Abstract
There is widespread interest in efficient characterization of differences between tumor and normal samples. Here, we demonstrate an effective methodology for genome-scale characterization of tumors. Using matched normal and tumor samples from liver cancer patients, as well as non-cancer-related normal liver tissue, we first determined changes in gene expression as monitored on RNA expression arrays. We identified several hundred mRNAs that were consistently changed in the tumor samples. To characterize the mechanisms responsible for creation of the tumor-specific transcriptome, we performed ChIP-chip experiments to assay binding of RNA polymerase II, H3me3K27, and H3me3K9 and DNA methylation in 25,000 promoter regions. These experiments identified changes in active and silenced regions of the genome in the tumor cells. Finally, we used a “virtual comparative genomic hybridization” (vCGH) method to identify copy number alterations in the tumor samples. Through comparison of RNA Polymerase II binding, chromatin structure, DNA methylation, and copy number changes, we suggest that the major contributor to creation of the liver tumor transcriptome was changes in gene copy number.
Keywords: liver tumors, RNA profiling, ChIP-chip, comparative genomic hybridization
INTRODUCTION
Liver cancer is the 5th most common cancer worldwide and the third most common cause of cancer mortality (1) and hepatocellular carcinoma (HCC) accounts for ~90% of primary liver cancers. Most cases of liver cancer occur in either sub-Saharan Africa or in Eastern Asia (China alone accounts for 50% of the world’s cases). The major risk factor differs in different areas. In most high-risk areas, the dominant risk factor is chronic hepatitis B virus (HBV) infection or consumption of Afloxatoxin B-contaminated food. Many of the high-rate Asian countries now vaccinate all newborns against HBV and thus HCC rates are decreasing in these areas. In contrast, rates of liver cancer in the low-risk areas, such as North and South America and Northern Europe, are increasing, perhaps due to an increased prevalence of hepatitis C virus (HCV) infection in these areas. Accordingly, in the United States, HCC is the fastest growing cause of cancer-related death in men.
In attempts to characterize molecular changes that occur during the development of HCC, several previous studies have compared RNA expression levels in normal vs. tumor liver using techniques such as directed analysis of the expression of candidate genes, serial analysis of gene expression (SAGE) assays, differential display, and hybridization of cDNA arrays (2–8). Gene expression alterations involving loss of checkpoint control have been commonly observed (see (1) for a review). In particular, down-regulation of the p53 and RB tumor-suppressor pathways are affected at multiple levels. For example, the vast majority of human HCC over express gankyrin, which inhibits both Rb and p53 checkpoint function (9, 10).
One approach toward the treatment of HCC is to attempt to reverse transcriptional deregulation in the tumor cells, thus restoring the normal liver transcriptome and the normal liver phenotype. For example, it has been shown that adenovirus-delivered gankyrin-specific inhibitory RNAs can reduce cell growth and enhance apoptosis in HCC cell culture lines (11). However, treatment of HCC in patients is more likely to involve therapies other than use of siRNAs. One potential therapeutic option is to target the transcriptional machinery and/or chromatin structure that is responsible for deregulated expression of genes such as gankyrin. The success of this approach depends upon a thorough understanding of the mechanisms by which genes are up or down regulated in the liver tumor tissue.
The expression of protein-coding genes can be regulated at different steps, including transcription initiation and elongation or mRNA processing, transport, stability and translation. In a normal cell, much of the regulation is thought to occur at the level of transcription initiation. Changes in transcription initiation in a tumor cell could be achieved by modulating the activity of a promoter that is already active. For example, a highly expressed gene could be down-regulated by loss of an upstream transcriptional regulator (due to deletions or mutations in the tumor genome) or a modestly expressed gene could be upregulated by the inappropriate tumor-specific expression of a transcriptional regulator that is normally not active in that differentiated cell type. Promoters regulated in such a manner generally show changes in the amount of general transcription factors, such as RNA polymerase II, bound to the core promoter region and/or to downstream regions of the gene. Another mechanism by which transcription initiation can be achieved is via changes in the chromatin structure of a gene region (see (12) for a review). For example, in differentiated cells many genes (in particular those which are expressed in pluripotent cells or in other highly differentiated cell types) are silenced by certain chromatin modifications. Specifically, DNA methylation and trimethylation of lysine 9 or lysine 27 of histone H3 (H3me3K9 or H3me3K27) are marks for silenced chromatin. Changes in chromatin structure have been previously associated with tumor development and thus gene expression of a tumor cell could be regulated by transitions of a particular gene region from an active to a silenced state, or vice versa. Finally, tumor cells can utilize another mechanism for altering the levels of particular genes that is not generally used in normal cells. Many tumors show amplification or deletion of certain regions of the genome. Such genomic alterations (in particular copy number changes observed at a low frequency in a certain tumor type) can be a consequence of the genomic instability that is known to be associated with tumor development. However, specific regions that display amplification or deletion at a high frequency in a tumor type may reflect a causal relationship between gene expression and tumor development.
Described below are our studies in which we have characterized the mechanisms by which the tumor-specific transcriptome is regulated in a human liver tumor. Interestingly, we find that the majority of genes that display altered RNA expression in HCC (as compared to normal liver tissue) are regulated by changes in copy number and not by changes in the recruitment of the transcriptional machinery or by modulation of gene silencing.
MATERIALS AND METHODS
Tissue Samples
Human normal adjacent tissues and tumor liver tissues were obtained from the National Disease Research Interchange NDRI (Philadelphia, PA) and from the University of Wisconsin-Madison Hospital (Madison, WI). Samples were surgically collected and preserved by flash freezing in liquid nitrogen followed by storage at −70°C. Patient 1 (OD04962) was a 58 year-old black male. Patient 2 (OD11753) was a 77 year-old male. Patient 3 (165) was an 81 year-old white female. Normal liver tissue (not-cancer-related) was from Patient 4 (00-559) who was a 50 year-old female. Purified normal hepatocytes (Patient 5) were purchased from CellzDirect (Pittsboro, NC) as cryopreserved suspension hepatocytes that were purified by iso-density percoll centrifugation. The HuH7 cell line was purchased from ATCC; cells were grown at 37° in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS and 1% Penicillin/Streptomycin.
RNA arrays
RNA samples were prepared using Qiagen’s RNeasy Micro Kit (Qiagen) and then assayed using the Agilent Systems Bioanalyzer 2100 to ensure that high quality RNA was used for the array experiments. The Illumina TotalPrep RNA amplification kit from Ambion (AMIL1791) was used to generate biotinylated, amplified RNA for hybridization with the Illumina Sentrix Expression Beadchips Human 6-v1 or HumanRef-8v2. The Human 6-v1 gene expression beadchips consisted of a 6-array, 2 stripe format comprising approximately 48,000 probes/array. In this array, 24,000 probes were from refseq sequences and 24,000 from other genbank sequences. Human Ref-8 v2 arrays consist of 24,000 probes from refseq sequences on a 8-strip format array (see http://www.illumina.com/pages.ilmn?ID=197 for more details concerning the Illumina arrays). Arrays were processed as per manufacturer’s instructions, scanned at medium PMT settings as recommended by the manufacturer, and analyzed using Bead Studio Software v. 2.3.41. Hybridization signals were analyzed using BeadStudio Gene Expression Module v.3 (Illumina). All transcripts were normalized and ranked according to the detection P-value, which is calculated based on signals of negative controls. The detection P-value is calculated as 1-R/N, where is R is the rank of the gene signal relative to negative controls and N is the number of negative controls. P-value can be interpreted as probability of seeing a certain signal level without specific probe-target hybridization. Expressed genes are indicated as those having a P-value of 0. Data was normalized using the “average” method, which simply adjusts the intensities of two populations of gene expression values such that the means of the populations become equal. Differential expression was calculated using an algorithm provided by Bead Studio. Fold-enrichment values were used to obtain the list of candidates with greater than 1.5-fold change and a p-value=0. RNA expression analysis was performed by Hierarchical Clustering using Genesis 1.7.2 software(http://genome.tugraz.at/genesisclient/genesisclient_description.shtml), with the average linkage clustering as agglomeration rule (13). All of the genes from the Illumina platform were used for the clustering analysis.
