Abstract
To characterize methicillin-resistant Staphylococcus aureus (MRSA) strains circulating in the community, we identified predictors of isolating community MRSA and genotyped a sample of MRSA collected from a community-based, high-risk population. Computerized databases of the Community Health Network of San Francisco and the Clinical Microbiology Laboratory were searched electronically for the years 1992–1999 to identify community-onset infections caused by MRSA. Sequential analyses were performed to identify predictors of MRSA strains. The majority (58%) of infections were caused by strains traceable to the hospital or to long-term care facilities. Injection drug use was associated with infections that were not associated with health care settings. Genotypes for 20 of 35 MRSA isolates recovered from injection drug users did not match any of >600 genotypes of clinical isolates. In a nonoutbreak setting, the hospital was the main source of community MRSA; however, the presence of genetically distinct and diverse MRSA strains indicates MRSA strains now also originate from the community.
The global emergence of drug-resistant bacteria is a pressing public health problem. Firmly established in hospitals worldwide, methicillin-resistant Staphylococcus aureus (MRSA) has now emerged as a significant community-acquired pathogen [1–14]. Several features appear to distinguish community from nosocomial MRSA strains: (1) absence of hospital-associated risk factors; (2) susceptibility to most antibiotics other than β-lactams; (3) distinct genotypes that do not match S. aureus strains commonly found in hospitals; (4) presence of type 4 staphylococcal chromosomal cassette mec (SCCmec) (the element that contains the methicillin-resistance determinant), not typical of nosocomial MRSA strains; and (5) the presence of genes encoding for toxins such as Pantone-Valentine leukocidin and the many staphylococcal enterotoxins [2, 15–21].
Community strains of MRSA may arise in either of 2 ways: hospital strains may be carried into the community, where they then spread person to person [1], or community MRSA may arise de novo when the methicillin-resistance gene complex is acquired by a methicillin-susceptible strain [2, 22]. Anecdotal reports, case series, and studies of outbreaks suggest the latter is occurring, but these may not accurately represent strains circulating in the community. To obtain evidence concerning the origins of community-acquired strains of MRSA in San Francisco, California, we took a novel combination approach that used a large database of clinical MRSA isolates to identify a target population at risk for community MRSA. A community-based sample of this target population was assessed for MRSA nasal carriage, and a molecular analysis of MRSA isolates was performed to determine their genotypes.
MATERIALS AND METHODS
Clinical Epidemiology
Setting
The Community Health Network (CHN) of San Francisco is a publicly funded health care delivery system with sites of care including San Francisco General Hospital (SFGH; a university-affiliated 550-bed public teaching hospital and the regional trauma center), 13 neighborhood health centers, a 1000-bed publicly funded long-term care facility (LTCF), a 150-bed long-term care mental health facility, and a 17,000-visit-per-year home health care network.
S. aureus bacterial culturing and antibiotic susceptibility testing
The clinical microbiology laboratory of SFGH performs all clinical antimicrobial susceptibility tests for the CHN. MIC determinations were performed with the Microscan Walkaway instrument (Dade International) in accordance with NCCLS guidelines [23].
Electronic data collection
Antimicrobial susceptibility data were obtained from existing clinical databases for all positive S. aureus culture results processed by the CHN centralized clinical laboratory at SFGH. The total number of bacterial cultures processed annually and the inpatient hospital census by year were obtained to determine secular trends in the volume of patients served and changes in bacterial culture ordering practices by health care providers.
Electronic records of previous hospitalizations and clinic visits were obtained for all CHN patients with cultures positive for S. aureus for the years 1992–1999 from existing CHN-wide databases. Patient registration records were also examined for previous residence in the county LTCF.
Statistical analysis
To adjust for potential bias arising from multiple S. aureus isolates cultured from a single patient, only data for the initial isolate for each calendar year was included in the analysis. When both a methicillin-resistant and susceptible organism were isolated from the same patient during a calendar year, preference was given to the methicillin-resistant isolate.
Fisher’s exact test was used to test for significant associations between categorical variables. Univariate and multiple logistic regressions were applied to estimate the OR associated with risk factors for methicillin resistance among S. aureus isolates and for multidrug resistance among MRSA isolates. The χ2 test for trend was used to evaluate evidence for increasing proportion of methicillin resistance among S. aureus isolates by calendar year.
