Abstract
During mouse development, the sophisticated vascular network of the lung is established from embryonic day (E)≈10.5 and continues to develop postnatally. This network is composed of endothelial cells enclosed by vascular smooth muscle, pericytes, and other mesenchymal cells. Recent in vivo lineage labeling studies in the developing heart and intestine suggest that some of the vascular smooth muscle cells arise from the surface mesothelium. In the developing lung, the Wilm's tumor 1 gene (Wt1) is expressed only in the mesothelial cells. Therefore, we lineage-labeled the mesothelium in vivo by using a Wt1-Cre transgene in combination with either Rosa26RlacZ, Rosa26RCAG-hPLAP, or Rosa26REYFP reporter alleles. In all three cases, cells derived from lineage-labeled mesothelium are found inside the lung and as smooth muscle actin (SMA) and PDGF receptor-beta positive cells in the walls of pulmonary blood vessels. To corroborate this finding, we used 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate, succinimidyl ester “mixed isomers” (CCFSE) dye to label mesothelial cells on the surface of the embryonic lung. Over the course of 72-h culture, dye-labeled cells also appear within the lung mesenchyme. Together, our data provide evidence that mesothelial cells serve as a source of vascular smooth muscle cells in the developing lung and suggest that a conserved mechanism applies to the development of blood vessels in all coelomic organs.
Keywords: lineage tracing, blood vessel, embryo, pleura
The function of the lung as a gas-exchange organ requires a precisely organized pulmonary vascular system. Although important remodeling occurs postnatally, the basic vascular network is set up early in development and is required for viability at birth. Our understanding of the molecular mechanisms regulating the formation of the pulmonary vascular system has advanced in recent years (1–3). Still, many important unanswered questions remain. One concerns the origin of the endothelial, smooth muscle cells, and pericytes that make up the blood vessels.
The embryonic lateral splanchnic mesoderm was considered to be the major source of the smooth muscle and pericytes that surround the endothelial cells of blood vessels growing into visceral organs. Recent studies of blood vessel formation in organs such as the heart and gut have added a new major source, the mesothelium (4–9). The mesothelium is a simple squamous epithelium lining the coelomic cavity and the organs housed within it. Recent cell lineage labeling studies on the developing heart provide evidence that the surface epicardial mesothelium undergoes epithelial-mesenchymal transition (EMT) and migrates into the myocardium where it differentiates into various cell types, including endothelium, smooth muscle cells, and cardiomyocytes (5, 8, 10, 11). In addition, lineage tracing and other studies show that the serosal mesothelium of the gut also contributes the majority of vascular smooth muscle cells (7, 12).
The embryonic and adult lungs are also encased by a thin layer of mesothelial cells. These cells are part of the pleura that provide vital protection and a smooth lubricated surface for movement of this organ. During development, the mesothelium has an important role in regulating the overall size and morphogenesis of the lung through interactions with submesothelial mesenchyme. For example, Fgf9 produced by the mesothelium signals to the underlying mesenchyme to stimulate proliferation and Fgf10 expression, which in turn signals to airway epithelium to regulate branching (13–15). In addition, lungs from Fgf9 null mutant embryos are defective in blood vessel development (2). Whereas there is strong evidence in favor of the mesothelium having an important signaling role in the embryonic lung, whether it contributes to the formation of the pulmonary vascular system is unknown. Here, we take advantage of the Wt1-Cre transgenic mouse line utilized by Wilm et al. (7) to lineage label the mesothelial cells during development. We demonstrate that lineage-labeled cells appear in the lung and give rise to vascular smooth muscle cells that populate the walls of vessels in both the proximal and distal lung. In addition, our data raise the possibility that other nonvascular mesenchymal cells are derived from the mesothelium. These findings potentially have implications for understanding some development defects in the lung and pathological conditions such as idiopathic pulmonary fibrosis.
Results and Discussion
Formation of the Lung Mesothelium.
