Abstract
Cysteine-rich secretory proteins (CRISPs) are widely distributed, and notably occur in the mammalian reproductive tract and in the salivary glands of venomous reptiles. Most CRISPs can inhibit ion channels, such as the cyclic nucleotide-gated ion channel, potassium channel, and calcium channel. Natrin is a CRISP that has been purified from snake venom. Its targets include the calcium-activated potassium channel, the voltage-gated potassium channel, and the calcium release channel/ryanodine receptor (RyR). Immunoprecipitation experiments showed that natrin binds specifically to type 1 RyR (RyR1) from skeletal muscle. Natrin was found to inhibit both the binding of ryanodine to RyR1, and the calcium-channel activity of RyR1. Cryo-electron microscopy and single-particle image reconstruction analysis revealed that natrin binds to the clamp domains of RyR1. Docking of the crystal structure of natrin into our cryo-electron microscopy density map of the RyR1 + natrin complex suggests that natrin inhibits RyR1 by stabilizing a domain-domain interaction, and that the cysteine-rich domain of natrin is crucial for binding. These findings help reveal how natrin toxin inhibits the RyR calcium release channel, and they allow us to posit a generalized mechanism that governs the interaction between CRISPs and ion channels.
INTRODUCTION
Cysteine-rich secretory proteins (CRISPs) are single-chain polypeptides that are characterized by the presence of 16 strictly conserved cysteine residues. Ten of these 16 cysteine residues are clustered in the carboxyl-terminal region. The CRISPs are widely distributed, and notably occur in the mammalian reproductive tract and in the salivary glands of venomous reptiles. The CRISPs are categorized into four subclasses, according to sequence homology and tissue specificity. CRISP-1 was first identified in mammalian epididymis, where it associates with the sperm surface during epididymal maturation and mediates sperm-egg fusion, and it is also known as acidic epididymal glycoprotein (1,2). CRISP-2, also known as testis-specific protein-1 (Tpx-1), was found in testes and spermatozoa, and is thought to mediate the interaction between spermatogenic cells and Sertoli cells (3,4). CRISP-3 was detected in various cell types, including salivary gland, pancreas, prostate, and B-cells. A large number of CRISP-3 family members are widely distributed in reptile venoms (5,6). CRISP-4 proteins are only expressed in the epididymis, from which they are secreted into the epididymal lumen and are able to interact with sperm (7).
Ryanodine receptors (RyRs) are the major calcium-release channels in striated muscle (8). They are highly enriched in both cardiac and skeletal muscles, or more precisely, in sarcoplasmic reticulum (SR) membrane. The SR is the major storage site of cellular calcium. Ryanodine receptors play a central role in excitation-contraction coupling, a process in which neuron-induced depolarization of the plasma membrane causes RyR molecules to release Ca2+ from the SR. The resulting increase in cytoplasmic [Ca2+] activates myofilaments so as to generate muscle contraction. To date, two CRISPs were shown to affect RyR channel functions. Helothermine, a well-known CRISP-3 protein that was isolated from the Mexican beaded lizard, can block a variety of ion channels, including voltage-gated Ca2+ channels, voltage-gated K+ channels, and RyRs (9–11). Helothermine inhibits both skeletal-muscle RyR1 and cardiac-muscle RyR2 (9). The carboxyl-terminal cysteine-rich domain (CRD) of Tpx-1 was demonstrated to inhibit RyR2, although it activates RyR1 (12).
In addition to helothermine, many CRISP-3 proteins were identified in venoms from a range of snakes, and were shown to inhibit specific K+ or Ca2+ ion channel activities (13–16). Recently, the structures were determined and the functions were explored for several CRISP-3 proteins, i.e., natrin (17,18), stecrisp (19), and triflin (20). Natrin is a 221-amino-acid toxin protein (molecular mass 25 kDa) purified from the venom of the snake Naja naja atra. Targets of this toxin include the calcium-activated potassium channel BKCa (17) and the voltage-gated potassium channel Kv1.3 (18). Here, we show that natrin binds specifically to the skeletal-muscle RyR1, and inhibits RyR1 calcium-channel activity. To gain insights into the structural basis of channel inhibition by natrin, we determined the binding site of natrin in the three-dimensional structure of RyR1, using cryo-electron microscopy (cryo-EM) and single-particle image processing. Docking of the x-ray crystal structure of natrin into the cryo-EM density map of the RyR1 + natrin complex indicated that the CRD and linker domain of natrin is essential for binding, and that the natrin molecule binds across two structural domains of RyR1 that are important for channel regulation.