Definitions
A hospital-associated S. aureus isolate was defined as one cultured from a clinical specimen obtained >72 h after hospital admission. A community-associated S. aureus isolate was defined as one cultured during the first 72 h of a patient’s hospital admission or from an outpatient. MRSA isolates resistant to ⩾3 non–β-lactam antibiotics classes were classified as multidrug-resistant MRSA (R-MRSA). The duration of hospitalization before collection of each S. aureus isolates was calculated, and patients were grouped into 5 mutually exclusive categories: (1) no hospitalizations within the previous 3 years, (2) hospitalization within the previous 6 months, (3) hospitalization within 6–12 months, (4) hospitalization within >1–2 years, and (5) hospitalization within >2–3 years.
Community-Based MRSA Sample
A community-based sample of nasal S. aureus colonization was conducted for April–September 1999 in the context of the Urban Health Study, an ongoing community-based research and prevention program among active injection drug users (IDUs) in San Francisco [24].
Microbiological Studies
The anterior nares of each subject consenting to the study were sampled with a cotton swab dampened with normal saline. The swab was inoculated in the field onto a 5% sheep’s blood tryptic soy agar plate. After overnight incubation at 37°C, colonies resembling staphylococci were individually inoculated onto mannitol salt and blood agar and incubated overnight. Isolates were identified as S. aureus if they produced the appropriate color change on mannitol salt agar and a positive tube coagulase test result.
Susceptibility of nares isolates to ampicillin, ciprofloxacin, tetracycline, gentamicin, erythromycin, trimethoprimsulfameth-oxazole, clindamycin, linezolid, and vancomycin were determined on Mueller-Hinton agar (purchased from BBL) by the disk diffusion method in the SFGH Molecular Epidemiology Reference Laboratory [25]. Results were interpreted in accordance with the NCCLS guideline M7–A5 [23]. Susceptibility to methicillin was tested according to the recommendations of McDougal and Thornsberry [26].
Molecular Studies
mecA, the gene that determines methicillin resistance, was detected in either whole cells or lysostaphin-treated cell lysates by the presence of a 533-bp PCR amplification product that used sense and antisense primers—5′-AAAATCGATGGTAAA-GGTTGGC-3′ and 5′-AGTTCTGCAGTACCGGATTTGC-3′ respectively (GenBank accession no. D86934) [27]. SCCmec type was determined by multiplex PCR [28]. Control strains for SCCmec types 1, 2, and 3 were as follows: type 1, COL; type 2, N315 and 67-0; and type 3, SFGH clinical isolates 1349 and 2766 (confirmed using the method of Okuma et al. [2]). MRSA isolates were genotyped by PFGE of SmaI digests of chromosomal DNA [29, 30], spa (staphylococcal protein A) typing [31], and multilocus sequencing typing (MLST) [32]. All MRSA strains were evaluated by SCCmec type, spa typing, and PFGE. We then grouped strains according to matches on the basis of spa and PFGE. A random isolate within each group was then chosen for MLST sequence typing. PFGE patterns were compiled with the BioRad Molecular Analyst program and were compared with those in a genotype database composed of 600 MRSA clinical isolates collected 1996–1999 by the Molecular Epidemiology Research Laboratories from the clinical microbiology laboratories at SFGH.
RESULTS
Increasing prevalence of methicillin resistance
Between 1988 and 1999, there were 20,819 S. aureus–positive cultures, of which 12,159 were initial isolates. Isolates from samples obtained for culture ≤72 h after hospital admission or from an outpatient setting (i.e., community-associated isolates) were less likely to be methicillin resistant (12.1%) compared with samples obtained for culture >72 h after hospital admission (i.e., hospital-associated isolates) (27.4%) (OR, 2.1; 95% CI, 1.9–2.4; P <.0001). The proportion of S. aureus isolates that were resistant to methicillin increased significantly from 3.1% in 1988 to 26.9% in 1999 (P <.0001, by χ2 test for trend), increasing among hospital-associated isolates from 16% in 1993 to 42% in 1999 and from 7% in 1993 to 29% in 1999 in the community-associated isolates (figure 1).
Figure 1.