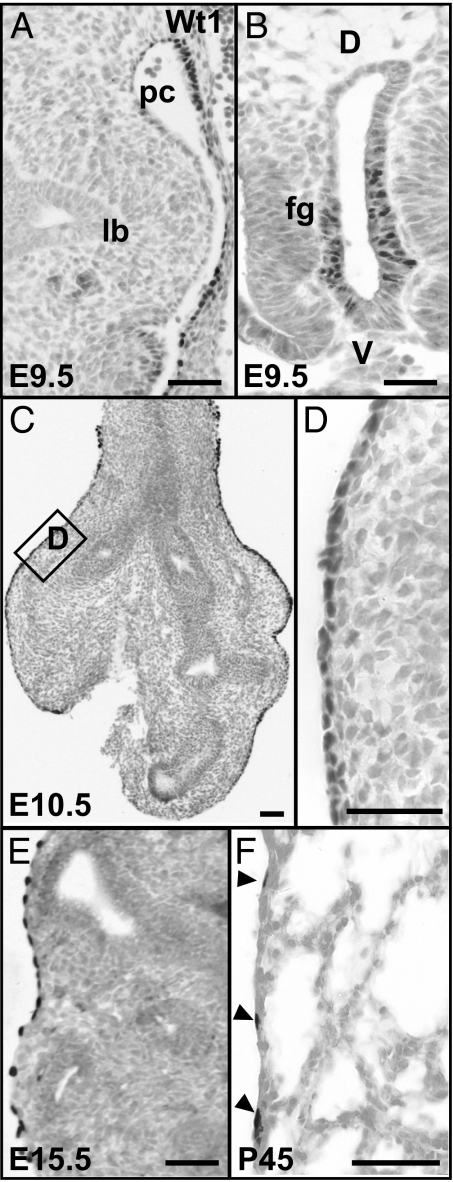
In the mouse, the development of the pulmonary vasculature matches lung branching morphogenesis from early E10.5 onwards and forms a dense capillary network encompassing the distal epithelium (16, 17). To establish a temporal link between the development of the mesothelium and the pulmonary vasculature, we examined the expression of Wt1, a marker for lung mesothelial cells (5, 7, 18), at selected stages of lung development (Fig. 1). At E9.5, the simple squamous epithelium covering the walls of the pericardio-peritoneal cavity shows strong reactivity to Wt1 antibody. By contrast, at this stage the primary lung buds lack Wt1 positive mesothelial cells (Fig. 1A). However, Wt1 protein is expressed by the epithelium of the ventral foregut, which gives rise to the future trachea after foregut separation (19) but not by the epithelium of the lung bud (Fig. 1 A and B). This foregut expression was confirmed by lineage-labeling by using Rosa26RlacZ (see below, data not shown). At E10.5, Wt1-positive mesothelial cells are tightly packed along the surface of the trachea and lung (Fig. 1 C and D). At this stage, expression of Wt1 in the endodermal epithelium of the trachea is no longer seen (Fig. 1C). Compared with the extensive cell proliferation that is observed inside the lung, both in mesenchymal and epithelial compartments, the proliferation of the mesothelium is rather limited, as shown by immunohistochemistry for phosphorylated Histone H3 (data not shown). Consistent with previous observations of mesothelial cells (15), we found that their internuclear distance increases during development, so that in the adult the cells are highly attenuated (Fig. 1 E and F). Significantly, at all stages examined, no expression of Wt1 is observed in the mesenchyme of the lung tissue (Fig. 1).
Fig. 1.
Expression of Wt1 protein in the developing and adult lung. (A) At E9.5, Wt1 protein is not detected in the primary lung buds, but in mesothelial cells along the wall of the pericardio-peritoneal cavity. (B) Wt1-positive cells in the ventral epithelium of the unseparated foregut. (C) At E10.5, Wt1-positive mesothelium now covers the surface of both the trachea and lungs. (D) A magnified view of the boxed region in C to show the Wt1-positive and tightly packed mesothelial cells. Wt1-positive mesothelial cells at E15.5 (E) and in the adult (arrowheads) (F). Note that at all times examined no Wt1-positive cells are present either in the epithelium or mesenchyme within the lung; lb, lung bud; pc, pericardio-peritoneal cavity; fg, foregut. (Scale bars, 50 μm.)
In Vivo Lineage Tracing of Lung Mesothelium.
