pentameric structure for a peptide with Phe at all a and d positions, Yoder et al. report a shorter sequence with the same core residues that forms a trimer [20,22]. Good agreement of calculations with experimental data in the study by Yadav et al. suggests that explanations for some unexpected changes may be accessible via computation [21]. The modeling in this case was aided by the availability of x-ray structures of both relevant topologies, however, and using these types of computational approaches to make predictions will be more difficult.
Coiled coils can also act as conformational switches in response to various stimuli. Kuhlman and colleagues used a computational approach to design a sequence that forms a coiled-coil homotrimer yet switches to the α/β zinc-finger fold in the presence of metals [23]. A peptide with similar properties was designed by hand in 2005 [24], and both designs show solution properties consistent with the intended folds. Coiled-coil peptides have also been designed to form fibers (both amyloid and non-amyloid) upon external triggering by pH or temperature, or simply upon mixing [25–28].
Interesting applications are suggested by studies where function is switched on or off using coiled coils. Loh and colleagues fused the bZIP domain of GCN4 and the enzyme barnase such that a topological constraint prevented them from existing in their folded states simultaneously. While folding of barnase normally prevailed, addition of AP-1 DNA drove GCN4 dimerization and switched off barnase activity [29]. Tanaka and colleagues inserted two out of three helices from a hetero-specific coiled-coil trimer into a circularly permuted version of RNaseT1. They showed that adding the missing helix could constitute the trimer, and with it the enzyme [30]. Reversible photo-activation of coiled-coil transcription factor binding to DNA has also been demonstrated [31]. These applications illustrate the potential utility of coiled coils for biomedical or materials applications, which will be enhanced as we understand sequence-structure relationships better.
Uncovering relationships between sequence, structure and stability
Interactions among a, d, e and g residues account for most structural specificity in coiled coils. Important determinants include electrostatic interactions involving e and g sites, packing complementarity of a and d positions, and hydrogen bonding requirements of buried or partially buried polar groups [4,13]. The influence of e/g electrostatics on coiled-coil stability has long been controversial and continues to attract attention [32,33]. A review by Bosshard and colleagues now clarifies many of the issues that have caused confusion in the literature, mostly due to poor definitions of reference states in energy comparisons [34].
Uncovering specificity-determining features by mining the PDB is becoming feasible now that the database is large and can be automatically surveyed for coiled coils [35]. Issues related to sampling remain severe, however, as the crystallized structures have many biases, and homo-oligomers are especially over-represented. Straussman et al. report that anti-parallel coiled coils in the PDB contain more charged residues in core positions than parallel ones, although it is not yet clear how to interpret this difference [36]. Kammerer et al. previously described a short sequence motif that is common in parallel trimers in the PDB but rare in other types of coiled coils, and mutational data support this motif favoring trimer formation [37].
Kennan and colleagues used non-natural amino acids to explore the influence of side-chain length on coiled-coil stability. Adding methylene groups to lysine and glutamate side chains at e and g positions in an ACID/BASE heterodimer significantly increased coiled-coil stability [38]. Modulating the length of guanidinium-functionalized side chains at a central a position had less effect. However, a difference in stability between pairing asparagine at a with aspartate vs glutamate at the opposing a’ was used to engineer four sequences that specifically segregated when mixed to give primarily two heterodimers [39]. Helix orientation in these designed dimers was not established.
Exciting advances relating coiled-coil sequences to stabilities and specificities come from systematic sets of quantitative measurements. Acharya et al. report a tour de force of 100 measurements of homo and heterotypic substitutions at a-a’-position pairs in a model bZIP-like coiled-coil heterodimer [40]. These data illustrate homotypic preferences for most hydrophobic residues and Asn, and heterotypic preferences for other polar or charged residues: Thr, Lys, Arg and Glu. The 100 measurements allow calculation of double-alanine coupling energies. Asparagine is particularly important for imparting specificity and shows large unfavorable coupling energies with all residues tested except Lys, Arg and itself. The a-a’ data can now be combined with measured preferences at g-e’ positions and relative stabilities of homotypic d-d’ interactions, generating a powerful dataset for analyzing sequence-stability relationships in leucine zippers [41].