MATERIALS AND METHODS
Purification of natrin
Natrin was purified from snake Naja naja atra, according to the published procedure (18).
Preparation of skeletal SR vesicles
Skeletal SR vesicles were prepared from New Zealand White rabbit skeletal muscle according to the published method, with modifications (21). Briefly, skeletal muscle was homogenized in 5 volumes (v/w) of homogenization buffer (0.3 M sucrose, 10 mM Na-HEPES, pH 7.0, 0.5 mM EDTA, 0.2 mM phenylmethylsulphonyl fluoride, and 1:1000 diluted protease inhibitory cocktail), and centrifuged at 11,500 × g for 20 min. The supernatant was then centrifuged at 110,000 × g for 1 h. The pellet from the last centrifugation was collected and re-suspended in buffer (0.3 M sucrose, 10 mM Na-HEPES, pH 7.0, 0.2 mM phenylmethylsulphonyl fluoride, and 1:1000 diluted protease inhibitor cocktail), aliquoted, quick-frozen in liquid nitrogen, and stored at −80°C. The protein concentration was determined by bicinchoninic-acid assay.
Purification of RyR1
The RyR1 was purified from 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) solubilized skeletal SR, as previously described (22,23).
Co-immunoprecipitation of natrin and RyR1
Skeletal SR (1 mg/mL) and natrin (50 μg/mL) were incubated in incubation buffer (25 mM NaCl, 20 mM Na-HEPES, pH 7.0) without Ca2+, or containing 10−7 M Ca2+, 10−5 M Ca2+, and 10−3 M Ca2+, respectively, for 2 h at 4°C, and then solubilized by CHAPS/soybean phospholipid, with a CHAPS/soybean phospholipids/protein weight ratio of 13:6.5:1. After incubation on ice for 30 min, samples were centrifuged at 110,000 × g for 30 min at 4°C. The supernatants were collected and divided into two aliquots. One microliter of monoclonal anti-RyR1 antibody 34C (Sigma-Aldrich, St. Louis, MO) and 5 μL anti-natrin serum (rabbit serum raised against natrin protein) were added to each aliquot. After incubation on ice for another 2 h, 20 μL Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added, and the samples were incubated overnight at 4°C. The beads were collected by centrifugation, and washed with incubation buffer. Proteins were resolved by electrophoresis in 3-15% linear gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis gels, transferred to polyvinylidene difluoride transfer membrane, and detected by peroxidase-coupled antibodies.
[3H]-ryanodine binding assay
Skeletal SR vesicles (250 μg/mL) or purified RyR1 (25 μg/mL) were incubated with 2 nM [3H]ryanodine in 0.2 M KCl, 20 mM Na-HEPES, pH 7.0, and 10 μM Ca2+, in a final volume of 100 μL, containing various concentrations of natrin and ryanodine (for a Lineweaver-Burk plot). After incubation overnight at 24°C, the binding reaction was quenched by rapid filtration through Whatman (Florham Park, NJ) GF/B glass fiber filters presoaked with 1% polyethyleneimine. The filters were then rinsed twice with 10 mL of ice-cold washing buffer containing 0.2 M KCl and 20 mM Na-HEPES, pH 7.0. The filters were air-dried and placed into 20-mL scintillation vials (Kimble, Vineland, NJ), and soaked in 5 mL scintillation solution (2.5 g (2,5-diphenyloxazole) and 150 mg (1,4-bis(5-phenyloxazol-2-yl) benzene) in 500 mL dimethyl benzene). The filters were counted after incubation overnight. Nonspecific binding was measured in the presence of a 1000-fold excess of unlabeled ryanodine. The experiments were repeated at least twice, and on two different skeletal SR vesicles and on purified RyR1 preparations.