ChIP assays and amplicon preparation
ChIP assays were performed as previously described (14) with the following modifications for liver tissues. Briefly, tissues were cut in small pieces with a razor blade, crosslinked in 1.5% formaldehyde for 15 minutes, processed in a Medimachine (BD Biosciences, San Jose, CA) using a 50 micron medicon to produce a liver cell suspension. Nuclear extracts were prepared and chromatin was sonicated using a Bioruptor Sonicator (Diagenode, Sparta, NJ). Each chromatin immunoprecipitation was performed using between 7–15 μg of chromatin. A detailed protocol for ChIP assays in liver tissue is available online at: http://www.genomecenter.ucdavis.edu/farnham/protocols/tissues.html. In addition a detailed protocol for ChIP miniaturization or “MicroChIP” is available (15). ChIP assays with the 5-Methylcytidine antibody (Eurogentec cat# BI-MECY-0100) were performed using the ChIP-IT Express kit (Active Motif, cat#53008). For these assays, genomic DNA was extracted by shaking cells in digestion buffer (100mM NaCl, 10mM TrisCl, pH 8, 25mM EDTA, pH 8, 0.5% SDS) and 0.1 mg/ml Proteinase K for 12–18 hours at 50°C and purified using a phenol-chlorophorm extraction method. Extracted DNA was sonicated to an average size of 800 bp, denatured at 95°C for 10 min, quickly chilled on ice and captured on magnetic beads following the protocol as described by the manufacturer. Antibodies used in this study include RNA Polymerase II (Covance 8WG16), H3me3K27 (Upstate 07-449), and H3me3K9 (Abcam 8898). The secondary rabbit anti-mouse IgG (cat# 55436) was purchased from MP Biomedicals. Standard PCR reactions using 2 uls of the immunoprecipitated DNA were performed. PCR products were separated by electrophoresis through 1.5% agarose gels and visualized by ethidium bromide intercalation. Amplicons, prepared using 50–80% of a ChIP sample, were generated using Sigma’s Whole Genome Amplification Kit 2; see our published ChIP protocol (16) and http://genomics.ucdavis.edu/farnham/ for details). Quality of the amplicons was monitored by PCR of positive and negative control regions (see Supplementary Figure S1).
DNA Microarrays
Amplicons were applied to 5 kb promoter arrays (see Supplementary Table S1 and www.nimblegen.com for details). The labeling and hybridization of DNA samples for ChIP-chip analysis was performed by NimbleGen Systems, Inc. Briefly, each DNA sample (1 μg) was denatured in the presence of 5′-Cy3- or Cy5-labeled random nonamers (TriLink Biotechnologies) and incubated with 100 units (exo-) Klenow fragment (NEB) and dNTP mix [6 mM each in TE buffer (10 mM Tris/1 mM EDTA, pH 7.4; Invitrogen)] for 2 h at 37°C. Reactions were terminated by addition of 0.5 M EDTA (pH 8.0), precipitated with isopropanol, and resuspended in water. Then, 13 ug of the Cy5-labeled ChIP sample and 13ug of the Cy3-labeled total sample were mixed together, dried down, and resuspended in 40 μl of NimbleGen Hybridization Buffer (NimbleGen Systems) plus 1.5 ug of human COT1 DNA. After denaturation, hybridization was carried out in a MAUI Hybridization System (BioMicro Systems) for 18 h at 42°C. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation, and scanned at 5-μm resolution using the GenePix 4000B scanner (Axon Instruments). Fluorescence intensity raw data were obtained from scanned images of the arrays using NIMBLESCAN 2.0 extraction software (NimbleGen Systems). For each spot on the array, log2-ratios of the Cy5-labeled test sample versus the Cy3-labeled reference sample were calculated. Then, the biweight mean of this log2 ratio was subtracted from each point; this procedure is approximately equivalent to mean-normalization of each channel.
Analysis of ChIP-chip data
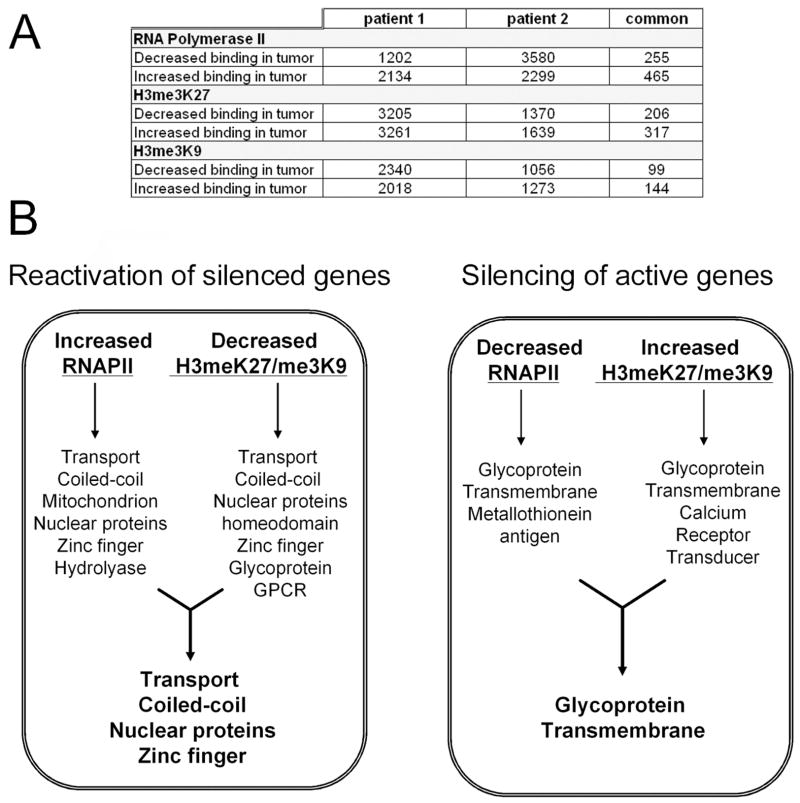
The human 5 kb promoter array consists of a set of two arrays encompassing a total of ~25,000 human promoters. We refer to the two arrays in the set as “promoter 1” and “promoter 2”. Most promoters encompass ~5 Kb of genomic sequence tiled with a 50 mer probe every ~100 nts. To analyze this data, we used a variant of the “Maxfour” approach previously described (17) for our data. However, we used a 10-point window instead of the 4-point window used previously. Further statistical exploration of this procedure will be presented elsewhere (Bieda et al., in preparation). As expected, we found that a comparison of repeated normal ChIP-chip experiments from the same samples showed an excellent correlation between experiments. However, there was a relatively small but clear deviation from the expected slope of 1 in these experiments, probably due to small variations in experimental procedures or arrays. Hence, we sought to normalize arrays to compensate for these effects in comparing tumor and normal samples. To do so, we relied on the expectation that a relatively small percentage of the ~25,000 promoters monitored in our experiments will differ in the tumor vs normal tissues. For one patient (patient 1), the slope of a fit of the tumor vs normal data for RNAPOLII was almost unity (1.03). In contrast, we found that for a second patient (patient 2), the slope of the tumor vs normal for RNAPOLII was 0.57, indicating a difference in the overall strength of ratio signal on the array. Hence, we corrected all values for these effects. To determine promoters that showed increased binding in tumor vs normal, we used the following parameters: the difference (i.e. tumor-normal) value had to exceed 0.2 and the absolute value of the ratio of the difference to the normal had to be at least 0.5. To calculate the list of promoters that, conversely, showed decreased binding in tumor vs normal, we used the same parameters, except that the difference was “normal-tumor” and