An efficient method for measuring relative stabilities in anti-parallel coiled-coil dimers has now been developed as well [42]. Relative stabilities for all pair-wise interactions among Leu, Ile, Val, Asn and Ala interacting at a-d’ positions have been reported. Further, strong sequence dependencies are apparent in the contributions of “vertical” interactions, i.e. interactions within the core along the coiled-coil axis, such as a’-a-a’ [43]. These measurements, along with earlier studies of anti-parallel interaction preferences [44], should be valuable for understanding determinants of anti-parallel coiled-coil dimer stability and specificity.
In another large-scale study, a coiled-coil peptide from GCN4 was tested for interaction with 589 single-mutation variants and 4320 double-mutation variants using cellulose-membrane arrays [45]. The utility of this very large data set for advancing our understanding is somewhat limited because the stoichiometries and helix orientations of most of the heterocomplexes formed are not known.
Calculation of specificity from sequence and structure
Principles governing coiled-coil stability and specificity would ideally be encoded in predictive models, and several such models have been proposed. A benchmark to assess performance is provided by a study from our lab that measured all pair-wise interactions among the leucine-zipper regions of most of the 53 human bZIP transcription factors [46]. These proteins homo and heterodimerize via a parallel dimeric coiled coil [41].
A machine-learning model trained on coiled-coil data not including the human bZIP interactions performs very well at predicting these, although it is somewhat difficult to interpret mechanistically [47]. A simpler sequence-based model for scoring different coiled-coil dimers involves summing up relevant interhelical coupling energies [40,41]. However, this does not work well for predicting bZIP pairs [48]. The most likely reason is that not all important coupling energies have been measured and, in particular, coupling of d-position residues is likely significant. With an artificial weight introduced to favor Leu-Leu pairing at d-d’ over all other possibilities, we found that a coupling-energy model does very well (G.G. and A.K., unpublished). Another simple model that also performs very well was proposed by Mason et al. [49]. Weights in this model were hand-selected based on the Vinson data and the authors’ experience.
Although it is noteworthy that extremely simple models can perform well, there are a number of caveats. For example, in the model by Mason et al., some unrealistic assumptions are made: a-a' and d-d' residue pair weights were assumed to be equivalent, and many weights were empirical. Although the model predicts bZIP association preferences well, this certainly does not mean that a-a’ and d-d’ interactions are equivalent or that the weights in the model are "correct". The risk of such models is that they will not perform well outside of the sequence space where they were validated or trained.
There has been progress in using structure-based modeling to understand coiled-coil interactions. We employed an ideal-backbone side-chain packing approach to model bZIP coiled-coil interaction preferences [48]. This performed similarly to the best sequence-based approaches, but only after several limitations of this type of modeling were addressed. We used a similar method to predict parallel vs. anti-parallel orientation for coiled-coil dimers that is ~81% accurate on known structures [50]. Ramos et al. used a molecular dynamics approach to dissect the oligomerization preferences of four coiled-coil sequences [51]. Each sequence was modeled in each of four oligomerization states. Impressively, the calculations correctly identified the native oligomerization state as having the lowest free energy for each sequence. However, the correct crystal-structure templates were used when modeling the native oligomers. Although some of the “memory” of the correct backbone was erased by molecular dynamics, the general applicability of this approach for cases where experimental structures are not available is not yet clear.
Coiled-coil partners for native proteins
It is highly desirable to be able to engineer custom peptides that interact with native coiled coils in a specific manner. Vinson and colleagues have described an elegant “A-ZIP” method for creating peptides that bind and inhibit native bZIP transcription factors [52,53]. A-ZIPs are based on bZIPs, but their basic DNA-binding region is replaced with an acidic extension. One limitation of the A-ZIP strategy is that the interaction specificity of the designs is inherited from the bZIP on which they are based.
In an attempt to make bZIP inhibitors by modifying the affinity and interaction specificity of existing bZIP coiled coils, Mason et al. used a selection approach. Peptides were selected from a combinatorial library to interact with cJun and cFos [49]. The best binders formed strong associations with their targets. However, other undesired complexes were also very stable. To address this, the authors modified their assay such that library members were selected in the presence of competitor peptides [54]. The resulting peptides bound to cJun and cFos stronger than they homo-dimerized or interacted with the each other’s target.
