Abstract
Identification of a causative pathogen is essential for the choice of treatment for most infectious diseases. Many FDA approved molecular assays; usually more sensitive and specific compared to traditional tests, have been developed in the last decade. A new trend of high throughput and multiplexing assays are emerging thanks to technological developments for the human genome sequencing project. The applications of microarray and ultra high throughput sequencing technologies for diagnostic microbiology are reviewed. The race for the $1000 genome technology by 2014 will have a profound impact in diagnosis and treatment of infectious diseases in the near future.
Keywords: Diagnostic Microbiology, Microarray, Next-generation sequencing
INTRODUCTION
Identification of the causative pathogen is essential for the choice of treatment for most infectious diseases. Microscopic examination in combination with staining and immunological techniques is usually simple and fast. Pathogen culture is also a useful tool for infectious disease diagnosis. However, not every pathogen can be identified by microscopy and many pathogens can not grow outside their hosts. Serology is not very useful for acute infections. Molecular diagnosis, which is generally more sensitive and specific, has become the method of choice to identify certain pathogens, particularly viruses [1].
During the last two decades, a number of scientific and technological advancements have been changing the landscape of diagnostic microbiology. The trend has been moving from antibody to nucleic acid based assays, from single to multiple pathogen detection, and from clinical laboratory to point-of-care tests [2]. Many more user-friendly commercial assays have been developed for molecular diagnostics (Table 1). In the meantime, tremendous efforts have been made to sequence genomes of both hosts and pathogens [3,4]. For example, there are nearly 3000 reference sequences (April, 2008) in the viral genome database at the NCBI (http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/viruses.html) compared to about 1000 reference viral genomes in August 2002 [5]. Such sequence information is very important to design multiplex molecular assays as well as new pathogen discovery. Furthermore, technologies co-evolved with the development of the Human Genome Project, such as microarrays and next-generation sequencers, are expected to have a great impact in the next revolution of pathogen diagnosis. This review will focus on these new aspects. Another important development in diagnostic microbiology is the introduction of microfluidics and nanotechnology based bionsensors [6-8], which will not be discussed here.
Table 1.
Molecular diagnostic tests for infectious diseases
|
Table 1 summarizes most of current molecular tests for pathogen diagnosis. The classification is somewhat arbitrary because these tests can be modified and combined. These methods have been well established and can be found in textbooks and other reviews [2,9-12], and will therefore not be discussed further. The majority of them have applied for FDA approved clinical tests. Companies manufacturing those clinical products are listed under each category in Table 1.
Notably, almost all of the current tests rely on prior knowledge of the pathogens and experience of a clinician. New approaches using microarray and next-generation sequencers have been developed for identifying previously unsuspected pathogens. The microarray technology is relatively well established and has been routinely used in many research laboratories, including that of the author. Tremendous interest in personalized genomics has driven an incredible amount of effort and resources into the new generation of sequencing technologies. This review will focus on real cases that are difficult to diagnose with current clinical tests. I will use my own experiences to illustrate the application of pathogen-specific microarrays for clinical diagnosis and will also review two recent pathogen discoveries that took advantage of the new sequencing technology.
MICROARRAY
Microarray is a very powerful and flexible technology that has been used for host and pathogen gene expression profiling and genotyping. One of the early applications in the field of infectious diseases is to study polymorphisms of the HIV protease gene to detect drug resistance. However, the more exciting and also more clinically relevant application is for unsuspected or unknown pathogen identification, which has been demonstrated in reports by ourselves and others [5,13-17]. By applying a strategy that will be described later, we helped to identify a novel coronavirus responsible for the SARS (severe acute respiratory syndrome) epidemic in 2003 [5,14].