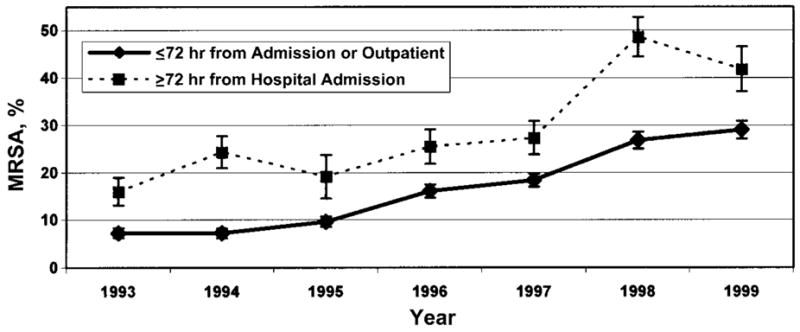
Percentage of strains of methicillin-resistant Staphylococcus aureus (MRSA), by year. Bars, SEMs.
MRSA were significantly more likely to be resistant to erythromycin, ciprofloxacin, clindamycin, gentamicin, or trimethoprim-sulfamethoxazole than were methicillin-susceptible S. aureus (MSSA) isolates (table 1). Thirty-three percent of MRSA isolates were resistant to ⩾3 antibiotics, compared with only 2% of MSSA isolates (P <.001). Among all MRSA isolates, community-associated MRSA isolates were less likely to be resistant to antibiotics than were hospital-associated MRSA isolates (P <.0001).
Table 1.
Staphylococcus aureus antibiotic resistance profile.
| Isolate, source | No. of patient isolates | Agent, % of resistant isolates
|
|||||
|---|---|---|---|---|---|---|---|
| Erythromycin | Tetracycline | Ciprofloxacin | Clindamycin | Gentamicin | TMP-SMZ | ||
| MSSA | |||||||
| Outpatient or person hospitalized for ≤72 h | 4532 | 29.9 | 17.2 | 2.0 | 2.3 | 1.5 | 7.4 |
| Person hospitalized for >72 h | 769 | 23.2 | 13.1 | 1.3 | 2.1 | 1.2 | 8.3 |
| MRSA | |||||||
| Outpatient or person hospitalized for ≤72 h | 811 | 67.3 | 12.2 | 44.3 | 43.5 | 26.6 | 15.8 |
| Person hospitalized for >72 h | 290 | 78.6 | 10.1 | 64.8 | 65.1 | 42.1 | 18.4 |
NOTE. MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; TMP-SMZ, trimethoprim-sulfamethoxazole.
Risk factors for methicillin resistance
Univariate risk factors for methicillin resistance among 4685 community-associated S. aureus isolates were more recent year during which the sample was obtained for culture (OR, 1.2; 95% CI, 1.2–1.3; P <.0001), previous residence in the LTCF (OR, 3.3; 95% CI, 2.7–4.1; P <.0001), previous hospitalization within the previous 3 years (OR, 1.7; 95% CI, 1.5–2.0; P <.0001), age of ⩾18 years (OR, 1.6; 95% CI, 1.01–2.5), homelessness (OR, 1.9; 95% CI, 1.5–2.4; P <.0001), and injection drug use (OR, 2.1; 95% CI, 1.5–3.0; P <.0001). Admission to the hospital during the previous year and admission to the LTCF were the only variables predictive of a MRSA isolate among patients with community-associated MRSA by multivariate analysis (table 2). Nonsignificant risk factors evaluated included sex, ethnicity, and history of recent outpatient care visit.
Table 2.
Multivariate ORs for risk of methicillin resistance and multidrug resistance among Staphylococcus aureus and methicillin-resistant S. aureus isolates recovered from cultures of samples obtained while the person was as an outpatient or within 72 h of hospital admission.