To determine the fate of lung mesothelial cells, we used the Cre/loxp lineage-labeling system to genetically label mesothelial cells and their descendents, whatever their fate. To trace the lineage derivatives of the mesothelium, we initially crossed Wt1-Cre mice to Rosa26RlacZ reporter mice to obtain offspring carrying both alleles. Lungs from these embryos and pups were collected and subjected to whole-mount X-Gal staining, followed by paraffin-embedding and sectioning. At all stages examined X-Gal positive endodermal epithelial cells were only seen within the trachea and main stem bronchi and never within the lobes of the lung. It appears that both ciliated and secretory (Clara) cells in the trachea are descended from Wt1-Cre expressing cells in the foregut (data not shown).
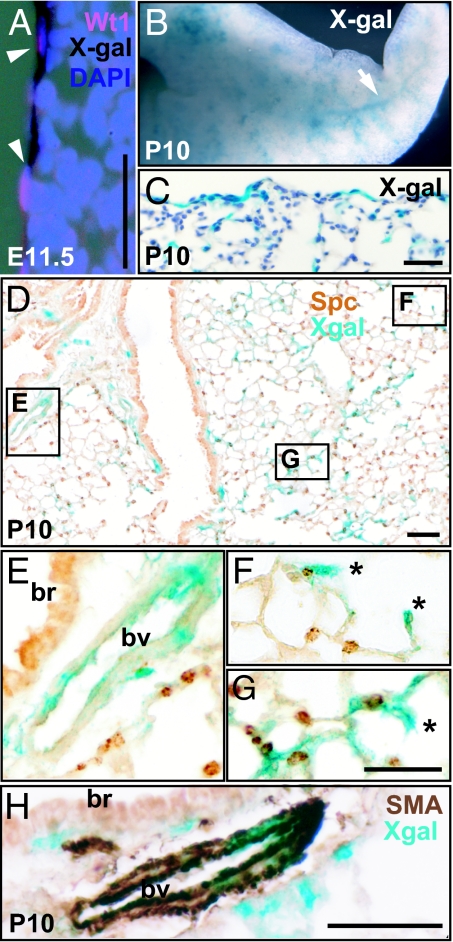
Examination of the surface mesothelium at E11.5 showed a positive correlation between Wt1 expression and X-Gal staining (Fig. 2A). Only a few cells positive for X-Gal are present within the lung proper at this time (data not shown), suggesting that the lineage-labeled mesothelial cells have just started to migrate into the lung. Significantly, it is at this stage (E11.5) that the first alpha-SMA positive cells are detected in the vascular wall of both arteries and veins (17), indicating a temporal link between the ingression of mesothelial cells and the maturation of the pulmonary vasculature. The influx and incorporation of mesothelial-derived cells into lung tissue is clearly demonstrated at postnatal day (P)10 by whole-mount staining, which showed an extensive network of X-Gal positive cells throughout all lobes (Fig. 2B). At no times examined (E11.5, E15.5, and P10) did we detect any X-Gal positive cells in Rosa26RlacZ lungs that have no Wt-Cre transgene. Analysis of sections showed that, consistent with the labeling system, most of the mesothelial cells are labeled with X-Gal on the lung surface. Significantly, as suggested from whole-mount views many X-Gal positive cells are also present within the lung tissue (Fig. 2 C and D). Some of these X-Gal positive cells are incorporated into the walls of pulmonary vessels. As shown in Fig. 2 D and E, some of these vessels are close to bronchi and have relatively thick walls, characteristics of arteries. To identify the X-Gal positive cells, we performed immunohistochemistry with an antibody against alpha-SMA. As shown in Fig. 2H, at P10 the X-Gal colocalizes with alpha-SMA in mural cells in the major blood vessels. Our quantitative analysis of P10 lungs revealed that, on average, 24.7% of SMA-positive cells of the pulmonary vasculature colabel with X-Gal (from a total of 811 SMA-positive cells counted from four Wt1-Cre;Rosa26RlacZ mice). By contrast, no colocalization of X-Gal and SMA was noticed in the airway smooth muscle cells underneath the bronchial epithelium. This finding is in line with observations in the developing gut where mesothelium only contributes to the vascular but not to the visceral smooth muscle (7).
Fig. 2.