the ratio was the difference/tumor value. Our goal was to identify promoters that showed changes in binding in the normal vs the tumor samples. Because tumor samples are often heterogeneous, we reasoned that many of the instances of differences in binding in the normal vs tumor samples might be modest. Therefore, we chose “very generous” parameters to identify the initial set of promoters that potentially showed changes in binding in the tumor samples. We realized that many of these potential changes would be false positives due to the generous cut-offs used in the array analysis. In fact, we confirmed this by analyzing two technical replicates of the normal to tumor comparison from patient 1. For example, our parameters identified 2134 promoters that showed increased binding of RNAPII in the tumor in replicate A and 2299 promoters that showed increased binding in replicate B. However, only 465 of these promoters showed the same direction change in both of the replicate experiments. This analysis tells us that the low overlap between patient 1 and patient 2 is to be expected due to the generous parameters and the nature of microarray hybridization. To deal with this issue, but still allow us to identify modest changes in binding in the tumor samples, we used generous cut-off parameters but then required that the promoters show the same directional change in two independent patients.
vCGH analysis
To analyze data, custom programs in perl, bash, and R were produced in a Linux environment. We used 2 normal cell arrays and 2 tumor cell arrays for human tissue data. For each tumor, the genomic DNA channels of the arrays were quantile normalized and a single “merged” gDNA vector of values was produced using the median values. We calculated the average of all probes for a single promoter so that the promoter was represented by a single value. The same procedure was applied to the normal cell arrays. After calculation of the ratio of tumor/normal, the data was smoothed using a 21-point mean filter. Data was displayed using the snapCGH package in R. Confirmations of the amplified or deleted regions identified using the ChIP-chip assays were performed using PCR, which we have found is a reliable and cost-effective method for analysis of DNA. For these experiments, the number of PCR cycles was kept low to ensure that the signals were within the linear range of the assay, providing a semi-quantitative analysis.
DAVID analysis
Functional annotations were performed using the program DAVID 2.1 (18); see also http://apps1.niaid.nih.gov/david/. The same parameters were used for all analyses presented in this study. These parameters were Gene Ontology (GO) Molecular Function term, level 2; Interpro name in the Protein Domains section; and SP_PIR_Keywords in the Functional Categories section. Before performing the analyses, hypothetical genes and genes with no known function were removed from the list. After performing the analyses, all categories that represented less than 4% of the total number of genes were eliminated. In addition, redundant terms (e.g. transcriptional regulation and transcription factor activity) and non-informative terms (e.g. multi-gene family) were also eliminated.
RESULTS
Characterization of the tumor-specific transcriptome
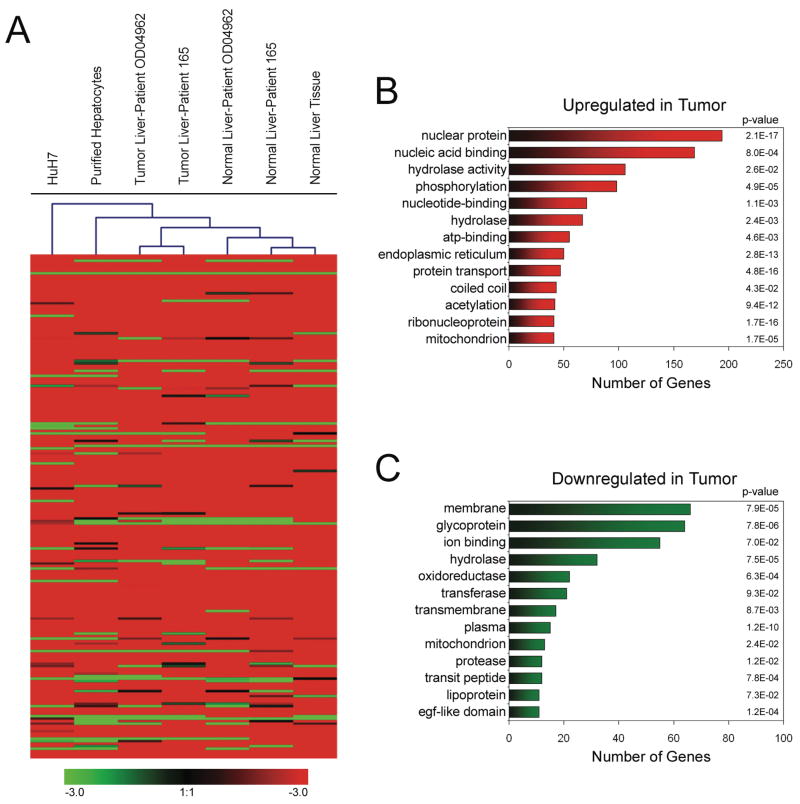
The first question we addressed was whether a section of liver tissue classified as normal, but taken from a patient having liver cancer, displayed gene expression patterns that were more closely related to the tumor sample from the same patient or to a normal sample from a person who did not have liver cancer. Using Illumina RNA expression arrays, we characterized RNA from two sets of matched normal and tumor liver samples, plus an additional sample of liver tissue from a patient who did not have liver cancer, a sample of purified normal hepatocytes, and Huh7 hepatablastoma cells (a summary of all the arrays used in this study is provided as Supplementary Table S1; a table providing all RNA expression data is provided as Supplementary Table S2). The results were then analyzed in two ways. First, a cluster diagram was prepared which groups the samples into the most closely related subsets (Figure 1A). We found that normal tissues all clustered together and the tumor tissues all clustered together. Thus, non-cancerous tissue from a patient with HCC is more closely related to other normal tissue than to tumor from the same patient. Interestingly, purified normal hepatocytes and Huh-7 hepatoma cells showed RNA profiles quite different than the tissue samples. This is most likely due to the fact that only ~80% of the cells in the liver are hepatocytes; the liver also contains other cell types, such as Kupffer cells. Thus, in contrast to the purified hepatocytes and Huh7 cells, the gene expression profiles of the tissue reflect the transcriptome of both hepatocytes and Kupffer cells. As a second method of analysis, we identified RNAs that were deregulated in the two NAT (normal liver associated with a tumor) samples as compared to liver from a patient who did not have liver cancer (Normal) and as compared to the two tumor samples from the matched patients (Tumor). The numbers of mRNAs that showed increased or decreased levels in these pairwise comparisons are shown in Supplementary Table S3; the lists of genes resulting from each comparison are provided as Supplementary Tables S4–21). We found that 2324 and 1587 mRNAs were upregulated in the tumors of patient 1 and patient 2, respectively; with 865 genes in common in the two sets. We also found that 934 and 686 mRNAs were downregulated in the tumors of patient 1 and patient 2, respectively; with 274 mRNAs in common in the two sets.