General microarray technologies and their applications in microbiology have been extensively reviewed [18-21]. One of the most obvious advantages for microarray technology is that hundreds to thousands of probes can be deposited on a tiny surface area. However, this alone is not enough for early pathogen detection, which may result in more efficient interventions. Methods that can be universally applied for various known and even unknown causative agents with great sensitivity are critical for this goal. Nucleic acid amplification, usually through broad spectrum PCR, is required. The sample is typically amplified by either pathogen-specific multiplex or degenerate PCR before array hybridization “Fig. (1)”. The second strategy is by randomly primed amplification “Fig. (2)”, which allows detecting a wider variety of pathogens but has lower sensitivity compared to pathogen-specific approach. Two examples are described here.
Fig. (1). The HPV typing array printed in quadruplicate.
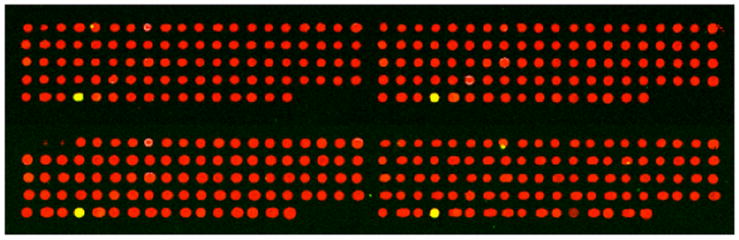
Each replicate block of the HPV typing array consists of 96 spots. In this case, HPV genomic DNA from the SiHa HPV16 infected cell line was amplified by PCR using modified B-GP5+/B-GP6+ primer pairs and labeled with a green dye. The red spots represent HPV types not present in the cell line and result from the red labeled Probe-70 binding to Spike-70. The four yellow spots represent detection of HPV16 in quadruplicate due to the mixing of the green labeled HPV16 DNA and the red labeled Probe-70.
Fig (2). Cluster analysis of 4 sputums and 1 HeLa sample.

The features are clustered according to similarity of the signal intensity patterns in the 5 microarrays. All measles and coronavirus OC43 probes and partial list of RSV, enterovirus probes are shown. Most of the features from the same virus are clustered together. Every sample has a very distinct virus signature except for the last lane, which is the HeLa sample. The HeLa sample is hybridized to HPV sequences, but the HPV features are not shown in this figure.
Multiplexing assay with pathogen-specific approach
More than 100 HPV types have been identified [22]. The HPV associated diseases are type-specific [23]. For example, there are current vaccines against HPV-16 and 18 that cause cervical cancer and HPV-6 and 11 that cause genital warts. Many types are related to skin lesions [24], such as the association of HPV-5 and 8 with skin cancers in patients having epidermodysplasia verruciformis.
We have recently modified the original Virochip protocols [5] to build an oligonucleotide microarray platform capable of identifying 37 mucosotropic HPVs that include 14 high-risk types and 23 low-risk types “Fig. (1)” [25]. The probe is located in a 150 bp L1 fragment that is bordered by GP5+ and GP6+ primers [26]. Specific oligonucleotide probes (30mer) for a given HPV type are printed multiple times in a particular spot with multiple spots for each HPV type. Prior to printing, every HPV-specific oligonucleotide probe is mixed with a Spike-70 oligonucleotide probe at the ratio of 50:1 for any given spot on the glass slide. Probe-70 is labeled red and is the reverse complement sequence to Spike-70. Probe-70 is mixed with HPV DNA amplified from samples. Prior to array hybridization the HPV DNA is labeled with a green dye. Since all spots contain probes for Spike-70, the majority of spots show up in the red channel when scanned “Fig. (1)”. However, if the spot contains probes for an HPV type in the sample then the green signal in the HPV labeled DNA merges with the red signal from the Spike-70:Probe-70 hybridization to yield a yellow spot “Fig. (1)”. Therefore, the inclusion of Spike-70 in each spot enables every spot to be detected in the red channel even though it represents an HPV type not in the sample. This is extremely useful for data processing and scanning of the chip.