| Outcome, exposure | No. of patient isolates | OR (95% CI) | P | % MRSA or % R-MRSAa |
|---|---|---|---|---|
| Risk of methicillin resistance among S. aureus (N = 4685) | ||||
| Previous LTCF residence | 55 | 2.1 (1.1–3.9) | .02 | 27.3 |
| Previous hospitalization, duration | ||||
| 0–6 months | 1061 | 3.5 (2.9–4.3) | <.0001 | 23.3 |
| >6–12 months | 234 | 2.2 (1.5–3.2) | <.0001 | 15.8 |
| >12–24 months | 185 | 1.5 (0.9–2.4) | .11 | 11.4 |
| >24–36 months | 68 | 0.7 (0.3–2.1) | .57 | 5.9 |
| None in previous 3 years | 3137 | Reference | … | 7.8 |
| Risk of multidrug resistance among MRSA (N = 553) | ||||
| Previous LTCF residence | 15 | 3.5 (1.2–10.5) | .02 | 60.0 |
| Previous hospitalization, duration | ||||
| >0–6 months | 247 | 1.7 (1.1–2.5) | .009 | 37.7 |
| >6–12 months | 37 | 0.4 (0.2–1.1) | .083 | 13.5 |
| >12–24 months | 21 | 0.3 (0.1–1.4) | .12 | 9.5 |
| >24–36 months | 4 | 0.9 (0.1–9.6) | .98 | 25.0 |
| None in previous 3 years | 244 | Reference | … | 25.8 |
NOTE. LTCF, long-term care facility; MRSA, methicillin-resistant S. aureus.
For the first outcome, the percentages (% MRSA) refer to the percentage of S. aureus isolates that were MRSA, and for the second outcome, the percentages (% R-MRSA) refer to the percentage of MRSA isolates that were multidrug resistant.
Risk factors for multidrug-resistant S. aureus
To determine whether multidrug resistance was associated with a nosocomial MRSA isolate, multivariate analysis was repeated for the 553 community-associated MRSA isolates. Previous LTCF residence and previous hospitalization within 0–6 months were associated with an increased risk (table 2) for R-MRSA, confirming the relationship between multiple resistances and nosocomial source, suggesting that nosocomial isolates accounted for a considerable proportion of the community MRSA group (47% of MRSA with LTCF or hospitalization within 6 months, 58% of MRSA with LTCF or hospitalization within 2 years).
Because multidrug resistance was a marker for a nosocomial strain, to define risk factors for “true” community MRSA (i.e., those purged of endemic hospital clones), univariate analysis was performed for community-associated MRSA, this time excluding community cases with R-MRSA isolates or with admission to the hospital or LTCF within the previous year. Factors associated with methicillin resistance in univariate analysis among these patients with community MRSA according to the more stringent definition were recent year of culture (OR, 1.4; 95% CI, 1.3–1.5; P <.0001), homelessness (OR, 1.7; 95% CI, 1.1–2.6; P =.015), and injection drug use (OR, 2.3; 95% CI, 1.1–4.8; P =.02). The proportion of these “true” community MRSA isolates among all positive S. aureus cultures (n =2979) increased significantly, from 3.3% in 1993 to 17.7% in 1999 (P < .001).
Community-based sample of IDUs
A previous study from our group [4] found that an urban poor, largely homeless population had a low MRSA prevalence of 2.8% and that the vast majority of these isolates were associated with hospital or LTCF contact. Among this homeless population, IDUs had significant risk for community-associated MRSA. In the community-based sample of IDUs, 172 (25.3%) of 683 subjects had S. aureus isolated from nasal swab cultures, 42 of which were MRSA, for an overall prevalence of 6.1% and a prevalence of MRSA of 24.4% among those colonized with S. aureus.
Genotypes of community-acquired MRSA
Genotypes were determined for 35 of the MRSA isolates (table 3) obtained from the community-based sample of IDUs. These comprised 9 distinct PFGE groups, 8 spa types, and 6 sequence types (STs) or clonal complexes (CCs). Five isolates had a unique PFGE type (U) (figure 2) not closely related to one another or to the other isolates.
Table 3.
Genotypes and SCCmec types of methicillin-resistant Staphylococcus aureus isolates from a community-based sample of injection drug users.
