Abstract
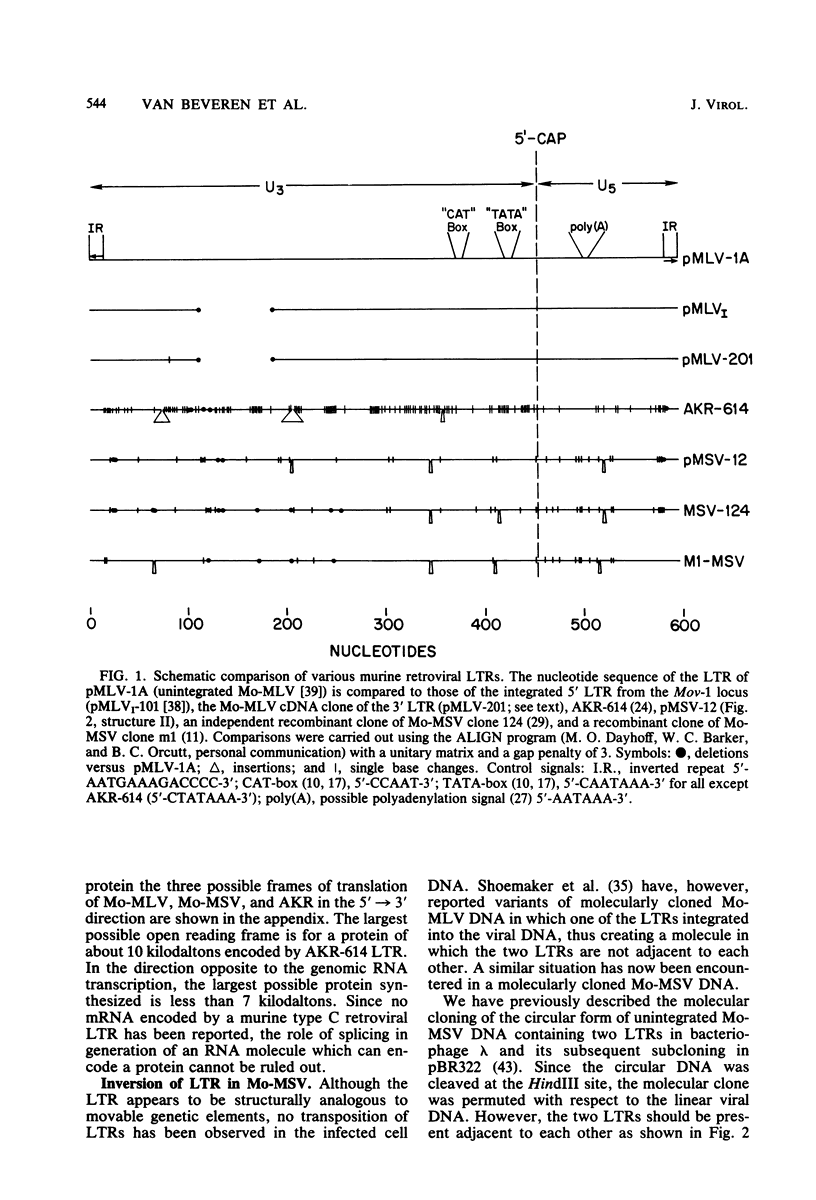
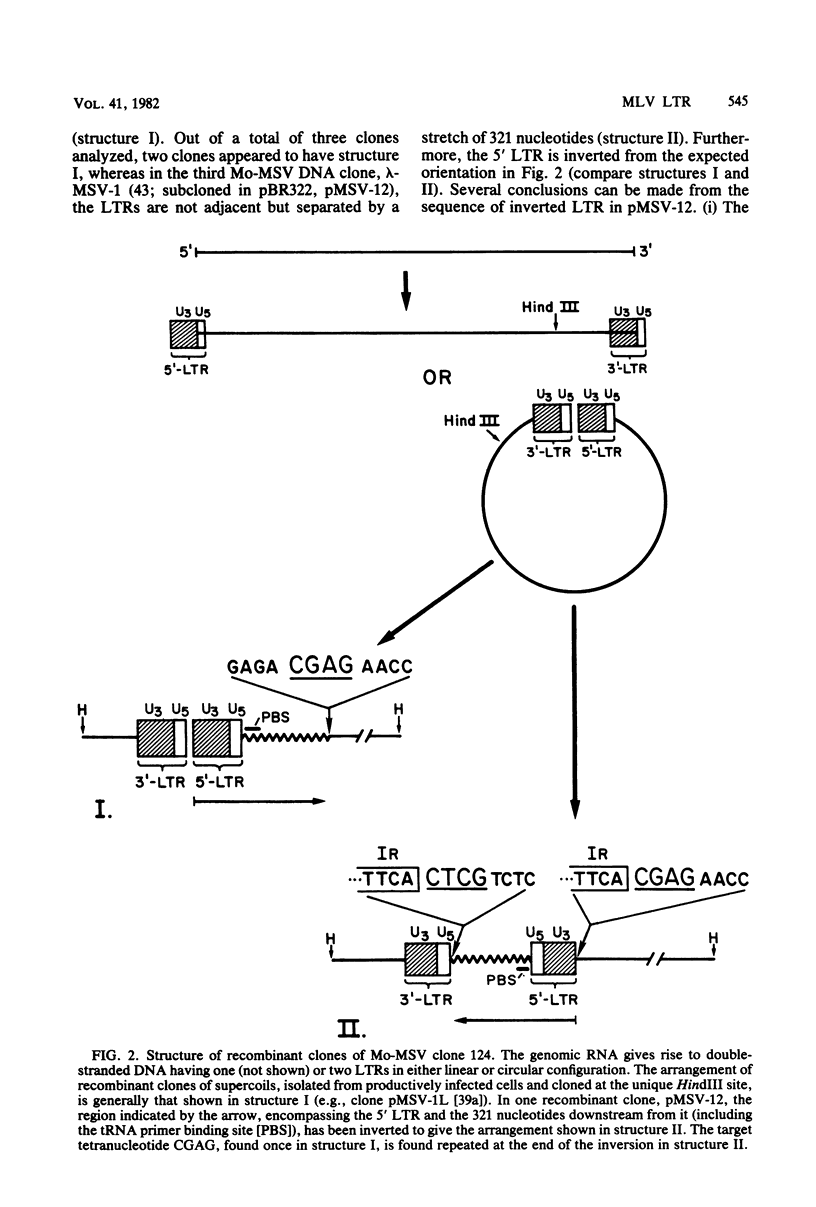
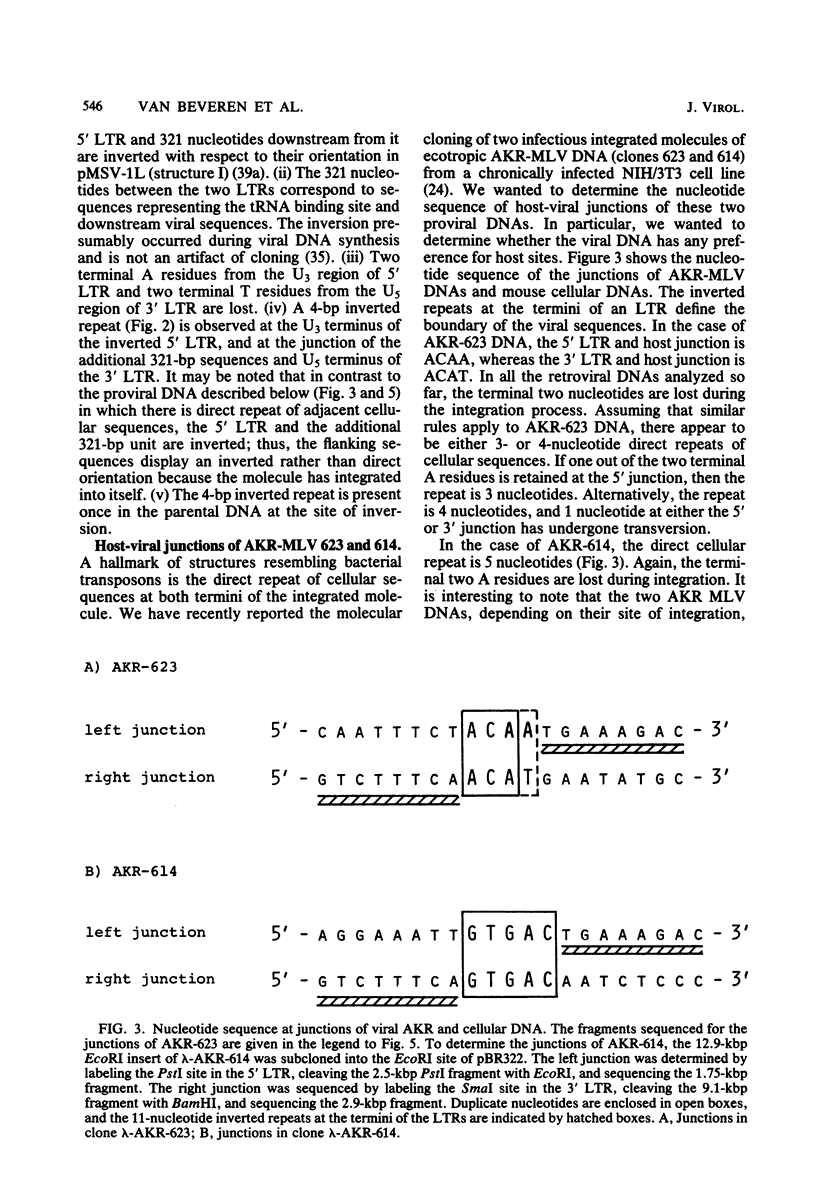
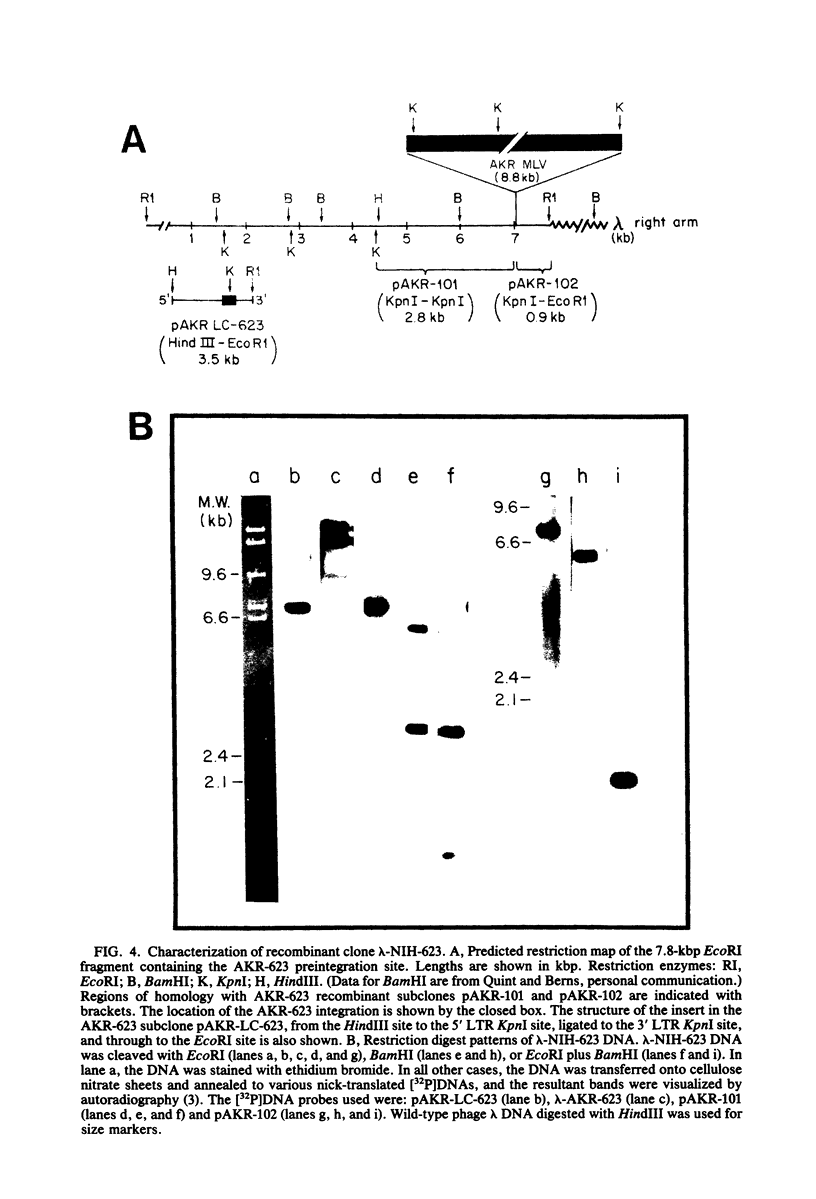
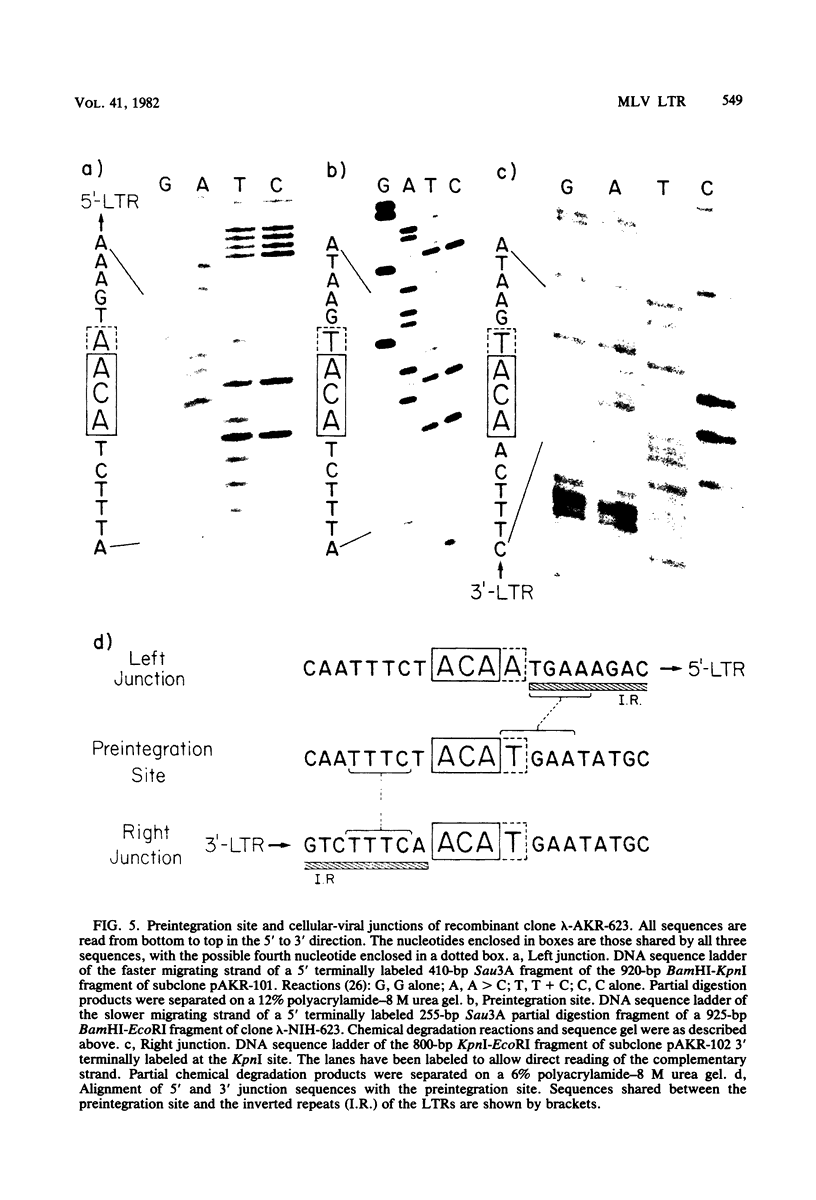
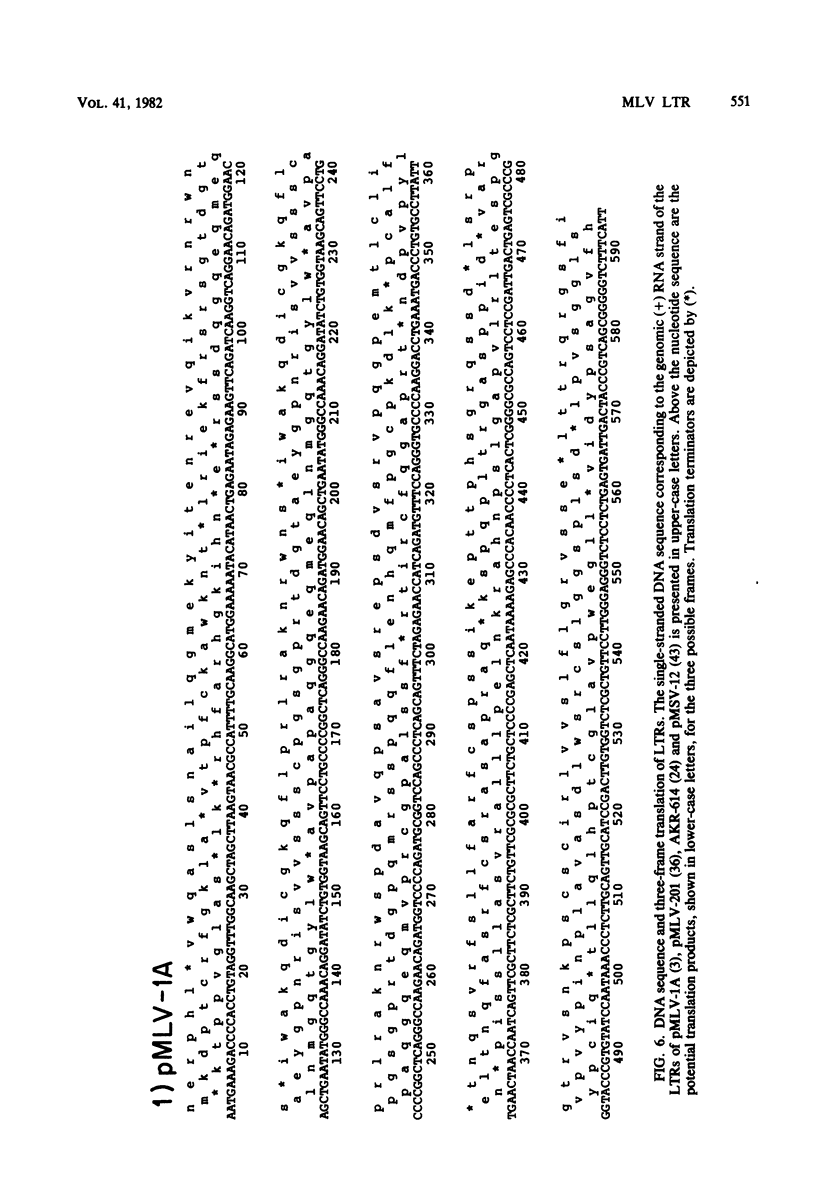
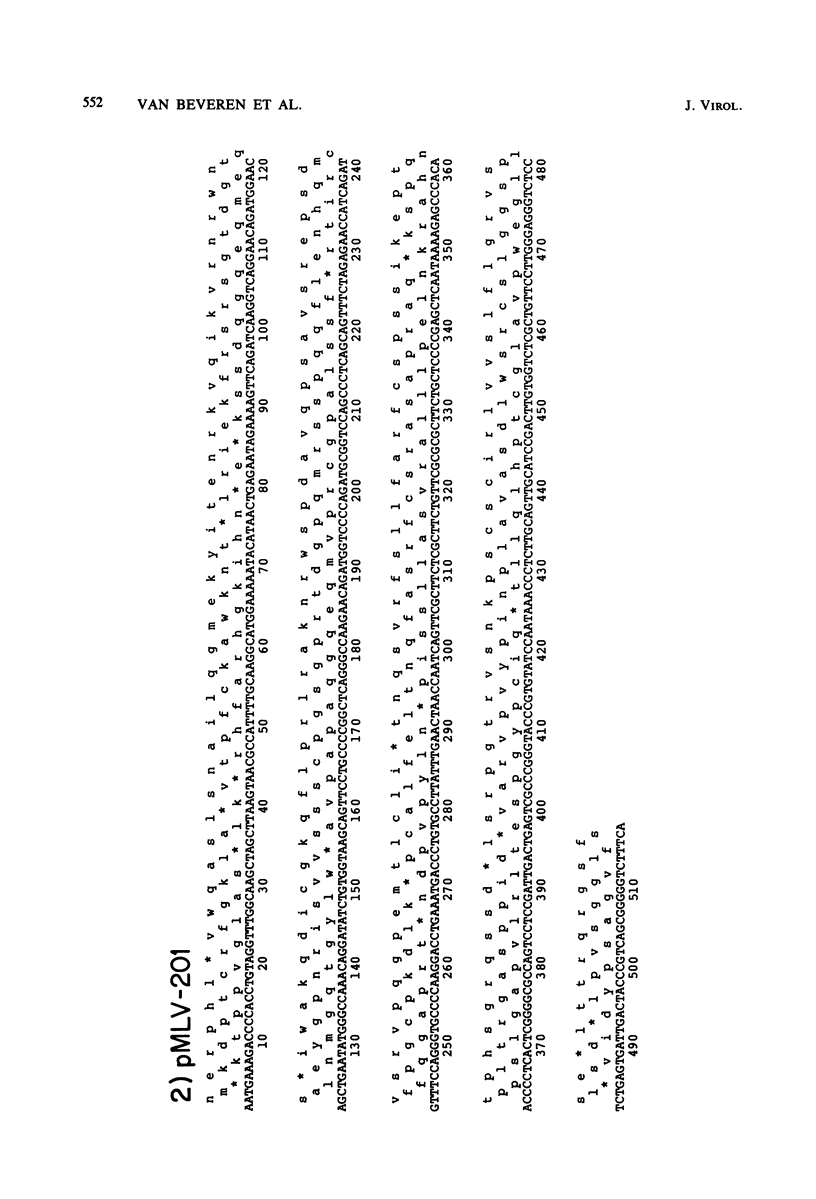
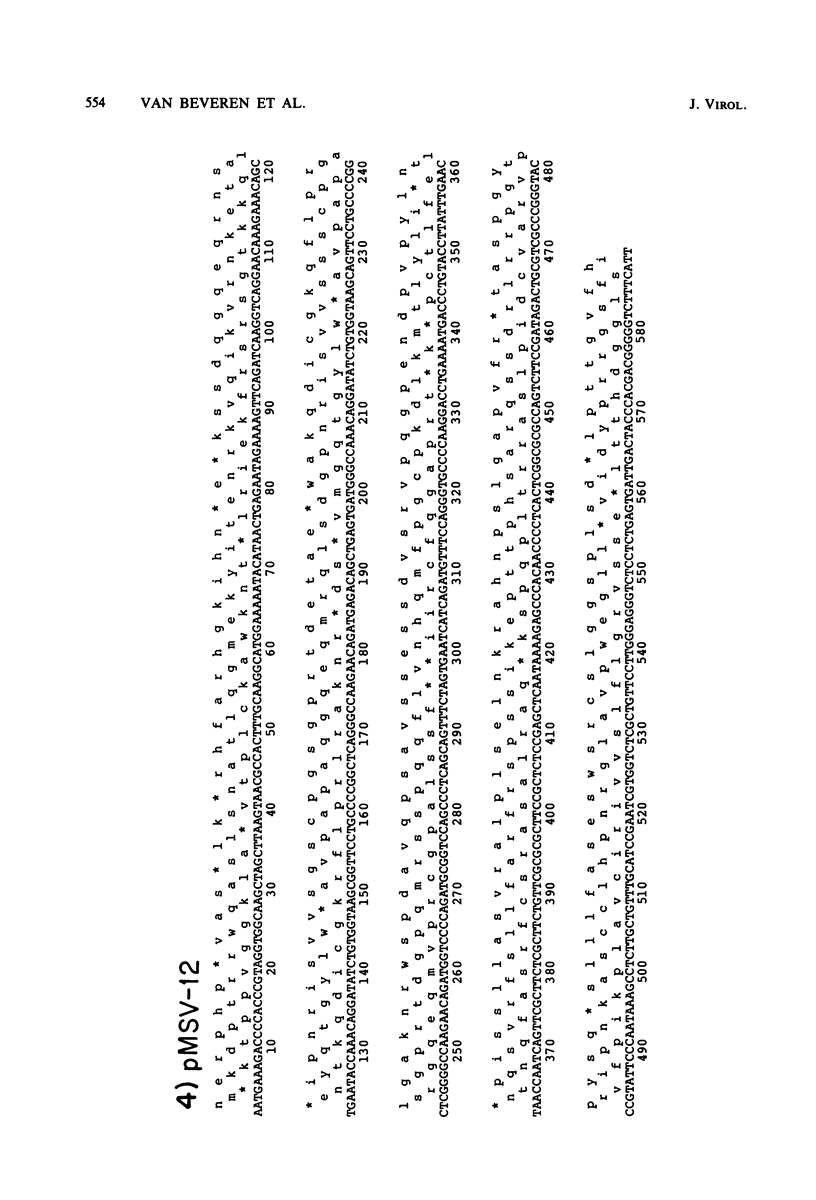
The nucleotide sequence of the long terminal repeat (LTR) of three murine retroviral DNAs has been determined. The data indicate that the U5 region (sequences originating from the 5' end of the genome) of various LTRs is more conserved than the U3 region (sequences from the 3' end of the genome). The location and sequence of the control elements such as the 5' cap, "TATA-like" sequences, "CCAAT-box," and presumptive polyadenylic acid addition signal AATAAA in the various LTRs are nearly identical. Some murine retroviral DNAs contain a duplication of sequences within the LTR ranging in size from 58 to 100 base