The protocol originally used to PCR amplify around 40 HPV types with GP5+ and GP6+ primers has been modified to fit Virochip procedures. This is accomplished by a pair of modified primers, B-GP5+ and B-GP6+, which sequences are 5’ extended with a primer B sequence (GTTTCCCAGTCACGATC) [5]. An additional PCR program (round C) was added to label and further amplify the first round of PCR products with primer B as previously described [5]. This preserves the ratio of amplicons derived from different types of HPV when multiple infections are present. The sensitivity of our HPV typing array in combination with the amplification protocol is 1-2 copies of the HPV genome as assessed using the SiHa cell line. This cell line only contains 1-2 integrated copies of the HPV 16 genome [27]. This method has been used to unambiguously identify the correct HPV type in the following cell lines: HeLa (HPV 18), Caski (HPV 16), SiHa (HPV 16) and ME180 (HPV 68). In addition, diverse HPV types have been already been identified in DNA samples extracted from anal Pap smears of HIV infected patients [25].
Comprehensive Virochip for early diagnosis – a case study
The same unbiased approach to identify the unknown SARS virus [5,14] was taken to examine the potential etiological pathogen for the second example when the author was at the University of California, San Francisco (UCSF). The Virochip was designed to include the most highly conserved 70mer sequences from nearly 1000 fully sequenced reference viral genome in the NCBI (as of August 2002). Patients’ clinical samples were prepared and amplified by the previously published randomly priming protocol [5].
During the time of SARS epidemics in 2003, a 22 year-old male who had returned from a trip to foreign country, visited UCSF clinic, fearing that he had been infected by the SARS virus. He experienced fever, sore throat, cough, sneezing, coryza, fatigue, and generalized body ache for 5 days. His body temperature was 39.6° C at the time of his visit.
His sputum was collected, and the sample was immediately lysed with RLT buffer (Qiagen), frozen at −80° C and transported on dry ice to the research lab for a Virochip analysis [5]. A complete procedure took about 24 hours. A clear measles virus signature was evident after 24 hours of Virochip procedure “Fig. (2)”.
Independent follow-up with this patient revealed that skin rash developed the following day. An infectious disease pediatrician was consulted. Measles virus infection was suspected since the patient had not been vaccinated with MMR (measles, mumps, and rubella) in the past. His chest X ray did not show evidence of infiltrates. Blood and urine tests showed mild lymphopenia and pyuria. A serology test revealed that this patient was CMV IgM negative; rubella antibody negative; parvovirus B19 IgG positive but IgM negative; EBV anti-EBNA positive and IgM anti-VCA negative; RPR nonreactive, and heterophile agglutinin negative. However, the patient was positive for measles IgM and IgG. Other tests, including routine blood culture and a pharyngeal swab for group A streptococcus cultures was negative. Furthermore, measles virus was confirmed from urine and nasal swab cultures approximately 2 weeks later.
Three additional patients were seen at the clinic, and sputum samples were also collected. The initial analyses for these samples also revealed clear virus signatures (Coronavirus Oc43, RSV, Enterovirus). Hierarchical cluster analysis [28] revealed 4 distinct gene clusters correlated with the microarray results using the samples collected from these four patients “Fig. (2)”. The HeLa cell control sample (harboring human papillomavirus 18) “Fig. (2), lane 5” had almost no detectable signal on these features, but very strong signals on those corresponding to papillomavirus samples (not shown).
The case showed a sharp contrast between the unbiased and straightforward Virochip approach and current standard clinical practice. Nevertheless, while the Virochip assay is powerful, it is not always revealing. The timing and location of collecting clinical samples, judged by clinicians, are also very important for this approach.
MASSIVE PARALLEL (NEXT-GENERATION) SEQUENCING
Demand for nucleic acid sequence information has been growing ever since the completion of the reference sequence for the Human Genome Project. In 2004, the National Human Genome Research Institute (NHGRI) challenged scientists to achieve a $100,000 human genome (3 Gb/haploid genome) by 2009 and a $1,000 genome by 2014 [29]. To stimulate interest in this area, the X Prize Foundation has established the $10 Million Archon X Prize for Genomics [30].