| Prototype strain | PFGE type
(no. of isolates)a |
SFGH relatedb | spa Repeat sequences | SCCmec type | ST/CC |
|---|---|---|---|---|---|
| J50 | A (5) | No | Z1D1M1D1N1K1B1 | NT | 59/– |
| J52 | B (10) | No | Z1D1M1D1M1N1K1B1 | IV | 59/– |
| JY39 | C (11) | Yes | Y1H1G1F1M1B1Q1B1L1O1 | IV | 8/8 |
| M36 | D (4) | Yes | Y1H1G1F1M1B1Q1B1L1O1 | IV | 8/8 |
| M17 | U (1) | No | T1J1M1B1M1D1M1G1G1M1K1 | IV | 5/5 |
| M25 | U (1) | No | X1A1K1A1O1M1Q1 | NT | 30/30 |
| M45 | U (1) | No | T1J1M1B1M1D1M1G1M1K1 | NT | 5/5 |
| J28 | U (1) | No | U1J1F1E1 | IV | 1/1 |
| J35 | U (1) | No | U1J1F1Q1P1L1M1 | IV | 12/12 |
NOTE. CC, clonal complex of which the multilocus sequencing typing (MLST) sequence type is a member; NT, nontypeable pattern of SCCmec with PCR products not classifiable into one of the 4 main types; PFGE, pulsed-field gel electrophoresis; SFGH, San Francisco General Hospital; ST, MLST sequence type.
PFGE type indicates groups of isolates with identical or closely related PFGE patterns. The PFGE types of isolates in this study correspond to the following PFGE clone groups published in Diep et al. [33]: types A and B, clone group P; types C and D, clone group C; unique strains M17 and M45, clone group D; strain M25, clone group Z; strain J28, clone group K. Strain J35, which is a unique isolate in this study, belongs to a recently identified type among CHN isolates, clone group F.
This indicates a PFGE type that is closely related to one identified from the MERL database of MRSA clinical isolates collected during 1996–1999.
Figure 2.
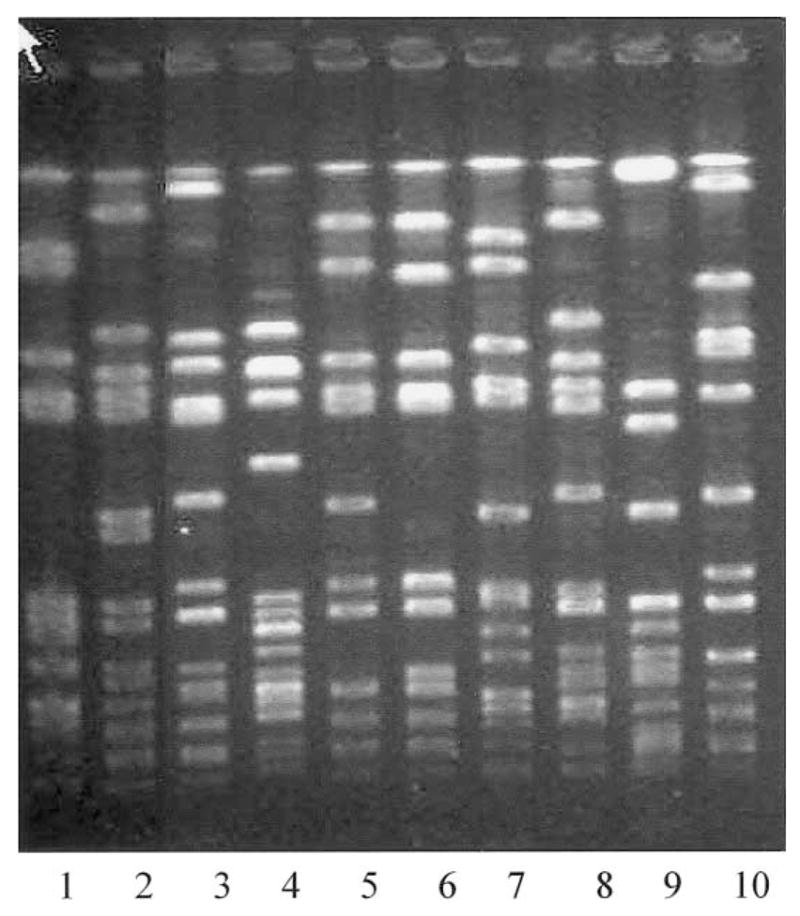
PFGE of prototype strains isolated from a community-based sample of injection drug users. Strains in each lane are as follows: 1, J50; 2, J28; 3, JY39; 4, M36; 5, M17; 6, M45; 7, J52; 8, J35; 9, M25; 10, reference laboratory strain.