pairs. A variant of molecularly cloned Moloney murine sarcoma virus DNA in which one of the two LTRs integrated into the viral DNA was also analyzed. A 4-base-pair duplication was generated at the site of integration of LTR in the viral DNA. The host-viral junction of two molecularly cloned AKR-murine leukemia virus DNAs (clones 623 and 614) was determined. In the case of AKR-623 DNA, a 3- or 4-base-pair direct repeat of cellular sequences flanking the viral DNA was observed. However, AKR-614 DNA contained a 5-base-pair repeat of cellular sequences. The nucleotide sequence of the preintegration site of AKR-623 DNA revealed that the cellular sequences duplicated during integration are present only once. Finally, a striking homology between the sequences flanking the preintegration site and viral LTRs was observed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Benz E. W., Jr, Dina D. Moloney murine sarcoma virions synthesize full-genome-length double-stranded DNA in vitro. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3294–3298. doi: 10.1073/pnas.76.7.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns A. J., Lai M. H., Bosselman R. A., McKennett M. A., Bacheler L. T., Fan H., Maandag E. C., van der Putten H. V., Verma I. M. Molecular cloning of unintegrated and a portion of integrated moloney murine leukemia viral DNA in bacteriophage lambda. J Virol. 1980 Oct;36(1):254–263. doi: 10.1128/jvi.36.1.254-263.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Retroviruses. Annu Rev Biochem. 1978;47:35–88. doi: 10.1146/annurev.bi.47.070178.000343. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Blechl A. E., Denniston-Thompson K., Faber H. E., Richards J. E., Slightom J. L., Tucker P. W., Smithies O. Cloning human fetal gamma globin and mouse alpha-type globin DNA: preparation and screening of shotgun collections. Science. 1978 Dec 22;202(4374):1279–1284. doi: 10.1126/science.725603. [DOI] [PubMed] [Google Scholar]