Lineage-labeled mesothelium and its descendents in Wt1-Cre;Rosa26RlacZ lungs. (A) Colocalization of X-Gal staining (black) and Wt1 protein (red) on the surface of E11.5 lung. Nuclei are counterstained with DAPI. (B) Whole mount X-Gal staining of P10 lung. The arrow indicates the network of X-Gal stained cells within the lung. (C and D) Sections after whole-mount X-Gal staining to show that X-Gal positive cells are located both on the surface and within the lung tissue. (D–G) Lung section stained with antibody against SftpC. (E–G) Magnified views of the boxed regions in D. (E) X-Gal positive cells are incorporated into the artery close to the bronchus. (F and G) Two examples of X-Gal cells in alveoli possibly pericyte, endothelial cell, or myofibroblast (indicated by *). (H) Colocalization of alpha-SMA (brown) and X-Gal (blue) in the artery wall next to a bronchus. Note that smooth muscle cells underneath the bronchus do not colocalize with X-Gal; br, bronchus; bv, blood vessel. (Scale bars, 50 μm.)
Also, we observed X-Gal positive cells located within the P10 alveoli, including developing secondary septae. Based on their location and morphology at the resolution shown, these cells could potentially be alveolar smooth muscle cells, alveolar myofibroblasts, microvascular pericytes, or endothelial cells of capillaries (Fig. 2 F and G). We also noted the presence of X-Gal positive mesenchymal cells of unknown identity underneath the airway and throughout the lung tissue between blood vessels (Fig. 2H).
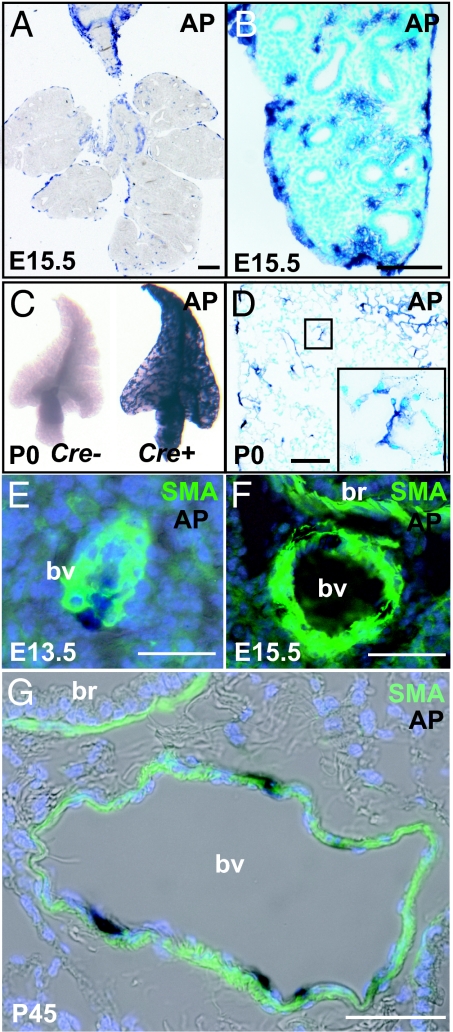
To confirm our finding by using the Rosa26RlacZ reporter allele, we used two other reporter mouse lines. The first was Rosa26RCAG-hPLAP. This line carries a conditional reporter allele based on the expression after recombination of human placental alkaline phosphatase (hPLAP) driven by the strong chicken actin gene (CAG) promoter. After staining, hPLAP-positive cells are easily identified both on the surface and within the Wt1-Cre;Rosa26RCAG-hPLAP lung at E15.5 (Fig. 3 A and B). Whole-mount staining of P0 Wt1-Cre;Rosa26RCAG-hPLAP lungs gives strong overall expression of hPLAP (Fig. 3C). By contrast, no AP-positive signals were seen in lungs carrying Rosa26RCAG-hPLAP but no Wt1-Cre transgene (Fig. 3C; data not shown). The AP-positive cells are readily detected within the lung (Fig. 3D). Similar to the results obtained with Rosa26RlacZ reporter mouse line, some of these AP-positive cells are distributed in the vessel walls and in the alveoli, including secondary septae. Colocalization with SMA after AP staining demonstrates these AP-positive cells are part of the walls of blood vessels at different embryonic stages (Fig. 3 E and F). At P0, a total of 33.8% of alpha-SMA positive cells in blood vessels are also reactive for AP staining (from counting a total of 3,119 cells from four mice). This colocalization of SMA and AP staining is also seen in the vasculature of the adult lung, as shown by representative section in Fig. 3G. This example shows a thin-walled vessel with the characteristic morphology of a vein.