Three “next-generation” sequencing platforms are currently available (as of April, 2008). The Roche 454 sequencer was introduced in 2004. The current GS-FLX model produces an average 250 bp per read, a combined throughput of about 100 Mb of sequence data per 7-hour run. The Illumina/Solexa Genome Analyzer, introduced in 2006, produces 1.3 Gb per 4-day run with 30-40 bp per read. The Applied Biosystems SOLiD sequencer was commercially released in late 2007. Each SOLiD run requires about 5 days and produces 3–4 Gb of sequence data with an average read length of 25–35 bp. All of these platforms apply the short-gun sequencing approach. While the total amount of the sequence data is impressive, the individual read is short, especially for the later two platforms. This poses a challenging bioinformatic task for sequence realignment and re-assembling. The technological bases of the current and future platforms have been extensively reviewed [31-37] and will not be discussed further here. While the cost of such a test is currently too expensive to become a clinical test, the speed of the technological advancement may make it an affordable test in the near future. In this section, I will discuss two exciting studies that took advantage of the next-generation sequencing platform to identify novel pathogens.
Fatal arenavirus infection associated with organ transplantation
Recently, three clusters of arenavirus transmission via organ transplantation have been documented [38,39]. In total, 10 out of 11 recipients linked to 3 individual donors died of an unexplained infectious disease 1-11 weeks after transplantation. The donor of each cluster did not have a history of acute infections. The prototype arenavirus, lymphocytic choriomeningitis virus (LCMV) was implicated in two clusters [38] and a novel arenavirus is responsible for the last cluster [39]. Notably, the only survivor was treated with ribavirin and lower level of immunosuppressants when the LCMV was identified as the etiological agent. The primary host of LCMV is wild house mouse, Mus musculus. LCMV infection usually causes asymptomatic or mild illness in immunocompetent humans. The mortality rate of recognized LCMV infection is less than 1%. Congenital LCMV infection is teratogenic [40]. The severity of the LCMV associated human diseases may be associated with immunity. Prior to the aforementioned transplant cases, two similar fatal infections were reported when LCMV was injected intravenously in attempt to regress lymphomas that were resistant to standard treatments [41].
Extensive microbiological studies were performed for the 3rd cluster [39]. These studies included bacterial and viral cultures; PCR for herpesviruses 1-8, lysavirus, influenza A and B viruses, respiratory syncytial virus, picornavirus, adenovirus, human parainfluenza virus, flavivirus, alphavirus, hantavirus, polyomavirus, Crimean–Congo hemorrhagic fever virus, Rift Valley fever virus, toxoplasma, Mycobacterium tuberculosis, and Mycoplasma pneumoniae as well as viral and panmicrobial oligonucleotide microarray analysis [17]. They did not reveal any candidate pathogens. A pooled RNA sample from 2 of the 3 recipients were amplified for massive parallel sequencing that yielded about 100,000 individual sequence reads, ranged from 45 to 337 nucleotides, by the Roche/454 GSL FLX sequencer. Subsequent sequence data processing revealed 14 sequence fragments that were consistent with Old World arenaviruses which are closest to LCMV. Primers for quantitative PCR were designed for detecting this novel arenavirus, which was present in 22 of 30 specimens from all of the recipients. The viral sequence was identical in all samples, indicating a common infectious source.