Figure 1. Analysis of the HCC transcriptome.
(A) RNA Hierarchical Clustering. RNA was prepared from the indicated samples of normal and tumor liver, from purified hepatocytes, and from Huh-7 hepatoma cells and analyzed using Illumina Sentrix Expression Beadchips. Genesis 1.7.2 software (http://genome.tugraz.at/genesisclient/genesisclient_description.shtml) was used to cluster the different samples into related subgroups. Red indicates higher expression in the tumor sample. Although all the genes were used for the cluster analysis, only the highest expressed genes are shown. (B) Gene ontology analysis of genes upregulated in HCC. (C) Gene ontology analysis of genes downregulated in HCC.
Using the program DAVID (http://david.abcc.ncifcrf.gov/), we determined the functional classification of the set of up-regulated and down-regulated genes based on their Gene Ontology descriptions. In addition to classifying genes into different sets, the DAVID program provides a P-value that indicates the probability that the set of genes was identified by chance based on the number of genes in the genome that fall into that particular category. The most highly represented categories and their P-values for the disregulated genes are shown in Figure 1B and 1C. Interestingly, we found that ribonucleoproteins involved in protein synthesis were highly upregulated in the liver tumors whereas genes involved in liver functions (such as metallothionein) were downregulated. Using more limited analyses, others have previously shown that ribosomal protein mRNAs are upregulated (19) and that metallothionein is downregulated (20) in HCC. Thus we were confident that we had identified a high confidence set of mRNAs that are deregulated in HCC.
Our next goal was to determine the mechanisms by which these mRNAs are up or down regulated in an HCC. As discussed above, there are several different mechanisms by which the expression level of mRNAs can be controlled, including a) changes in preinitiation complex formation (as monitored by levels of RNAPII bound to the promoter region), b) changes in chromatin silencing (as monitored by levels of DNA methylation or modified histones, specifically H3me3K27 and H3me3K9), and c) changes in gene copy number. We have investigated the role of these three mechanisms in development of a liver tumor using ChIP-chip technology; a summary of all the arrays used for the ChIP experiments is provided in Supplementary Table S1, the ChIP-chip data for RNAPII, H3me3K27, H3me3K9, and 5-meC is provided as Supplementary Table S22, and the lists of genes showing changes in binding of RNAPII, H3me3K27, H3me3K9, and 5-meC are provided as Supplementary Table S24–26.
Reproducibility of ChIP-chip experiments
Although ChIPs assays are now a fairly standard experimental methodology, most studies have used cell lines grown in culture for the starting material. Because we are using small amounts of frozen tumor samples, we first modified the ChIP protocol for use with small numbers of cells (15) and then adapted the MicroChIP protocol for use with tumor samples (see Materials and Methods). After amplification (see Materials and Methods), the samples were hybridized to microarrays. For all the ChIP-chip experiments performed in this study, we used the NimbleGen two array set that allowed analysis of ~5 kb regions (corresponding to ~4.2kb upstream to 0.8kb downstream of the transcription start sites) from ~25,000 promoters, with promoters being represented by ~50 probes spaced ~100 bp apart. After array normalization, the hybridization signals were divided by the total input signals to provide a fold-enrichment value for each 50-bp oligomer on the array. To identify binding sites on these promoter arrays, we used a modified Maxfour analysis program (see Materials and Methods). To demonstrate the reproducibility of our MicroChIP protocol for tissue samples, we performed two independent RNAPII and H3me3K27 ChIP-chip experiments using a matched set of normal and tumor liver samples. We found that these experiments led to highly reproducible results (Supplementary Figure S2A, S2B). In addition, we found that, in accord with expectations (21), the silencing mark of H3me3K27 was almost entirely mutually exclusive with RNAPOLII in both normal and tumor samples (Supplementary Figure 2C). Thus, we are confident that the modifications made to the ChIP assay provide high quality, reproducible ChIP-chip data.
Identification of active promoters
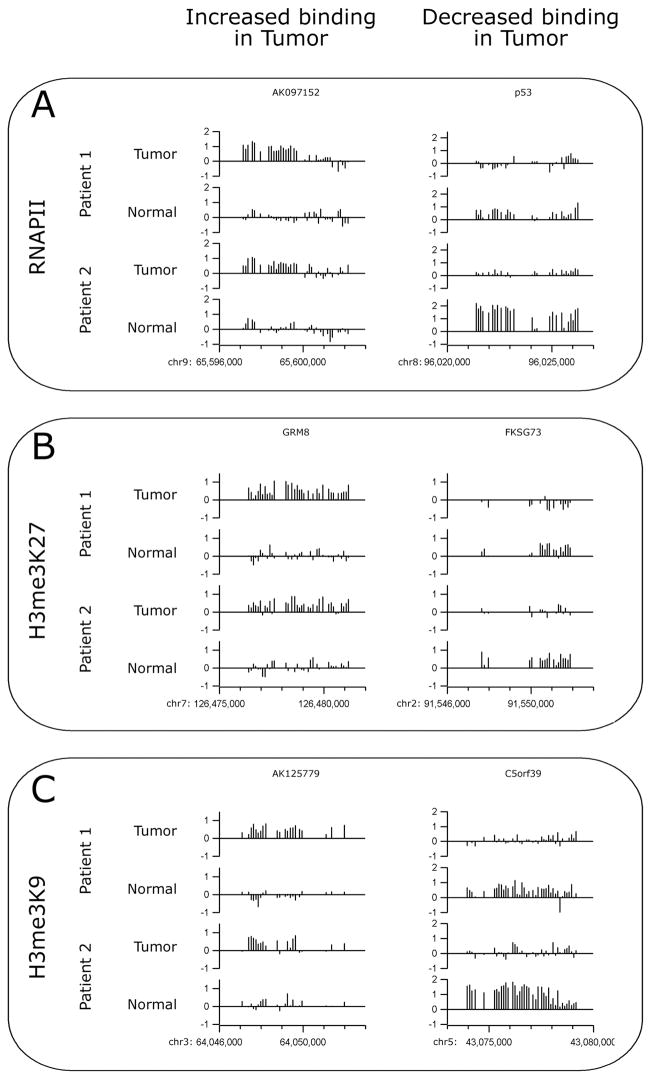
As a first step toward characterizing the mechanisms responsible for changes in gene expression in the tumor samples, we compared levels of RNAPOLII bound to the promoter regions in matched normal and tumor liver samples from two different HCC patients. Using an antibody to RNAPOLII, ChIP samples were prepared from the 4 samples using between 7 and 15 ug of chromatin (4–8 × 105 cells). Amplicons were prepared from the ChIP samples and the quality of the amplicons was assayed by analysis of positive (the promoter for the largest subunit of RNAPOLII) and negative (ZNF44 and EVX1) control regions prior to array hybridization (see Supplementary Figure S1). The amplicons were then labeled and hybridized to promoter arrays; promoters were ranked using the Maxfour analysis program (as described above) and the values for each promoter in the normal and tumor samples were compared. Comparison of repeated normal samples from the same patient revealed an excellent overall correlation. However, the slope of a linear fit could deviate significantly from 1, indicating that some arrays simply showed an overall stronger ratio signal than other arrays. We applied a simple procedure to estimate and compensate for intensity effects (see MATERIALS AND METHODS for detailed description). To generate a generous list of promoters that were increased in tumor vs normal, we set a minimum difference between the tumor and normal samples and also a minimum enhancement value (see MATERIALS AND METHODS). Examples of the hybridization data for binding of RNAPII are shown in Figure 2A. The height of the bars indicates the fold enrichment of each oligomer from a particular promoter region in two sets of matched normal and tumor (two different patients) ChIP-chip experiments; examples shown represent cases in which binding is increased (or decreased) in both sets of matched samples. A summary of the changes of RNAPII binding in the normal vs. tumor tissues is shown in Figure 3A. Using our generous parameters, we found that of the ~25,000 promoters examined, ~8% showed increased binding of RNAPII in the tumor and ~5% showed decreased binding of RNAPII in the tumor. For patient 2, ~9% showed increased binding of RNAPII and ~14% showed decreased binding of RNAPII in the tumor sample. However, only 1–2% of the promoters showed consistent increases or decreases in RNAPII binding in both tumor samples (as compared to their matched normal samples).