Fig. 3.
Localization of lineage-labeled mesothelial cells and their descendants in Wt1-Cre;Rosa26RCAG-hPLAP lungs. (A and B) Sections of E15.5 lung after whole-mount staining for AP. Note that AP-positive cells are present both on the surface and inside the lung. (C) Whole-mount view of a lobe of AP-stained Wt1-Cre;Rosa26RCAG-hPLAP P0 lung (Left) and Rosa26RCAG-hPLAP control lung, which has no Wt1-Cre transgene (Right). (D) Section of P0 lung after whole-mount AP staining. Inset shows AP-positive cells in the alveoli. (E–G) Colocalization of SMA and AP in some of the vascular smooth muscle cells at E13.5 (E), E15.5 (F), and P45 (G); br, bronchus; bv, blood vessel. [Scale bars: 100 μm (A–D) and 50 μm (E–G).]
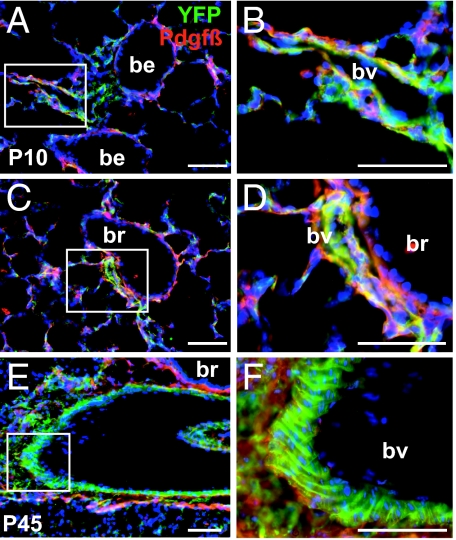
Last, we used the Rosa26REYFP reporter strain to map the localization of descendants of Wt1-Cre expressing cells in the lung of postnatal mice (P10 and P45). The availability of antibodies against eYFP enabled us to use immunohistochemistry to localize the lineage label and markers of smooth muscle cells [SMA (data not shown) and PDGF receptor-beta]. As shown in Fig. 4, lineage-labeled cells expressing PDGF receptor-beta are present in the thin walls of blood vessels associated with small bronchioles, as well as in the relatively thick walls of vessels associated with larger bronchi, which are presumed arteries. These findings further support the conclusion that mesothelial cells give rise to smooth muscle cells in both arteries and veins. As in the other studies, lineage-labeled mesenchymal cells are also seen within the alveoli and throughout the interstitial tissue of the lung (Fig. 4).
Fig. 4.
Lineage-labeled cells in Wt1-Cre;Rosa26REYFP lungs. (A–F) Immunohistochemistry of sections of P10 (A–D) and P45 (E–F) lungs from Wt1-Cre;Rosa26REYFP mice with antibody to GFP (green) and PDGF receptor-beta (red). (B, D, and F) Magnified view of the boxed regions in A, C, and E, respectively. Note the presence of lineage-labeled cells that also express PDGF receptor-beta on their surface in the relatively thin walls of vessels closely associated with bronchioles (A–D) and in the thicker walls of a vessel (presumed artery) associated with a larger bronchus (E and F). Note also in F the typical organization of lineage-labeled smooth muscle in the thick mural wall of the presumptive artery. In all sections lineage-labeled cells are absent from the population of airway smooth muscle. All nuclei are counterstained with DAPI; be, bronchiole; br, bronchus; bv, blood vessel. (Scale bars, 50 μm.)
The Fate of Dye-Labeled Mesothelium in the Cultured Embryonic Lung.