Fifteen isolates (43%) (PFGE types C and D) were closely related to PFGE genotypes previously identified among >600 clinical isolates collected 1996–1999 at the CHN. All 15 were SCCmec type 4. These strains belonged to CC8, an archetypal nosocomial MRSA genotype [2, 34]. Twenty isolates (57%) had genotypes not found in the 1996–1999 collection (although many have since been identified among strains collected after 1999), and they were genotypically more diverse, constituting 7 PFGE patterns and 7 spa types. Thirteen isolates were SCCmec type 4, and 7 were nontypeable. Two isolates, M17 and M45, had similar spa types and were members of CC5, another archetypal MRSA nosocomial genotype [2, 34]. Stain J28 was a member of CC1, the same as that of the Minnesota and North Dakota community MRSA strains [2, 35]. Strain M25 was a member of CC30, a common type among community MRSA isolates in Australia [2]. PFGE groups A and B both were ST59 and strain J35 was ST12/CC12. These are uncommon MRSA sequence types—they have not previously been reported among community strains and are rarely found in nosocomial collections [2, 34, 36].
Consistent with a community phenotype, isolates with genotypes not found in the database tended to be drug susceptible, with 13 of 20 susceptible to all non–β-lactam antibiotics versus 5 of 15 isolates whose genotypes were in the database, although this difference was not statistically significant. Hospitalization within the previous 12 months was relatively uncommon in this community-based sample of IDUs. Only 9 patients (21%) had been hospitalized (all at SFGH) within the previous year, although the majority (29 [69%] of 42) reported ⩾1 visit to the emergency department within the previous year, usually for treatment of skin or soft-tissue infections.
DISCUSSION
MRSA, which has until recently been regarded as almost exclusively a hospital-associated pathogen, has been increasingly identified as a cause of community-onset infections. Some have argued that, because of the dramatically increasing prevalence of MRSA in the hospital, the parallel epidemic in the community is attributable to individuals returning to the community with MRSA from health care facilities [6, 37]. However, other evidence—both epidemiological and from molecular typing studies—have suggested that community MRSA strains are not simply feral hospital strains.
The present investigation used a serial, cross-sectional analysis of electronic databases to identify predictors of isolating a community MRSA strain. The clinical epidemiology identified 2 reservoirs for community strains of MRSA. The majority of community-onset MRSA infections identified from CHN electronic databases were almost certainly caused by strains acquired from hospitalization or residence in an LTCF, because these were the only predictors of MRSA identified by multivariate analysis of all MRSA isolates.
When the analysis was performed with community cases purged of those likely to be of health care facility origin, 3 risk factors were identified: homelessness, injection drug use, and year that the sample was obtained for culture. In a previously published community-based study in San Francisco, homelessness was not confirmed to be a significant risk factor for MRSA and MRSA genotypes that were identified and could be epidemiologically linked to acquisition in the hospital [4]. The community-based survey of IDUs, however, confirmed the predicted high prevalence of MRSA in this population, and the MRSA isolates recovered from this population had the hallmarks of community strains: they were largely susceptible to non–β-lactam antibiotics; type 4 SCCmec, recently described in other community MRSA isolates [15], predominated; and hospital admission among MRSA carriers within the prior year was lacking.
The molecular typing studies of these community isolates, mirroring the clinical epidemiology, identified 2 potential reservoirs for community MRSA isolates. Clearly, health care facilities were an important contributor, because nearly one half of the MRSA isolates had PFGE genotypes matching those of MRSA clinical isolates in a database of strains collected during 1996–1999. All were members of CC8, which is common among nosocomial MRSA worldwide. These isolates are probably feral hospital-endemic clones.
Community MRSA isolates not matching those within the PFGE database included 2 that were MLST sequence and clonal complex type 5. This CC is historically hospital associated and common among SFGH nosocomial isolates as well. The remaining isolates from the community-based sample were sequence and CC types reported as community-outbreak strains from other locations and not common among nosocomial isolates (CC1 and CC30) or uncommon nosocomial isolates and not previously reported as community isolates (ST59 and CC12).
The striking genetic feature of the community MRSA isolates, regardless of presumed source and despite diversity of genotype, was the predominance of SCCmec type 4, which was present in 89% of isolates, including the isolates whose genotypes were represented in the database of CHN strain collection. Type 4 SCCmec, although it may be the predominant community type, should not be considered unique to community isolates, as others have observed [34]. Indeed, there is recent evidence of the increasing prevalence of type 4 SCCmec within both community and nosocomial strains of MRSA in San Francisco and identification of nosocomial MRSA containing type 4 SCCmec [38, 39].