Merkel cell polyomavirus (MCV)
Chang, Moore and their colleague, who discovered Kaposi’s sarcoma herpesvirus (Human Heperpervius-8) in 1994 [42], reported the likely 7th genuine human oncogenic virus, Merkel cell polyomavirus (MCV) in early 2008 [43]. Merkel-cell carcinoma (MCC) is a rare but aggressive skin cancer of neuroendocrine origin [44]. The cell of origin is thought to be the Merkel cell, which is a mechanoreceptor in the basal layer of the epidermis. In white populations, the estimated incidence is 0.23 per 100,000, 10 times greater than in black populations [45]. Most cases occur in elderly and only 5% of cases happen before the age of 50 [45]. Sun exposure and immunosuppression have been implicated in MCC development. The risk of MCC in renal transplantation was about 0.13 per 1000 person-years [46]. MCC has also been reported in patients with HIV infection [47-57] (relative risk at 13.4 [47]) and with chronic lymphatic leukemia [58-61]. Several similarities between MCC and Kaposi’s sarcoma led the group to suspect that MCC might also have an infectious origin.
Central to the discovery of MCV is a technique called Digital Transcriptome Subtraction (DTS), which is conceptually simple but technically demanding. Similar to the approach described in the previous section, cDNA libraries derived from MCC tumors were subjected to high throughput sequencing by a Roche/454 sequencer. Near 400,000 sequence reads were generated from two libraries. The majority (99.4%) of the sequences derived from human origin are subtracted. Only one of the remaining 2395 cDNA had high homology to the T antigen of two polyomaviruses. One additional cDNA was subsequently identified to be part of the MCV sequence. Subsequently, it was found that 80% (8/10) of the MCC had integrated MCV in the human genome. Monoclonal viral integration was revealed the patterns of Sothern blot analysis. Only 8-16% of control tissues had low copy number of MCV infection.
These two examples show a very promising future for diagnostic microbiology. While it is too costly to apply this approach routinely, it is justified when an epidemic infection of unknown pathogen, like SARS in 2003, occurs. We may expect such ultra high throughput sequencing a routine clinical test when $1000 genome becomes a reality, which is a goal set by NIH before 2014. However, clinicians and scientists will face challenges to determine if a microbial or viral sequence present in the final data is an etiological agent since there are many commensals and opportunistic pathogens in our body. It is notable that the pathogen sequences are very rare compared to the total number of sequences in these two studies (14/100,000 and 2/400,000 respectively). Furthermore, a completely novel sequence may not be initially informative. As shown in the case of MCV, the presence of a novel virus is hinted at by a sequence that is homologous to known polyomaviruses. Another sequence was only identified as part of the MCV genome when the complete viral sequence was known.
CONCLUSION
The rapid technological advancement in the area of genomics, bioinformatics and nanotechnology is expected to change our views of the interaction between humans and surrounding microorganisms (commenals and pathogens) in the near future. The technology may lead to discoveries of new pathogens and may promote researchers to develop new interventions (vaccines and drugs) since the pathogens will be easily identified and monitored. In the meantime, the FDA has to develop new rules for accommodating the innovation to benefit more patients.
Acknowledgments
I am grateful to Stuart Ibsen and Dietrich Dehlinger for critical reading of the manuscript. The author is partially supported by NIH grants, AI36214-12S1 and AI074521-01.