These experiments illustrate a need for methods that optimize both affinity and specificity. An appealing approach to this problem was pioneered by Havranek and Harbury [55], but applying this broadly confronts obstacles. We recently developed a framework for specificity design that is flexible and computationally efficient. Our method, called CLASSY, uses the technique of cluster expansion to dramatically accelerate design calculations [56]. CLASSY is based on optimizing the stability of a design•target complex while imposing a constraint on the stability difference between the design•target complex and design•undesired partner complexes. We have applied it to design peptides intended to bind each of the 20 families of human bZIP coiled coils while interacting minimally with the remaining 19 families. Many designs indeed bind their targets with high specificity (G.G., A. Reinke and A.K., unpublished).
Conclusions
The coiled-coil energy landscape is degenerate, in that small changes in sequence can lead to large changes in structure. This presents challenges and opportunities to protein engineers. Despite a great body of knowledge, we cannot yet predict coiled-coil topologies from sequence, or reliably design coiled-coil complexes other than the simplest folds. Future coiled-coil designs and structures will surely continue to surprise us. As a model for studying structural specificity, and as a scaffold with high functional potential, coiled coils remain as compelling as ever. New experimental and computational methods with the capacity to consider multiple states will advance both our basic understanding of coiled-coil structural biology and the practical application of coiled-coil engineering to problems in biomedical and materials science.
Acknowledgements
We thank members of the Keating laboratory, especially J. Apgar, N. Zizlsperger, A. Reinke and O. Ashenberg for work in this area and comments on the manuscript. Funding for work on coiled coils in the lab is provided by the NIH (GM67681) and the NSF (CAREER award MCB0347203 to AK, and equipment grant 0216437).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lupasq AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 2.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 3.Betz SF, Bryson JW, DeGrado WF. Native-like and structurally characterized designed alpha-helical bundles. Curr Opin Struct Biol. 1995;5:457–463. doi: 10.1016/0959-440x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 4.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 5.Crick FH. The Fourier Transform of a Coiled-Coil. Acta Cryst. 1953;6:685–689. [Google Scholar]
- 6.Harbury PB, Tidor B, Kim PS. Repacking protein cores with backbone freedom: structure prediction for coiled coils. Proc. Natl. Acad. Sci. U S A. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupas A. Coiled coils: new structures and new functions. Trends in Biochemical Sciences. 1996;21:375–382. [PubMed] [Google Scholar]
- 8.Bromley EH, Channon K, Moutevelis E, Woolfson DN. Peptide and protein building blocks for synthetic biology: from programming biomolecules to self-organized biomolecular systems. ACS Chem Biol. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose A, Schraegle SJ, Stahlberg EA, Meier I. Coiled-coil protein composition of 22 proteomes--differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol. 2005;5:66. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose A, Meier I. Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins. Cell Mol Life Sci. 2004;61:1996–2009. doi: 10.1007/s00018-004-4039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolfson DN, Ryadnov MG. Peptide-based fibrous biomaterials: Some things old, new and borrowed. Curr Opin Chem Biol. 2006;10:559–567. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 14.Deng YQ, Liu J, Zheng Q, Eliezer D, Kallenbach NR, Lu M. Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat. Structure. 2006;14:247–255. doi: 10.1016/j.str.2005.10.010.* This paper and reference [15] report the structural consequences of mutating a GCN4 leucine-zipper peptide to impose a 3-3-1 hydrophobic repeat. When 3 charged g positions are mutated to either valine or alanine, a similar anti-parallel tetramer structure results. An a-position asparagine forms inter-helical hydrogen bonds and is exposed to solvent. Both tetramers are very stable in solution, with the melting temperature of the valine variant over 90 °C. This suggests that charged residues at g and e positions act as a negative design element in native GCN4.
- 15.Liu J, Deng YQ, Zheng Q, Cheng CS, Kallenbach NR, Lu M. A parallel coiled-coil tetramer with offset helices. Biochemistry. 2006;45:15224–15231. doi: 10.1021/bi061914m.* When 3 e-position residues in a GCN4-derived coiled coil are mutated from charged residues to valine, an extremely unusual parallel tetramer results. The packing resembles two parallel trimers that are joined by sharing two helices. The result is a core that is packed more tightly than in more canonical 4-fold symmetrical parallel tetramers. A continuous network of hydrogen bonds connects four a-position asparagines.