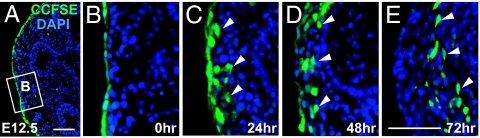
Previous studies on heart and gut mesothelial cells have suggested that these cells undergo EMT before migration and differentiation into vascular smooth muscle cells (7, 10, 11). To confirm our finding that the mesothelium gives rise to the mesenchymal cells within the developing lung, we took advantage of an in vitro culture system with embryonic lung at E12.5. We first labeled the mesothelium with the lipophilic dye CCFSE (see Material and Methods), which has been well established to mark only surface cells (4, 7). As expected, at the time of CCFSE application, 97% of the covering mesothelial cells were labeled with the fluorescent dye (Fig. 5 A and B). It is of particular note that no submesothelial mesenchymal cells are labeled with CCFSE at this time (Fig. 5B). After 24 h of culture, CCFSE-marked cells were still observed on the surface but were also present in the submesothelial mesenchyme (Fig. 5C). After another 24–48 h of culture, some of these dye-labeled cells were present even deeper within the lung tissue (up to 70 μm from the surface; see Fig. 5 D and E).
Fig. 5.
In vitro culture of embryonic lung with dye-labeled mesothelium. (A–E) Distribution of CCFSE-labeled cells after culturing for indicated periods of time (A, 0 h). (B) A magnified view of the boxed region in A. Note that the majority of the surface mesothelial cells are labeled with dye. (C) After 24 h of culture, some dye-labeled cells are present beneath the surface. After culture for 48 (D) and 72 h (E), some dye-labeled are present within the lung tissue. Arrows indicate dye-labeled cells within the lung tissue. All nuclei are counterstained with DAPI. (Scale bars, 50 μm.)
Conclusions
This article provides evidence that, during embryonic development, mesothelial cells covering the surface of the lung migrate into the organ and give rise to various cell types, including vascular smooth muscle cells. This evidence is based on the behavior of live mesothelial cells labeled by distinctly different techniques: by vital dye and by three different genetic reporter alleles (Rosa26RlacZ, Rosa26RCAG-hPLAP, and Rosa26REYFP). Our data from genetic lineage tracing suggest that, on average, 30% (24.7–33.8%) of the smooth muscle cells in the walls of the blood vessels are derived from mesothelial cells. At present, we do not know why this value is lower than seen in the heart and gut (5, 7, 8). One possibility is that the overall lineage labeling of mesothelial cells is less efficient than it appears from analysis of tissue sections (Figs. 2 and 3). Another possibility is that there are two alternative sources of smooth muscle, the mesothelium and the splanchnic mesoderm-derived mesenchyme (3, 20–22). More detailed lineage tracing studies and analysis of the phenotypes of labeled and unlabeled cells in the walls of the blood vessels will be needed to distinguish between these possibilities.
Our genetic lineage labeling studies also raise the possibility that some mesenchymal cells outside the walls of the blood vessels, potentially interstitial fibroblasts, alveolar myofibroblasts, and even a few endothelial cells, are derived from Wt1-positive mesothelium. However, we cannot exclude at this time the possibility that nonsmooth muscle cells are derived from CD34-positive hematopoietic progenitor cells circulating from the bone marrow. These progenitors have been shown to express low levels of Wt1 (23, 24). Even if this origin turned out to be the case, our results would be evidence for a significant contribution of bone marrow-derived cells to both the embryonic and uninjured adult lung.
Last, our data here address only the developmental potential of mesothelial cells in the embryonic lung. It will be critical to know whether mesothelial cells on the surface of the adult lung are multipotent and able to give rise to mesenchymal cells (e.g., during injury and repair, or in response to pathological conditions). A recent report showed that overexpression of Tgfβ1 in the mesothelium of the rat lung results in pleural fibrosis, which subsequently expands into the lung parenchyma (25). One possibility, which could be tested by combining Tgfβ1 treatment with the use of an inducible lineage-labeling Wt1-CreERT2 allele (5), is that the cytokine induces local EMT in mesothelial cells that then move into the lung and proliferate as matrix-secreting fibroblasts.
In conclusion, our data provide evidence that the mesothelium of the lung contributes to mesenchymal cells within the organ, and in particular, to the smooth muscle cells of the pulmonary vasculature. Thus, the surface mesothelium represents a previously unappreciated progenitor cell population for the embryonic lung. This result, along with previous findings in the heart and gut (5–8, 10, 11), suggests that there is a common mechanism linking the development of the surface mesothelium of all coelomic organs with the development and maturation of the internal vasculature. Our findings also have important implications for understanding some development defects in the lung and pathological conditions such as idiopathic pulmonary fibrosis.