Abbreviation
- CMV
Cytomegalovirus
- EBV
Epstein-Barr virus
- FDA
Food and Drug Administration
- HPV
Human Papillomavirus
- LCMV
Lymphocytic choriomeningitis virus
- MCC
Merkel cell carcinoma
- MCV
Merkel cell polyomavirus
- NCBI
National Center for Biotechnology Information
- NIH
National Institutes of Health
- PCR
Polymerase chain reaction
- RSV
Respiratory syncytial virus
- SARS
Severe acute respiratory syndrome
References
- 1.Leland DS, Ginocchio CC. Clin Microbiol Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson BH, Nicholson JK. Annu Rev Public Health. 2005;26:281–302. doi: 10.1146/annurev.publhealth.26.021304.144522. [DOI] [PubMed] [Google Scholar]
- 3.Markowitz VM. Curr Opin Biotechnol. 2007;18:267–272. doi: 10.1016/j.copbio.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Pallen MJ, Wren BW. Nature. 2007;449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin CD, Linder V, Sia SK. Lab Chip. 2007;7:41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 7.Lazcka O, Del Campo FJ, Munoz FX. Biosens Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 8.Jain KK. Clin Chem. 2007;53:2002–2009. doi: 10.1373/clinchem.2007.090795. [DOI] [PubMed] [Google Scholar]
- 9.Nolte FS, Caliendo AM. In: Manual of Clinical Microbiology. 9. Murray P, Baron EJ, Jorgensen J, Landry ML, Pfaller M, editors. ASM Press; Washington, D.C: 2007. pp. 218–244. [Google Scholar]
- 10.Procop GW. Clin Infect Dis. 2007;45(Suppl 2):S99–S111. doi: 10.1086/519259. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Rothman RE. Lancet Infect Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, 3rd, Smith TF. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YT, Beck S, Greenhow T, Wang D, Urisman A, DeRisi JL, Ganem D. The 41st Annual Meeting of IDSA (Infectious Diseases Society of America) Clinical Infectious Diseases; San Diego: 2003. pp. 196pp. LB–193. [Google Scholar]
- 14.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 15.Chiu CY, Rouskin S, Koshy A, Urisman A, Fischer K, Yagi S, Schnurr D, Eckburg PB, Tompkins LS, Blackburn BG, Merker JD, Patterson BK, Ganem D, DeRisi JL. Clin Infect Dis. 2006;43:e71–76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu CY, Alizadeh AA, Rouskin S, Merker JD, Yeh E, Yagi S, Schnurr D, Patterson BK, Ganem D, DeRisi JL. J Clin Microbiol. 2007;45:2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palacios G, Quan PL, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Zhai J, Renwick N, Hui J, Hegyi H, Grolla A, Strong JE, Towner JS, Geisbert TW, Jahrling PB, Buchen-Osmond C, Ellerbrok H, Sanchez-Seco MP, Lussier Y, Formenty P, Nichol MS, Feldmann H, Briese T, Lipkin WI. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant PA, Venter D, Robins-Browne R, Curtis N. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- 19.Clewley JP. J Clin Virol. 2004;29:2–12. doi: 10.1016/j.jcv.2003.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodrossy L, Sessitsch A. Curr Opin Microbiol. 2004;7:245–254. doi: 10.1016/j.mib.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Call DR. Crit Rev Microbiol. 2005;31:91–99. doi: 10.1080/10408410590921736. [DOI] [PubMed] [Google Scholar]
- 22.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 23.Bernard HU. J Clin Virol. 2005;32(Suppl 1):S1–6. doi: 10.1016/j.jcv.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Akgul B, Cooke JC, Storey A. J Pathol. 2006;208:165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- 25.Lu Q, Smith D, Richman DD, Liu YT. AACR Annual Meeting. Los Angles, CA: 2007. [Google Scholar]
- 26.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. J Clin Microbiol. 2002;40:779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner JD. J Gen Virol. 1999;80(Pt 7):1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.http://www.genome.gov/12513210 (2004).
- 30.http://genomics.xprize.org/.