- 16.Liu J, Zheng Q, Deng Y, Li Q, Kallenbach NR, Lu M. Conformational specificity of the Lac repressor coiled-coil tetramerization domain. Biochemistry. 2007;46:14951–14959. doi: 10.1021/bi701930d. [DOI] [PubMed] [Google Scholar]
- 17.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 18.Sales M, Plecs JJ, Holton JM, Alber T. Structure of a designed, right-handed coiled-coil tetramer containing all biological amino acids. Protein Science. 2007;16:2224–2232. doi: 10.1110/ps.062702907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Zheng Q, Deng YQ, Cheng CS, Kallenbach NR, Lu M. A seven-helix coiled coil. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15457–15462. doi: 10.1073/pnas.0604871103.** This paper describes the crystal structure of a remarkable parallel coiled-coil heptamer. Several unique features include a one-amino-acid axial offset between adjacent helices, which results in an exactly one-heptad shift between the first and the seventh helices. This feature leads to a- and d-position residues forming two continuous screws in the core of the structure. The a-position asparagines in the center of the coiled coil form a string of hydrogen bonds, which is terminated only in the interface between the 1st and 7th helices. The authors report that substitution of this asparagine with valine, serine or threonine causes unfolding of the molecule in solution, while substitution with glutamine appears to retain the same structure.
- 20.Liu J, Zheng Q, Deng YQ, Kallenbach NR, Lu M. Conformational transition between four and five-stranded phenylalanine zippers determined by a local packing interaction. Journal of Molecular Biology. 2006;361:168–179. doi: 10.1016/j.jmb.2006.05.063.* A change of just a single amino acid per helix led to a dramatic change of structure in this study. Mutation of a core phenylalanine to a methionine switched the oligomerization state and also caused the two most N-terminal heptads of the pentamer to become disordered, as shown in x-ray structures of both coiled coils. The authors discuss how the folding pathway of the peptides may be perturbed by the mutation.
- 21.Yadav MK, Leman LJ, Price DJ, Brooks CL, Stout CD, Ghadiri MR. Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution. Biochemistry. 2006;45:4463–4473. doi: 10.1021/bi060092q.** The surprising discovery that a peptide assumed to form a parallel tetramer instead formed an anti-parallel tetramer structure led to an analysis of 12 mutational variants of the parent peptide GCN4-pLI. GCN4-pLI is parallel, while the point mutant Glu20Cys is anti-parallel. Glu20Ser was crystallized as both a parallel and an anti-parallel tetramer. Computational analyses using replica exchange suggested a balance between packing preferences for the parallel orientation being offset by electrostatic preferences for anti-parallel arrangements. The glutamate at position 20 in GCN4-pLI reduces the electrostatic preference such that a parallel structure is formed. Without this charge, the parallel and anti-parallel structures are predicted to be similarly favorable, in agreement with the experiments.
- 22.Yoder NC, Kumar K. Selective protein-protein interactions driven by a phenylalanine interface. Journal of the American Chemical Society. 2006;128:188–191. doi: 10.1021/ja055494k. [DOI] [PubMed] [Google Scholar]
- 23.Ambroggio XI, Kuhlman B. Computational design of a single amino acid sequence that can switch between two distinct protein folds. Journal of the American Chemical Society. 2006;128:1154–1161. doi: 10.1021/ja054718w.* RosettaDesign was used to search for a sequence compatible with both a trimeric coiled-coil backbone and a 2Cys-2His zinc-finger. The best-scoring sequence was synthesized and characterized. Results from circular dichroism spectroscopy, cobalt absorption spectroscopy and analytical ultracentrifugation were consistent with the peptide reversibly switching between the intended folds upon addition of zinc or cobalt.