Figure 2. Examples of normal and tumor ChIP-chip hybridization patterns.
Examples of the hybrdization patterns for promoters that show increased or decreased binding of RNAPII (A), H3me3K27 (B), or H3me3K9 (C) are shown for the normal and tumor samples for both patients. Each vertical bar represents the enrichment of a single probe as a log2 ratio value (y axes) between the enriched ChIP sample and the Input sample.
Figure 3. Analysis of active vs. silenced promoters.
(A) Shown are the numbers of promoters that show changes in binding of RNAPII, H3me3K7, or H3me3K9 in the normal vs tumor samples. (B). Shown are the gene ontology categories that show coordinate changes in RNAPII and H3me3K27 or H3me3K9 binding.
Identification of silenced promoters
Changes in gene expression can also occur due to changes in chromatin structure. Regions of the genome bound by histone H3 trimethylated on lysine 9 or lysine 27 have been termed silenced chromatin, and represent regions of the genome whose compacted structure inhibits binding of site specific transcription factors and the general transcriptional machinery. To determine if changes in gene expression in the liver tumors was a consequence of changes in gene silencing, we performed ChIP assays using antibodies that specifically recognize H3me3K27 or H3me3K9. The ChIP samples were amplified and the quality of the amplicons was assayed by analysis of positive (EVX1 for H3me3K27 and ZNF44 for H3me3K9) and negative (the largest subunit of RNAPOLII) control regions (Supplementary Figure S1). The amplicons were then labeled and hybridized to promoter arrays; promoters were ranked using the modified Maxfour analysis program and the values for each promoter in the normal and tumor samples were compared as described above. Examples of the hybridization data for binding of H3me3K27 and H3me3K9 are shown in Figure 2B and 2C and a summary of the changes is shown in Figure 3A. We found that of the ~25,000 promoters examined, ~13% showed increased binding of H3me3K27 in the tumor and ~13% showed decreased binding of H3me3K27 in the tumor. For patient 2, ~6% showed increased binding of H3me3K27 and ~6% showed decreased binding of H3me3K27 in the tumor sample. However, again only ~1% of the promoters showed consistent increases or decreases in H3me3K27 binding in both tumor samples (as compared to their matched normal samples). We found even fewer changes in H3me3K9 binding. Of the ~25,000 promoters examined, ~8% showed increased binding of H3me3K9 in the tumor and ~10% showed decreased binding of H3me3K9 in the tumor. For a second patient, ~5% showed increased binding of H3me3K9 and ~4% showed decreased binding of H3me3K9 in the tumor sample. However, less than ~1% of the promoters showed consistent increases or decreases in H3me3K9 binding in both tumor samples (as compared to their matched normal samples).
Although very few promoters showed consistent changes in binding of RNAPII, H3me3K27, or H3me3K9 in both of the patients, we could identify a small set of promoters that showed increased RNAPII and decreased H3me3K27 or H3me3K9 or decreased RNAPII and increased H3me3K27 or H3me3K9 in both of the patients. Interestingly, the sets of active genes that were silenced in the tumors (i.e. they showed a decrease in RNAPII binding and an increase in H3me3K27 and/or H3me3K9 binding) and the sets of silenced genes that were reactivated in the tumors (i.e. they showed an increase in RNAPII binding and a decrease in H3me3K27 and/or H3me3K9 binding) were highly enriched in specific gene ontology categories. Specifically, genes in the glycoprotein and transmembrane categories were silenced in the liver tumors whereas genes in the categories of nuclear proteins, zinc fingers, transport, and coiled coil domains were reactivated in the tumors (Figure 3B). A detailed ontology analysis of the genes regulated by consistent changes in binding of RNAPII, H3me3K27 and H3me3K9 in the tumor tissues is shown in Supplementary Figure S3.
Analysis of copy number changes
Copy number variation is a hallmark of many cancers. Information on the prevalence and exact locations of these changes is of wide interest and may prove to be useful for differential diagnosis and, more fundamentally, may give insight into the differing patterns of various cancers. One widely used approach to assessing these changes is comparative genomic hybridization (CGH). Recently, high density tiled arrays have been used with CGH to discover quite small changes in genomes (22). However, a high resolution CGH analysis of the entire human genome would be an expensive undertaking, requiring 38 arrays (at a density of 380,000 probes/array) to 10 arrays (at a density of 2.1 million probes/array). In our ChIP-chip promoter array experiments, we apply both precipitated experimental sample and input genomic DNA to the arrays. Hence, we have genome-scale array data for genomic DNA from normal and tumor tissue. We reasoned that this genomic DNA data should reveal copy number changes in the tumor samples. In Supplementary Figure S4, we demonstrate how use of the total genomic DNA channels from promoter ChIP-chip studies can be used to perform an in silico CGH experiment, an approach we term “virtual CGH” (vCGH). Briefly, we extract the hybridization signals from the Input channel of the normal and the tumor ChIP-chip experiments. The data from the two normal (or two tumor) ChIP-chip experiments is quantile normalized and merged to produce a median value. We then divide the tumor signals by the normal signals (so that amplified regions give values greater than 0 and deleted regions give values less than 0), and then plot the resultant values using a windowing function; further details are provided in Materials and Methods.