Our results indicate that a large proportion of community MRSA strains in San Francisco are feral descendentis of hospital endemic clones that over time have adopted a community phenotype of multiple-drug susceptibility. Others strains appear truly to be community-adapted residents. It is possible that these clones also originally were endemic in hospitals, but, being relatively unfit for an environment of heavy antibiotic exposure, they prefer the more salutary, less antibiotic-selective community setting. Alternatively, these may have recently arisen by horizontal transfer of type 4 SCCmec into a methicillin-susceptible background, suggested by the fact that they are members of clonal complexes not common among MRSA, community or nosocomial. The genetic diversity of community MRSA isolates and the presence of nontypeable (perhaps novel) SCCmec types also demonstrate mobility and plasticity of SCCmec [22]. It is the smallest of the 4 known SCCmec elements, 21–24 kb in size (compared with 35 to >60 kb in size of types 1, 2, and 3); it is small enough to be packaged in a bacteriophage and horizontally transmitted, whereas the other 3 types are too large. The complete absence of the other 3 SCCmec types, which are readily found in collections of nosocomial strains [2, 34, 35] in the community isolates, is surprising and suggests a strong counterselection for these elements or strains carrying them in the community or a strong selective advantage for type 4 strains.
Given that the epidemiology and individual strain prevalence of MRSA is known to exhibit significant regional variation, caution must be used in generalizing these specific results to other geographic locations. However, as the trends in the emergence and spread of community-acquired MRSA in San Francisco have been mirrored by community-acquired MRSA trends in other US and international settings, it is likely that the general mechanisms and findings in such locations will be similar, once investigated.
The findings presented have important therapeutic implications. If community strains continue to spread and increase in prevalence independent of a hospital source, then empirical approaches to therapy for S. aureus infection will have to take this into account. In addition, mathematical modeling suggests that the most effective method of controlling drug-resistant bacteria is to reduce their input into hospitals [40]. To the extent that MRSA input in the community is independent of a hospital reservoir, it will be much more difficult—if not impossible—to control.
Acknowledgments
Financial support. Doris Duke Foundation (to E.D.C. and D.R.B.), Pharmacia (to F.P.-R.), and National Institutes of Health/National Institute of Allergy and Infectious Diseases (grant RO1 AI46610–01; to E.D.C.). The Urban Health Study was supported by grants R01-DA09532, R01-DA12109, and R01-DA11241-02S1 from the National Institute on Drug Abuse.
References
- 1.Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36:131–9. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- 2.Okuma K, Iwakawa K, Turnidge JD, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–94. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dufour P, Gillet Y, Bes M, et al. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin Infect Dis. 2002;35:819–24. doi: 10.1086/342576. [DOI] [PubMed] [Google Scholar]
- 4.Charlebois ED, Bangsberg DR, Moss NJ, et al. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin Infect Dis. 2002;34:425–33. doi: 10.1086/338069. [DOI] [PubMed] [Google Scholar]
- 5.Hussain FM, Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus colonization in healthy children attending an outpatient pediatric clinic. Pediatr Infect Dis J. 2001;20:763–7. doi: 10.1097/00006454-200108000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Chambers HF. The changing epidemiology of Staphylococcus aureus. Emerg Infect Dis. 2001;7:178–82. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shopsin B, Mathema B, Martinez J, et al. Prevalence of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the community. J Infect Dis. 2000;182:359–62. doi: 10.1086/315695. [DOI] [PubMed] [Google Scholar]
- 8.Gorak EJ, Yamada SM, Brown JD. Community-acquired methicillin-resistant Staphylococcus aureus in hospitalized adults and children without known risk factors. Clin Infect Dis. 1999;29:797–800. doi: 10.1086/520437. [DOI] [PubMed] [Google Scholar]
- 9.US Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. JAMA. 1999;282:1123–5. [PubMed] [Google Scholar]
- 10.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 11.Boyce JM. Are the epidemiology and microbiology of methicillin-resistant Staphylococcus aureus changing? [editorial; comment] JAMA. 1998;279:623–4. doi: 10.1001/jama.279.8.623. [DOI] [PubMed] [Google Scholar]