- 24.Cerasoli E, Sharpe BK, Woolfson DN. ZiCo: a peptide designed to switch folded state upon binding zinc. J Am Chem Soc. 2005;127:15008–15009. doi: 10.1021/ja0543604. [DOI] [PubMed] [Google Scholar]
- 25.Ciani B, Hutchinson EG, Sessions RB, Woolfson DN. A designed system for assessing how sequence affects alpha to beta conformational transitions in proteins. J Biol Chem. 2002;277:10150–10155. doi: 10.1074/jbc.M107663200. [DOI] [PubMed] [Google Scholar]
- 26.Kammerer RA, Kostrewa D, Zurdo J, Detken A, Garcia-Echeverria C, Green JD, Muller SA, Meier BH, Winkler FK, Dobson CM, et al. Exploring amyloid formation by a de novo design. Proc Natl Acad Sci U S A. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kammerer RA, Steinmetz MO. De novo design of a two-stranded coiled-coil switch peptide. Journal of Structural Biology. 2006;155:146–153. doi: 10.1016/j.jsb.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Engineering nanoscale order into a designed protein fiber. Proc Natl Acad Sci U S A. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha JH, Butler JS, Mitrea DM, Loh SN. Modular enzyme design: Regulation by mutually exclusive protein folding. Journal of Molecular Biology. 2006;357:1058–1062. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuzawa S, Mizuno T, Tanaka T. Activating an enzyme by an engineered coiled coil switch. Chemistry-a European Journal. 2006;12:7345–7352. doi: 10.1002/chem.200600007. [DOI] [PubMed] [Google Scholar]
- 31.Woolley GA, Jaikaran ASI, Berezovski M, Calarco JP, Krylov SN, Smart OS, Kumita JR. Reversible photocontrol of DNA binding by a designed GCN4-bZIP protein. Biochemistry. 2006;45:6075–6084. doi: 10.1021/bi060142r. [DOI] [PubMed] [Google Scholar]
- 32.Matousek WM, Ciani B, Fitch CA, Garcia-Moreno B, Kammerer RA, Alexandrescu AT. Electrostatic contributions to the stability of the GCN4 leucine zipper structure. Journal of Molecular Biology. 2007;374:206–219. doi: 10.1016/j.jmb.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier M, Burkhard P. Statistical analysis of intrahelical ionic interactions in alpha-helices and coiled coils. Journal of Structural Biology. 2006;155:116–129. doi: 10.1016/j.jsb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Bosshard HR, Marti DN, Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit. 2004;17:1–16. doi: 10.1002/jmr.657. [DOI] [PubMed] [Google Scholar]
- 35.Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 36.Straussman R, Ben-Ya'acov A, Woolfson DN, Ravid S. Kinking the coiled coil - Negatively charged residues at the coiled-coil interface. Journal of Molecular Biology. 2007;366:1232–1242. doi: 10.1016/j.jmb.2006.11.083. [DOI] [PubMed] [Google Scholar]
- 37.Kammerer RA, Kostrewa D, Progias P, Honnappa S, Avila D, Lustig A, Winkler FK, Pieters J, Steinmetz MO. A conserved trimerization motif controls the topology of short coiled coils. Proc Natl Acad Sci U S A. 2005;102:13891–13896. doi: 10.1073/pnas.0502390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan SJ, Kennan AJ. Variable stability heterodimeric coiled-coils from manipulation of electrostatic interface residue chain length. Journal of the American Chemical Society. 2007;129:10255–10260. doi: 10.1021/ja073717w. [DOI] [PubMed] [Google Scholar]
- 39.Diss ML, Kennan AJ. Orthogonal recognition in dimeric coiled coils via buried polar-group modulation. J Am Chem Soc. 2008;130:1321–1327. doi: 10.1021/ja076265w. [DOI] [PubMed] [Google Scholar]
- 40.Acharya A, Rishi V, Vinson C. Stability of 100 homo and heterotypic coiled-coil a-a ' pairs for ten amino acids ( A, L, I, V, N, K, S, T, E, and R) Biochemistry. 2006;45:11324–11332. doi: 10.1021/bi060822u.** A heterodimerizing leucine zipper was used to study the effects of substitutions at a pair of opposing a positions. This coiled coil is established to retain its structure upon local mutation. Each of 100 heterodimers was characterized by thermal denaturation monitored by circular dichroism. Dimerization free energies and alanine double-mutant coupling energies were calculated, elegantly dissecting the importance of various core residues for coiled-coil stability and specificity. The data are especially valuable for rationalizing and predicting the interaction behavior of bZIP transcription factor coiled coils, and provide a rare benchmark for computational methods.
- 41.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadley EB, Gellman SH. An antiparallel alpha-helical coiled-coil model system for rapid assessment of side-chain recognition at the hydrophobic interface. Journal of the American Chemical Society. 2006;128:16444–16445. doi: 10.1021/ja067178r.* A thiol-thioester exchange assay that can be read out by HPLC was developed to quantify side-chain interaction preferences in a short anti-parallel coiled coil. The authors examined a-d’ interactions and report 24 free-energy differences relative to Ala-Ala for all possible pairs among Leu, Ile, Val, Asn and Ala. This assay provides an efficient way to gather a large amount of data describing anti-parallel coiled coils. See also reference [42].