Materials and Methods
Mouse Strains.
The Tg(WT1-Cre)AG11Dbdr (Wt1-Cre) transgenic mouse line has been described previously (7). The transgene is based on the WT280Cre YAC and is the same to that described for the WT280LZ YAC (26), except that the beta-Galactosidase ORF was replaced by a nuclear localization signal (NLS)-tagged Cre recombinase ORF (7). Wt1-Cre, Gt(ROSA)26Sor (Rosa26RlacZ), and Rosa26REYFP (Gt(ROSA)26Sortm1(EYFP)Cos) mouse lines were maintained on a (C57BL/6 × 129/SvEv) mixed background and genotyped as described previously (7, 19). The Rosa26RCAG-hPLAP reporter mouse was generated by first cloning the hPLAP cDNA (27) into the pBigT plasmid (28) immediately after the loxP-neo-4xpolyA-loxP cassette. The loxP-neo-4xpolyA-loxP-hPLAP-polyA (STOP-hPLAP) fragment was then inserted into a Rosa26R-acceptor plasmid (28). An 1.6-kb CAG promoter was subsequently cloned upstream of the STOP-hPLAP cassette. This targeting construct was electroporated into embryonic stem cells to generate the Rosa26RCAG-hPLAP knock-in mice. The primer set for genotyping Rosa26RCAG-hPLAP is 5′-CACTTGCTCTCCCAAAGTCG-3′, 5′-TAGTCTAACTCGCGACACTG-3′. All animal experiments were approved by the Duke University Institutional Animal Care and Use Committee.
Histology and Immunohistochemistry.
Lung tissues were fixed in 4% paraformaldehyde in PBS (PFA) for 4 h at 4°C, paraffin-embedded, and sectioned as previously described (19, 29). Immunohistochemistry for Wt1 (1:50 dilution, mouse monoclonal antibody, M3561; DAKO), alpha-SMA (1:500 dilution, mouse monoclonal antibody, A2547; Sigma), CD31 (PECAM) (1:500 dilution, rat monoclonal antibody, 550274; PharMingen), phosphorylated Histone H3 (1:500 dilution, rat monocolonal antibody, HTA28; Sigma), ProSurfactant protein C (SftpC 1:200 dilution, rabbit polyclonal antibody, AB3428; Chemicon), GFP (1:500 dilution, rat monoclonal antibody, 0440484; Nacalai Tesque), and PDGF receptor-beta (1:100 dilution, mouse monoclonal antibody, 14–1402-81; eBioscience) was performed according to standard procedures. For Wt1 staining, antigen retrieval was performed by boiling sections in 10 μM sodium citrate at full power for 5 m, and MOM kit (Vector Laboratories) was used according to the manufacturer's instructions.
Whole-Mount X-Gal Staining and AP Staining.
Whole-mount X-Gal staining was performed according to standard protocols (29). AP staining was visualized with substrate BCIP/NBT following standard procedure. After whole-mount staining, samples were dehydrated, paraffin-embedded, and sectioned as previously described (19).
In Vivo Labeling of Embryonic Lungs and their Culture.
E12.5 embryos were separated from extraembryonic tissues but allowed to remain attached to the placenta. CCFSE (Molecular Probes) was diluted to 24 μM in sterile PBS. The dye was then injected into the pleural cavity through a small opening in the lateral body wall covering the heart and lung. After injection, embryos were incubated for 1 h at 37°C and 5% CO2 in DMEM supplemented with 10% FBS. Subsequently, lungs were isolated and cultured on a coverslip in 3-ml serum free medium [DMEM/F12 (GIBCO), supplemented with Penicillin/Streptomycin antibiotics, Insulin (10 μg/ml), Transferrin (5 μg/ml), Selenium (0.065 μg/ml)], 40 ng/ml EGF, and 20 ng/ml FGF2 (R&D Systems). Cultures were harvested after 24, 48, and 72 h. At least two embryonic lungs at each time were fixed with 4% PFA for 15 m on ice and then paraffin-embedded and sectioned. Control explants without CCFSE treatment showed no difference in tissue integrity and viability (data not shown). The culture has been repeated for four times.