- 12.Pate KR, Nolan RL, Bannerman TL, Feldman S. Methicillin-resistant Staphylococcus aureus in the community. Lancet. 1995;346:978. doi: 10.1016/s0140-6736(95)91605-9. [DOI] [PubMed] [Google Scholar]
- 13.Berman DS, Eisner W, Kreiswirth B. Community-acquired methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1993;329:1896. doi: 10.1056/NEJM199312163292517. [DOI] [PubMed] [Google Scholar]
- 14.Hamoudi AC, Palmer RN, King TL. Nafcillin resistant Staphylococcus aureus: a possible community origin. Infect Control. 1983;4:153–7. doi: 10.1017/s0195941700058070. [DOI] [PubMed] [Google Scholar]
- 15.Ma XX, Ito T, Tiensasitorn C, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–52. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groom AV, Wolsey DH, Naimi TS, et al. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA. 2001;286:1201–5. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- 17.Naimi TS, LeDell KH, Boxrud DJ, et al. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin Infect Dis. 2001;33:990–6. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- 18.Suggs AH, Maranan MC, Boyle-Vavra S, Daum RS. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr Infect Dis J. 1999;18:410–4. doi: 10.1097/00006454-199905000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 20.Vandenesch F, Naimi T, Enright MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care–associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–84. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 22.Daum RS, Ito T, Hiramatsu K, et al. A novel methicillin-resistanc ecassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J Infect Dis. 2002;186:1344–7. doi: 10.1086/344326. [DOI] [PubMed] [Google Scholar]
- 23.NCCLS. Document M7–A5. 5. Wayne, PA: NCCLS; 2000. Methods for dilution antimicrobial susceptibility test for bacterial that grow aerobically, Approved standard. [Google Scholar]
- 24.Kral AH, Bluthenthal RN, Lorvick J, Gee L, Bacchetti P, Edlin BR. Sexual transmission of HIV-1 among injection drug users in San Francisco, USA: risk factor analysis. Lancet. 2001;357:1397–401. doi: 10.1016/S0140-6736(00)04562-1. [DOI] [PubMed] [Google Scholar]
- 25.NCCLS. Approved standard. 4. Villanova, PA: NCCLS; 1999. Performance standard for antimicrobial disk susceptibility tests. [Google Scholar]
- 26.McDougal LK, Thornsberry C. New recommendations for disk diffusion antimicrobial susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1984;19:482–8. doi: 10.1128/jcm.19.4.482-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killgore GE, Holloway B, Tenover FC. A 5′ nuclease PCR (TaqMan) high-throughput assay for detection of the mecA gene in staphylococci. J Clin Microbiol. 2000;38:2516–9. doi: 10.1128/jcm.38.7.2516-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–61. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maslow JN, Mulligan ME, Arbeit RD. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–62. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 30.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diep BA, Perdreau-Remington F, Sensabaugh GF. Clonal characterization of Staphylococcus aureus by multilocus restriction fragment typing, a rapid screening approach for molecular epidemiology. J Clin Microbiol. 2003;41:4559–64. doi: 10.1128/JCM.41.10.4559-4564.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fey PD, Said-Salim B, Rupp ME, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:196–203. doi: 10.1128/AAC.47.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feil EJ, Cooper JE, Grundmann H, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones ME, Mayfield DC, Thornsberry C, Karlowsky JA, Sahm DF, Peterson D. Prevalence of oxacillin resistance in Staphylococcus aureus among inpatients and outpatients in the United States during 2000. Antimicrob Agents Chemother. 2002;46:3104–5. doi: 10.1128/AAC.46.9.3104-3105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aires de Sousa M, Bartzavali C, Spiliopoulou I, Sanches IS, Crisostomo MI, de Lencastre H. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J Clin Microbiol. 2003;41:2027–32. doi: 10.1128/JCM.41.5.2027-2032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carleton H, Charlebois E, Perdreau-Remington F. Dramatic increase of staphylococcal chromosomal cassette mec (SCCmec) type IV in both a nosocomial and community setting [C2–1983]. Program and abstracts of the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy (Chicago); Washington, DC: American Society for Microbiology. 2003. [Google Scholar]
- 40.Levin BR. Minimizing potential resistance: a population dynamics view. Clin Infect Dis. 2001;33(Suppl 3):S161–9. doi: 10.1086/321843. [DOI] [PubMed] [Google Scholar]