- Bosselman R. A., Verma I. M. Genome organization of retroviruses. V. In vitro-synthesized Moloney murine leukemia viral DNA has long terminal redundancy. J Virol. 1980 Jan;33(1):487–493. doi: 10.1128/jvi.33.1.487-493.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Nunberg J. H., Kaufman R. J., Erlich H. A., Schimke R. T., Cohen S. N. Phenotypic expression in E. coli of a DNA sequence coding for mouse dihydrofolate reductase. Nature. 1978 Oct 19;275(5681):617–624. doi: 10.1038/275617a0. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmuir P., Brorein W. J., Jr, Simon M. A., Rubin G. M. Insertion of the Drosophila transposable element copia generates a 5 base pair duplication. Cell. 1980 Sep;21(2):575–579. doi: 10.1016/0092-8674(80)90495-x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Van Beveren C., Verma I. M. Identification of a RNA polymerase II initiation site in the long terminal repeat of Moloney murine leukemia viral DNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5411–5415. doi: 10.1073/pnas.78.9.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Goff S., Shields A., Yoshimura F., Mitra S., Baltimore D. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell. 1979 Apr;16(4):863–874. doi: 10.1016/0092-8674(79)90101-6. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Haegeman G., Fiers W. Characterization of the 5'-terminal cap structures of early simian virus 40 mRNA. J Virol. 1980 Sep;35(3):955–961. doi: 10.1128/jvi.35.3.955-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Jones M., Bosselman R. A., van der Hoorn F. A., Berns A., Fan H., Verma I. M. Identification and molecular cloning of Moloney mouse sarcoma virus-specific sequences from uninfected mouse cells. Proc Natl Acad Sci U S A. 1980 May;77(5):2651–2655. doi: 10.1073/pnas.77.5.