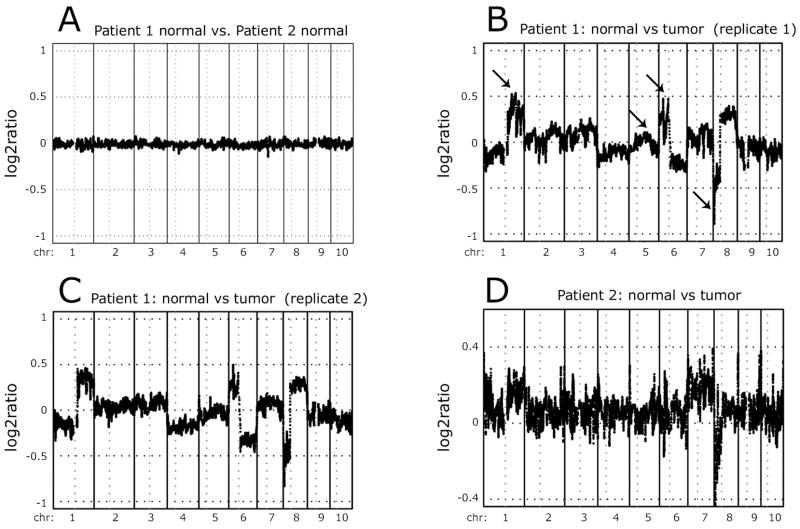
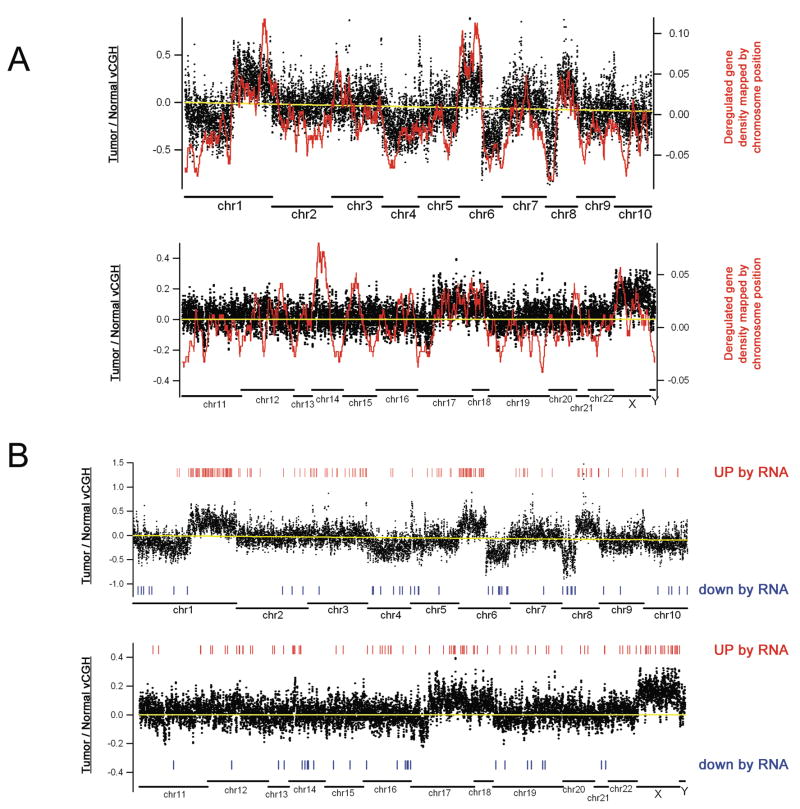
To demonstrate the reproducibility of our vCGH analysis method, we performed two control experiments. First, we used our vCGH procedure on the two normal tissue samples from the cancer patients; this analysis led to essentially no differences (Figure 4A). Second, we used the data from the two separate sets of ChIP-chip experiments performed on Patient 1 (the ChIP experiments shown in Supplementary Figure S2). Using the first set of Input hybridization data from promoter array 1 (representing chromosomes 1–10), comparison of the normal vs. tumor tissue identified a complex pattern of copy number differences. For example, we identified increased copies of promoters located at 1q, 6p, and 8q (Figure 4B) and throughout the entire X chromosome (see below). We also observed loss of promoters at 6q and 8p (Figure 4B). Importantly, a vCGH analysis using ChIP-chip data from a completely separate set of experiments for the same patient showed identical patterns (Figure 4C). For a second patient (patient 2), the differences were weaker (perhaps due to contaminating normal tissue in the tumor sample), but the general pattern was observed (Figure 4D). We confirmed the loss or gain of several of the identified regions using PCR analysis of DNA prepared from normal and tumor samples (Supplementary Figure S5). Others have seen similar gains and losses of these chromosomal regions in HCC (23, 24). For example, we observe gains of 1q, 6p, and 8q; these same regions have been reported as copy number gains in 59%, 24%, and 38% of HCC. Similarly, we observe losses of 8p and 17p; these same regions have been previously reported to be lost in 41% and 49% of HCC.
Figure 4. Analysis of copy number changes in HCC samples.
vCGH analysis was performed comparing the normal DNA from both patients (A), the tumor vs. normal DNA from two separate ChIP-chip experiments for patient 1 (B, C), and the tumor vs. normal DNA for patient 2 (D). The y axes indicate the log2 ratio of the tumor to normal signals. Values above 0 represent copy number gains in the tumor and values below 0 represent copy number losses in the tumor. In panel B, the arrows indicate several of the amplified (chromosome 1q and 6p), deleted (chromosome 8p), or control (chromosome 5q) regions analyzed in Figure S6.
In addition, our vCGH method can be applied to cell lines. In particular, vCGH can be used as a test of the “true identity” of a cell line. There has been an increasing realization that many cell lines are misidentified (25). A major concern is that many cell lines have been overtaken by HeLa cells, a hardy and fast-growing cell line that has been in culture for over 50 years. The contamination of other cell lines with HeLa cells does not necessarily occur in the investigators lab but rather the cell line may arrive from other sources already misidentified. Using Input DNA tracks from ChIP-chip experiments, we compared HeLa to human ES cells. Our vCGH analysis of the HeLa cells showed a clear pattern of copy number changes (see Supplementary Figure S6). We note that the well characterized amplification of 5p in HeLa cells (26) was clearly seen in our vCGH analysis data. Having established the vCGH pattern of HeLa, we then used total DNA signals from ChIP-chip data from Ntera2 testicular embryonal carcinoma cells. The Ntera2 cells showed a distinct characteristic pattern that reproduced copy number changes that are often associated with testicular germ cell tumors (27), indicating that the tumor cell lines can be distinguished from HeLa cells using vCGH.