Confocal Microscopy and Cell Counting.
Images were captured with a Leica ASMDW laser scanning confocal microscope. For cell counting, a total of 24 optical sections from lungs costained with X-Gal and alpha-SMA antibody (four lungs at P10) or AP staining and alpha-SMA antibody (four lungs at P0) were included. Cells were manually counted on a z-series of optical sections, and multiple optical sections were examined to distinguish cell boundaries.
Acknowledgments.
We thank members of the B.L.M.H. laboratory and Dr. David Brass in the Department of Pediatrics at Duke University Medical Center for critical reading and helpful suggestions. This work was supported by National Institutes of Health Grants HL071303 (to B.L.M.H.) and R01HL34318 (to D.B.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol. 2005;25:5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.deMello DE, Reid LM. Embryonic and early fetal development of human lung vasculature and its functional implications. Pediatr Dev Pathol. 2000;3:439–449. doi: 10.1007/s100240010090. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Pomares JM, et al. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- 5.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: A review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 7.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 8.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: Discontinuous formation of coronary vessels. Proc Natl Acad Sci USA. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 11.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi M, Bader DM, Wilm B. Serosal mesothelium retains vasculogenic potential. Dev Dyn. 2007;236:2973–2979. doi: 10.1002/dvdy.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 14.Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 15.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 16.Gebb SA, Shannon JM. Tissue interactions mediate early events in pulmonary vasculogenesis. Dev Dyn. 2000;217:159–169. doi: 10.1002/(SICI)1097-0177(200002)217:2<159::AID-DVDY3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Parera MC, et al. Distal angiogenesis: A new concept for lung vascular morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2005;288:L141–L149. doi: 10.1152/ajplung.00148.2004. [DOI] [PubMed] [Google Scholar]
- 18.Kumar-Singh S, et al. WT1 mutation in malignant mesothelioma and WT1 immunoreactivity in relation to p53 and growth factor receptor expression, cell-type transition, and prognosis. J Pathol. 1997;181:67–74. doi: 10.1002/(SICI)1096-9896(199701)181:1<67::AID-PATH723>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 19.Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: Current players and new roles for noggin and Bmps. Differentiation. 2006;74:422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- 20.deMello DE, Sawyer D, Galvin N, Reid LM. Early fetal development of lung vasculature. Am J Respir cell Mol Biol. 1997;16:568–581. doi: 10.1165/ajrcmb.16.5.9160839. [DOI] [PubMed] [Google Scholar]
- 21.Hall SM, Hislop AA, Haworth SG. Origin, differentiation, and maturation of human pulmonary veins. Am J Respir cell Mol Biol. 2002;26:333–340. doi: 10.1165/ajrcmb.26.3.4698. [DOI] [PubMed] [Google Scholar]
- 22.Hall SM, Hislop AA, Pierce CM, Haworth SG. Prenatal origins of human intrapulmonary arteries: Formation and smooth muscle maturation. Am J Respir cell Mol Biol. 2000;23:194–203. doi: 10.1165/ajrcmb.23.2.3975. [DOI] [PubMed] [Google Scholar]
- 23.Fraizer GC, Patmasiriwat P, Zhang X, Saunders GF. Expression of the tumor suppressor gene WT1 in both human and mouse bone marrow. Blood. 1995;86:4704–4706. [PubMed] [Google Scholar]
- 24.Menssen HD, Renkl HJ, Entezami M, Thiel E. Wilms' tumor gene expression in human CD34+ hematopoietic progenitors during fetal development and early clonogenic growth. Blood. 1997;89:3486–3487. [PubMed] [Google Scholar]
- 25.Decologne N, et al. TGF-beta1 induces progressive pleural scarring and subpleural fibrosis. J Immunol. 2007;179:6043–6051. doi: 10.4049/jimmunol.179.9.6043. [DOI] [PubMed] [Google Scholar]
- 26.Moore AW, et al. YAC transgenic analysis reveals Wilms' tumour 1 gene activity in the proliferating coelomic epithelium, developing diaphragm and limb. Mech Dev. 1998;79:169–184. doi: 10.1016/s0925-4773(98)00188-9. [DOI] [PubMed] [Google Scholar]
- 27.Leighton PA, et al. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- 28.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Que J, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]