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Rands E., Chattopadhyay S. K., Garon C. F., Hager G. L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc Natl Acad Sci U S A. 1980 Jan;77(1):614–618. doi: 10.1073/pnas.77.1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rands E., Lowy D. R., Lander M. R., Chattopadhyay S. K. Restriction endonuclease mapping of ecotropic murine leukemia viral DNAs: size and sequence heterogeneity of the long terminal repeat. Virology. 1981 Jan 30;108(2):445–452. doi: 10.1016/0042-6822(81)90451-7. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Aaronson S. A. Complete nucleotide sequence and organization of the Moloney murine sarcoma virus genome. Science. 1981 Oct 23;214(4519):445–450. doi: 10.1126/science.6170110. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Canaani E., Robbins K. C., Tronick S. R., Zain S., Aaronson S. A. Nucleotide sequence analysis of the transforming region and large terminal redundancies of Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5234–5238. doi: 10.1073/pnas.77.9.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Piatak M., Weissman S. M. Simian virus 40 early mRNA's. I. Genomic localization of 3' and 5' termini and two major splices in mRNA from transformed and lytically infected cells. J Virol. 1979 Apr;30(1):279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Galleshaw J. A., Jonas V., Berns A. J., Doolittle R. F., Donoghue D. J., Verma I. M. Nucleotide sequence and formation of the transforming gene of a mouse sarcoma virus. Nature. 1981 Jan 22;289(5795):258–262. doi: 10.1038/289258a0. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Heasley S., Kung H. J., Oppermann H., Smith V. C., Bishop J. M., Shank P. R. Kinetics of synthesis, structure and purification of avian sarcoma virus-specific DNA made in the cytoplasm of acutely infected cells. J Mol Biol. 1978 Mar 25;120(1):55–82. doi: 10.1016/0022-2836(78)90295-4. [DOI] [PubMed] [Google Scholar]
- Verma I. M., Lai M. H., Bosselman R. A., McKennett M. A., Fan H., Berns A. Molecular cloning of unintegrated Moloney mouse sarcoma virus DNA in bacteriophage lambda. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1773–1777. doi: 10.1073/pnas.77.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. The reverse transcriptase. Biochim Biophys Acta. 1977 Mar 21;473(1):1–38. doi: 10.1016/0304-419x(77)90005-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., de Crombrugghe B., Pastan I. Identification of a functional promoter in the long terminal repeat of Rous sarcoma virus. Cell. 1980 Dec;22(3):787–797. doi: 10.1016/0092-8674(80)90555-3. [DOI] [PubMed] [Google Scholar]
- van der Putten H., Quint W., van Raaij J., Maandag E. R., Verma I. M., Berns A. M-MuLV-induced leukemogenesis: integration and structure of recombinant proviruses in tumors. Cell. 1981 Jun;24(3):729–739. doi: 10.1016/0092-8674(81)90099-4. [DOI] [PubMed] [Google Scholar]