Mechanisms responsible for the tumor transcriptome
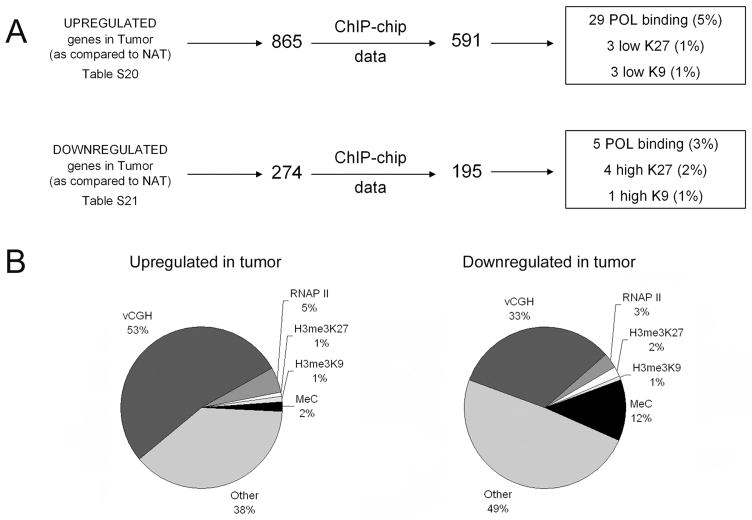
Having analyzed the sets of active and silenced promoters in normal and tumor cells and identified the promoters that show copy number changes in the tumor cells, we could return to the question posed earlier; i.e. what mechanisms are responsible for mediating the changes in levels of RNAs in the HCC samples. Because the RNA and ChIP-chip data were performed using different platforms, it was first necessary to determine which of the deregulated RNAs had promoters represented on the NimbleGen arrays. We found that the promoters of 591 of the 865 mRNAs that were commonly upregulated in both tumors and 195 of the 274 RNAs that were commonly downregulated in both tumors were present on the NimbleGen promoter arrays (Figure 5A). Interestingly, increased binding of RNAPII occurred at only 5% and decreased levels of silenced chromatin (as monitored by decreases in either H3me3K27 or H3me3K9) occurred at only 2% in the genes upregulated in the tumors. Similarly, decreased RNAPII binding occurred at only 3% and increased silencing occurred at only 3% of the genes downregulated in tumors (Figure 5B). In contrast, 53% of the promoters of the upregulated mRNAs and 33% of the promoters for the downregulated mRNAs displayed changes in copy number. Although copy number changes did appear to be responsible for a large percentage of the observed changes in gene expression, we noted that 40% and 61% of the up and down regulated genes, respectively, were not regulated by any of the tested mechanisms. Another mechanism which has been previously associated with changes in gene expression in human tumors is DNA methylation. To determine if DNA methylation was responsible for regulation of a subset of the tumor transcriptome, we performed ChIP experiments using an antibody that recognizes 5-methyl cytosine to precipitate methylated DNA from normal and tumor tissue of patient 1 (see Supplementary Table S22 for the ChIP-chip results monitoring DNA methylation in the normal and tumor samples). We found that 2% of the increased RNAs and 12% of the decreased RNAs were regulated by changes in DNA methylation (Figure 5B and Supplementary Table S26). Thus, DNA methylation was responsible for the down regulation of a significant number of genes in the tumor sample. However, changes in copy number remained the most predominant of the tested mechanisms for regulation of the tumor transcriptome (Figure 5B). In fact, mapping of the location of the deregulated mRNAs from patient 1 onto the vCGH pattern of this same patient clearly demonstrates the correlation of increased mRNA levels with increased copy number and of decreased mRNA levels with loss of genomic regions (Figure 6A). There are several regions that show a high density of upregulated RNAs in regions that do not show increases in gene copy number. These RNAs must have been increased by other mechanisms. For example, the first half of chromosome 14 shows a non-copy number-related increase in upregulated RNAs. In this region, there are 22 promoters that show an increase in RNAPII binding in the tumor sample (see Supplementary Table S23 for a list of the promoters that show changes in binding of RNAPII in the tumor samples). We also mapped the location of the mRNAs commonly deregulated in both tumor RNA samples onto the vCGH pattern of patient 1 (Figure 6B). Clearly, many of the commonly up- and down-regulated mRNAs map to regions having copy number changes.
Figure 5. Overview of the mechanisms regulating the HCC transcriptome.
(A) RNA and ChIP-chip platform comparisons. The RNAs which were commonly deregulated in the HCC samples were mapped to the NimbleGen arrays and then the number of each of these deregulated mRNAs that show changes in binding of RNAPolII, H3me3K27, or H3me3K9 was determined. (B) A pie chart demonstrating the percentages of deregulated mRNAs resulting from copy number changes (vCGH), changes in binding of RNAP II, H3me3K27, or H3me3K9, or changes in DNA methylation is shown for the mRNAs up and downregulated in liver tumors.
Figure 6. Mapping deregulated RNAs onto the vCGH pattern.
(A) The mRNAs that are upregulated or downregulated in patient 1 are shown as a density plot in relation to the vCGH pattern from the same patient. Deregulated mRNA density was calculated based on chromosomal location by a sliding-window average algorithm. Positive values on the right Y axis represent up-regulated genes and negative values represent down-regulated genes. (B) The mRNAs that showed increased or decreased copy number in both patient 1 and patient 2 are mapped regulative to the vCGH pattern of patient 1.
DISCUSSION
The goal of our study was to determine the extent to which several commonly used mechanisms of transcription regulation contribute to gene expression changes that occur during the development of a human liver tumor. Accordingly, we began our studies by identifying a set of transcripts that were disregulated in both of two independent liver tumors. We then examined changes in transcription initiation complex formation, gene silencing, and gene copy number for these deregulated genes. We found that, of these mechanisms, amplification and deletion of chromosomal regions was most often used to confer changes in mRNA expression in the liver tumor.
Although our study was not meant to be a comprehensive analysis of RNA changes in HCC, it is critical to note that we did identify sets of deregulated genes that a) fit into several interesting gene ontology categories and b) correspond to genes identified in previous studies of tumors. For example, we found that the most commonly downregulated functional categories of genes were those from the metallotheionein (e.g MT1A, MT1F, MT1G, MT1M, MT1X and MT2A) and cytochrome P45 (e.g CYP1A2, CYP2C9, CYP21 CYP3A5, and CYP3A7) families. This suggests that a dedifferentiation of the liver has occurred in the tumor cells. Similarly, Xu et al (3) performed an analysis of ESTs from Hepatitis B positive HCC samples and found that genes involved in liver function were downregulated. More specifically, Tao et al (20) performed a direct analysis of the metallotheionein family in HCC and found that many metallotheionein family members were downregulated and suggested that downregulated expression of this family may be a marker for hepatocarcinogenesis. We found that one of the most commonly upregulated categories of genes included ribonucleoproteins involved in protein synthesis and transport. Similarly, Kondoh et al (19) used differential display to identify 5 cDNAs that were upregulated in HCC; all of these corresponded to ribosomal protein mRNAs. Interestingly, they also demonstrated that the expression of these ribosomal protein mRNAs was high in three HCC cells lines irrespective of the growth state of the cell and suggested that activation of these genes is an important manifestation of the HCC phenotype. The other main category of genes that we discovered to be upregulated in the HCC samples was nuclear proteins (see Supplementary Figure S7 for categorized list of the nuclear proteins). Of the 194 nuclear proteins that were upregulated in the HCC samples, 25 are involved in chromosomal biology and biogenesis, 12 are involved in nucleocytoplasmic transport, 24 are involved in DNA replication and repair, 32 are involved in RNA metabolism, and 67 are transcriptional regulators. Of the 67 transcriptional regulators, 38 (> 50%) fit into the zinc ion binding/zinc finger category (Supplementary Figure S7). Several of the upregulated transcription factors have been previously associated with cancers. For example, FoxQ1 has been shown to be overexpressed in pancreatic cancer (28); Sox18 is upregulated in gastric, pancreatic, breast, and embryonal tumor cell lines (29), and DP1 has been previously shown to be increased in expression in HCCs (30, 31).