- 31.Mardis ER. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Rusk N, Kiermer V. Nat Methods. 2008;5:15. doi: 10.1038/nmeth1155. [DOI] [PubMed] [Google Scholar]
- 33.von Bubnoff A. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 34.Shaffer C. Nat Biotechnol. 2007;25:149. doi: 10.1038/nbt0207-149. [DOI] [PubMed] [Google Scholar]
- 35.Church GM. Sci Am. 2006;294:46–54. doi: 10.1038/scientificamerican0106-46. [DOI] [PubMed] [Google Scholar]
- 36.Metzker ML. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 37.Shendure J, Mitra RD, Varma C, Church GM. Nat Rev Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 38.Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A, Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR. N Engl J Med. 2006;354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 39.Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. N Engl J Med. 2008 doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 40.Jamieson DJ, Kourtis AP, Bell M, Rasmussen SA. Am J Obstet Gynecol. 2006;194:1532–1536. doi: 10.1016/j.ajog.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 41.Horton J, Hotchin JE, Olson KB, Davies JN. Cancer Res. 1971;31:1066–1068. [PubMed] [Google Scholar]
- 42.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 43.Feng H, Shuda M, Chang Y, Moore PS. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang CK, Toker C. Cancer. 1978;42:2311–2321. doi: 10.1002/1097-0142(197811)42:5<2311::aid-cncr2820420531>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 45.Miller RW, Rabkin CS. Cancer Epidemiol Biomarkers Prev. 1999;8:153–158. [PubMed] [Google Scholar]
- 46.Bordea C, Wojnarowska F, Millard PR, Doll H, Welsh K, Morris PJ. Transplantation. 2004;77:574–579. doi: 10.1097/01.tp.0000108491.62935.df. [DOI] [PubMed] [Google Scholar]
- 47.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Lancet. 2002;359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 48.Catlett JP, Todd WM, Carr ME., Jr Va Med Q. 1992;119:256–258. [PubMed] [Google Scholar]
- 49.Samarendra P, Berkowitz L, Kumari S, Alexis R. South Med J. 2000;93:920–922. [PubMed] [Google Scholar]
- 50.Solomon RK, Lundeen SJ, Hamlar DD, Pambuccian SE. Diagn Cytopathol. 2001;24:186–192. doi: 10.1002/1097-0339(200103)24:3<186::aid-dc1038>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 51.An KP, Ratner D. J Am Acad Dermatol. 2001;45:309–312. doi: 10.1067/mjd.2001.114732. [DOI] [PubMed] [Google Scholar]
- 52.Matichard E, Descamps V, Grossin M, Genin R, Bouvet E, Crickx B. Br J Dermatol. 2002;146:671–673. doi: 10.1046/j.1365-2133.2002.04592.x. [DOI] [PubMed] [Google Scholar]
- 53.Calza L, Beltrami C, Manfredi R, Colangeli V, Freo E, Chiodo F. Br J Dermatol. 2002;146:895–898. doi: 10.1046/j.1365-2133.2002.04646.x. [DOI] [PubMed] [Google Scholar]
- 54.Burack J, Altschuler EL. J R Soc Med. 2003;96:238–239. doi: 10.1258/jrsm.96.5.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colebunders R, Bottieau E, Van den Brande J, Colpaert C, Van Marck E. HIV Med. 2004;5:452–454. doi: 10.1111/j.1468-1293.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 56.Cone LA, Gade-Andavolu R, Lesnick RH, Aitken D, Bush WS, Potts BE. Aids. 2006;20:474–475. doi: 10.1097/01.aids.0000206511.85395.09. [DOI] [PubMed] [Google Scholar]
- 57.Manganoni MA, Farisoglio C, Tucci G, Venturini M, Marocolo D, Aquilano MC, El-Hamad I, Ferrari VD, Calzavara Pinton PG. AIDS Patient Care STDS. 2007;21:447–451. doi: 10.1089/apc.2006.0152. [DOI] [PubMed] [Google Scholar]
- 58.Ziprin P, Smith S, Salerno G, Rosin RD. Br J Dermatol. 2000;142:525–528. doi: 10.1046/j.1365-2133.2000.03370.x. [DOI] [PubMed] [Google Scholar]
- 59.Safadi R, Pappo O, Okon E, Sviri S, Eldor A. Leuk Lymphoma. 1996;20:509–511. doi: 10.3109/10428199609052438. [DOI] [PubMed] [Google Scholar]
- 60.Ben-David A, Lazarov A, Lev S, Nussbaum B. Dermatol Online J. 2005;11:16. [PubMed] [Google Scholar]
- 61.Barroeta JE, Farkas T. Diagn Cytopathol. 2007;35:293–295. doi: 10.1002/dc.20616. [DOI] [PubMed] [Google Scholar]