- 43.Hadley EB, Testa OD, Woolfson DN, Gellman SH. Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proc Natl Acad Sci U S A. 2008;105:530–535. doi: 10.1073/pnas.0709068105.** Using the assay of reference [43], the authors extended their studies to examine a-d’ interactions in two different contexts. Significant differences between energies measured in two environments implicated a role for “vertical” interactions (here a’-a-a’) in affecting anti-parallel coiled-coil stability. These results are supported by studies in a different, longer anti-parallel coiled coil, and by striking differences in the conformational heterogeneity of side chains in energetically preferred vs non-preferred combinations.
- 44.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 45.Portwich M, Keller S, Strauss HM, Mahrenholzx CC, Kretzschmar I, Kramer A, Volkmer R. A network of coiled-coil associations derived from synthetic GCN4 leucine-zipper arrays. Angewandte Chemie-International Edition. 2007;46:1654–1657. doi: 10.1002/anie.200603246. [DOI] [PubMed] [Google Scholar]
- 46.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 47.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. Journal of Molecular Biology. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036.** This paper reports the use of structure-based models to predict bZIP interaction preferences. Models were built using idealized fixed backbones and side-chain packing, followed by continuous side-chain relaxation and energy evaluation. A number of critical limitations of this type of modeling were elucidated and addressed. Several critical a-a’ interactions were modeled poorly and consequently replaced with terms derived from other studies to give a model with good predictive performance. The study highlights the predicted importance of core-to-edge interactions such as a-g’ and d-e’ for determining coiled-coil dimer stability and specificity.
- 49.Mason JM, Schmitz MA, Muller K, Arndt KM. Semirational design of Jun-Fos coiled coils with increased affinity: Universal implications for leucine zipper prediction and design. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8989–8994. doi: 10.1073/pnas.0509880103.** Protein-fragment complementation was used to select peptides that bind Fos or Jun out of libraries of ~105 members. All pair-wise mixtures that could be formed among the two selected peptides and all members of the Jun and Fos families were thermodynamically characterized, resulting in 45 melting temperatures. The selected peptides bound their targets strongly, although other combinations also produced stable interactions. The authors propose a weight-based model to rationalize their data that performs well for predicting interaction preferences measured in reference [46].
- 50.Apgar JR, Gutwin KN, Keating AE. Predicting helix orientation for coiled-coil dimers. Proteins-Structure Function and Bioinformatics. 2008 doi: 10.1002/prot.22118. in press.* This paper reports the development, testing and comparison of several methods for predicting coiled-coil dimer orientation specificity. Methods were tested on 131 examples of known structure. Explicit-structure methods involved modeling sequences on large numbers of parallel and anti-parallel backbones and evaluating the energies of these models using a variety of scoring functions. Implicit-structure methods used sequence input and involved scoring pairs of residue-residue interactions with different weights. The best methods of each type achieved ~81% prediction accuracy.
- 51.Ramos J, Lazaridis T. Energetic determinants of oligomeric state specificity in coiled coils. Journal of the American Chemical Society. 2006;128:15499–15510. doi: 10.1021/ja0655284. [DOI] [PubMed] [Google Scholar]
- 52.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. Embo J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerdes MJ, Myakishev M, Frost NA, Rishi V, Moitra J, Acharya A, Levy MR, Park SW, Glick A, Yuspa SH, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66:7578–7588. doi: 10.1158/0008-5472.CAN-06-1247. [DOI] [PubMed] [Google Scholar]
- 54.Mason JM, Muller KM, Arndt KM. Positive aspects of negative design: Simultaneous selection of specificity and interaction stability. Biochemistry. 2007;46:4804–4814. doi: 10.1021/bi602506p.* The assay of reference [49] was extended to allow for the simultaneous selection of specificity and stability by including competitor peptides in the selections. Two peptides produced in this way, one to bind Fos and one to bind Jun, were shown to associate with their respective targets stronger than they homodimerized or interacted with each other’s targets.
- 55.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 56.Grigoryan G, Zhou F, Lustig SR, Ceder G, Morgan D, Keating AE. Ultra-fast evaluation of protein energies directly from sequence. PLoS Comput Biol. 2006;2:e63. doi: 10.1371/journal.pcbi.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]