As noted above, gankyrin mRNA is overexpressed in most HCC samples (9, 10); we also observed increased gankyrin mRNA in both of the HCC samples we analyzed. Detailed analysis of the gankryin gene revealed that it was not increased in the tumor samples due to changes in RNAPII, H3me3K27, or H3me3K9 binding. Rather, the gankryin gene is amplified. In fact, the amplification ratio of the gankyrin gene was in the top 3% of all amplified promoters. Overexpression of gankyrin can lead to degradation of p53 and Rb, contributing to genomic instability and oncogenic transformation. Rb functions to suppress the E2F family, a key regulator of cell proliferation. Interestingly, we also observed an increase in levels of DP1, the obligate heterodimeric partner of E2F1-E2F6, in the tumor samples. Expression analysis indicates that E2F3 and E2F4 are expressed at similar levels in the normal and tumor tissues, whereas the other E2Fs are not expressed in any of the liver samples. The increased expression of DP1 and decreased levels of Rb protein (due to gankyrin) may synergize to allow enhanced E2F3- or E2F4-mediated activation of key proliferation-responsive genes in the liver tumors.
In summary, we found that changes in preinitiation complex formation and changes in gene silencing do not play major roles in the development of the tested liver tumors. Rather, many gene expression changes in the tumors were a direct result of changes in gene copy number. We also note that 38% and 49% of the up and down regulated genes, respectively, were not regulated by any of the mechanisms. Recent estimates suggest that ~50% of human genes have alternative promoters (32). The set of alternative promoters for human genes is not well characterized and thus these promoters are not well represented on the arrays. Therefore, it is possible that some changes in RNA levels in the liver tumors were due to differential usage of novel alternative promoters. It is also possible that changes in transcription elongation and/or transcript stability contribute to the regulation of the set of genes in the “other” category.
Although our analysis was limited to a small number of liver tumors, our results suggest that clinical treatments with chromatin modifying drugs, which have shown promise in the treatment of cancers of hematologic origin, may not be effective for HCC (see (12) for a review of the clinical applications of epigenetic drugs). The mechanisms regulating the tumor transcriptome may differ for different tumor types and therefore RNA levels, chromatin structure, and gene copy number should all be analyzed to provide the required information to allow a rationale design of tumor type-specific chemotherapeutic regimens. Finally, we have outlined an efficient and cost-effective method to analyze copy number changes at all the known promoters regions in the human genome; this analysis can be performed at no extra cost using the data from ChIP-chip experiments.
Supplementary Material
Acknowledgments
We thank the members of the Farnham lab for helpful discussion. The RNA was labeled and the Illumina beadchips were processed and scanned at the UC Davis Genome Center Expression Analysis Core Facility.
Financial support: This work was supported in part by Public Health Service grants CA45250 and DK067889 to PJF; MB was also supported in part by NSF grant 0225674.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and Molecular Carcinogenesis. Reviews in Basic and Clinical Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–39. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L, Hui L, Wang S, et al. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–81. [PubMed] [Google Scholar]
- 4.Graveel CR, Harkins-Perry SR, Acevedo LG, Farnham PJ. Identification and characterization of CRG-L2, a new marker for liver tumor development. Oncogene. 2003;22:1730–6. doi: 10.1038/sj.onc.1206309. [DOI] [PubMed] [Google Scholar]
- 5.Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ. Expression profiling and identification of novel genes in hapatocellular carcinomas. Oncogene. 2001;20:2704–12. doi: 10.1038/sj.onc.1204391. [DOI] [PubMed] [Google Scholar]
- 6.Lee J-S, Thorgeirsson SS. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134–43. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
- 7.Delpuech O, Trabut J-B, Carnot F, Feuillard J, Brechot C, Kremsdorf D. Identification, using cDNA macroarray analysis, of distinct gene expression profiles associated with pathological and virological features of hepatocellular carcinoma. Oncogene. 2002;21:2926–37. doi: 10.1038/sj.onc.1205392. [DOI] [PubMed] [Google Scholar]
- 8.Su W-H, Chao C-C, Yeh S-H, Chen D-S, Chen P-J, Jou Y-S. OncoDB. HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2006;35:D727–D31. doi: 10.1093/nar/gkl845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higashitsuji H, Higashitsuji H, Itoh K, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Higashitsuji H, Itoh K, Nagao T, et al. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–9. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Fu X, Chen Y, et al. Use of adenovirus-delivered siRNA to target oncoprotein p28GANK in hepatocellular carcinoma. Gastroenterology. 2005;128:2029–41. doi: 10.1053/j.gastro.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Sigalotti L, Fratta E, Coral S, et al. Epigenetic drugs as pleiotropic agents in cancer treatment: biomolecular aspects and clinical applications. J Cell Physiol. 2007;212:330–44. doi: 10.1002/jcp.21066. [DOI] [PubMed] [Google Scholar]
- 13.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 14.Weinmann AS, Bartley SM, Zhang MQ, Zhang T, Farnham PJ. The use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–32. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acevedo LG, Iniguez AL, Holster HL, Zhang X, Green R, Farnham PJ. Genome-scale ChIP-chip analysis using 10,000 human cells. Biotechniques. 2007;43:791–7. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–80. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krig SR, Jin VX, Bieda MC, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–12. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis GJ, Sherman BT, Hosack Da, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 19.Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Enhanced expression of S8, L12, L23a, L27, and L30 ribosomal protein mRNAs in human hepatocellular carcinomas. Anticancer Res. 2001;21:2429–33. [PubMed] [Google Scholar]
- 20.Tao X, Zheng JM, Chen XF, Zhang SH. Downregulated expression of metallothionein and its clinicopathological significance in hepatocellular carcinoma. Hepatol Res. 2007;37:820–7. doi: 10.1111/j.1872-034X.2007.00113.x. [DOI] [PubMed] [Google Scholar]
- 21.Squazzo SL, Komashko VM, O’Geen H, et al. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Research. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban AE, Korbel JO, Selzer R, et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci U S A. 2006;103:4534–9. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang J, Kim N-G, Piao Z, et al. Assessment of chromosomal losses and gains in hepatocellular carcinoma. Cancer Letters. 2002;182:193–202. doi: 10.1016/s0304-3835(02)00083-6. [DOI] [PubMed] [Google Scholar]
- 24.Patil MA, Gutgemann I, Zhang J, et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26:2050–7. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee R. Cases of Mistaken Identity. Science. 2007;315:928–31. doi: 10.1126/science.315.5814.928. [DOI] [PubMed] [Google Scholar]
- 26.Macville M, Schrock E, Padilla-Nash H, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Research. 1999;59:141–50. [PubMed] [Google Scholar]
- 27.Oosterhuis JW, Looijenga LHJ. Testicular germ-cell tumours in a broader perspective. Nature Reviews-Cancer. 2005;5:210–22. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 28.Cao D, Hustinx SR, Sui G, et al. Identification of novel highly expressed genes in pancreatic ductal adenocarcinomas through a bioinformatics analysis of expressed sequence tags. Cancer Biol Ther. 2004;3:1081–9. doi: 10.4161/cbt.3.11.1175. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh T, Katoh M. Expression of human SOX18 in normal tissues and tumors. Int J Mol Med. 2002;10:339–44. [PubMed] [Google Scholar]
- 30.Yasui K, Arii S, Zhao C, et al. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 113q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–84. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 31.Yasui K, Okamoto H, Arii S, Inazawa J. Association of over-expressed TFDP1 with progression of hepatocellular carcinomas. J Hum Genet. 2003;48:609–13. doi: 10.1007/s10038-003-0086-3. [DOI] [PubMed] [Google Scholar]
- 32.Kimura K, Wakamatsu A, Suzuki Y, et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.