Calcium efflux from skeletal SR vesicles
Calcium efflux measurements were carried out according to the published procedure (24). Briefly, skeletal SR vesicles (100 μg/mL) were added to a 2-mL solution containing 100 mM KH2PO4, pH 7.0, 4 mM MgCl2, 1 mM Na2ATP, and 0.5 mM antipyrylazo III. The temperature was maintained at 25°C. Extravesicular [Ca2+] was monitored at 710 nm, using a Hitachi U2010 spectrophotometer (Hitachi, Tokyo, Japan). Vesicles were loaded with Ca2+ by the addition of four aliquots of CaCl2, each initially increasing the extravesicular Ca2+ concentration by 7.5 μM over the previous preceding concentration. Sufficient time (2–5 min) was allowed between one addition of Ca2+ and the next, so that the uptake reduced the Ca2+ concentration to baseline level. Then 200 nM thapsigargin in H2O, 200 nM thapsigargin in buffer (30 mM KCl, 25 mM Na-HEPES, pH 7.0), or 200 nM thapsigargin with 0.5 μM natrin in buffer (30 mM KCl, 25 mM Na-HEPES, pH 7.0) were added to suppress Ca2+-ATPase activity and to allow extravesicular [Ca2+] to increase. The calcium efflux was measured for another 5 min, and ruthenium red (5 μM) was added to confirm that Ca2+ release was occurring through RyR1 channels. Finally, the Ca2+ ionophore A23187 (3 μg/mL) was added, to release all Ca2+ remaining in the vesicles.
Cryo-EM and image processing
Cryo-electron microscopy was carried out as described previously (25). Images were processed using the SPIDER/WEB software package (26). Three-dimensional reconstructions of RyR1 and the RyR1 + natrin complex were computed according to the projection matching method (25). The final three-dimensional reconstructions of the RyR1 control and the RyR1 + natrin complex were computed from 16,519 and 7959 particles, respectively. The resolution for both reconstructions was 21 Å, according to the Fourier shell correlation criterion, with a cutoff of 0.5 (27). An x-ray crystal structure of natrin (Protein Data Bank code 2GIZ) was docked into the cryo-EM density of the three-dimensional difference map manually, using the molecular graphics programs O (http://xray.bmc.uu.se/∼alwyn/index.html) and Chimera (http://www.cgl.ucsf.edu/chimera/). Various possible fitting orientations were evaluated by the correlation coefficient using the SPIDER program, and the best-fitting orientation was chosen using the highest correlation coefficient (28).
RESULTS
Previous studies showed that natrin toxin can block the calcium-activated potassium channel BKCa (17) and the voltage-gated potassium channel Kv1.3 (18). In this study, we address the following questions: Does natrin bind specifically to RyR1? If so, what is the functional effect of natrin-binding on the RyR1 calcium release channel? Where is the natrin-binding site in the three-dimensional structure of RyR1? Can the structural information obtained from the binding-site determination be used to explain any functional effect of natrin on RyR1?
Natrin binds specifically to RyR1
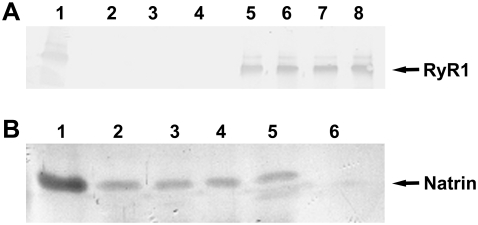
The specific binding between RyR1 and natrin toxin was tested in co-immunoprecipitation experiments. As shown in Fig. 1 A, RyR1s were precipitated by rabbit serum raised against natrin, and in Fig. 1 B, natrin proteins were precipitated by anti-RyR1 antibody. These co-immunoprecipitation experiments clearly demonstrated that natrin toxin binds specifically to RyR1.
FIGURE 1.
Co-immunoprecipitation of natrin and RyR1. (A) RyR1 precipitation by anti-natrin antiserum. The RyR1 bands are indicated. Lane 1, skeletal SR; lane 2, anti-natrin anti-serum; lane 3, natrin; lane 4, buffer (control); lanes 5–8, SR incubated with natrin at 0 M Ca2+, 10−7 M Ca2+, 10−5 M Ca2+, and 10−3 M Ca2+, respectively. (B) Natrin precipitation by anti-RyR1 antibody. Natrin bands are indicated. Lane 1, natrin; lanes 2–5, SR incubated with natrin at 0 M Ca2+, 10−7 M Ca2+, 10−5 M Ca2+, and 10−3 M Ca2+, respectively; lane 6, control buffer.
Natrin toxin reduces [3H]ryanodine binding to RyR1
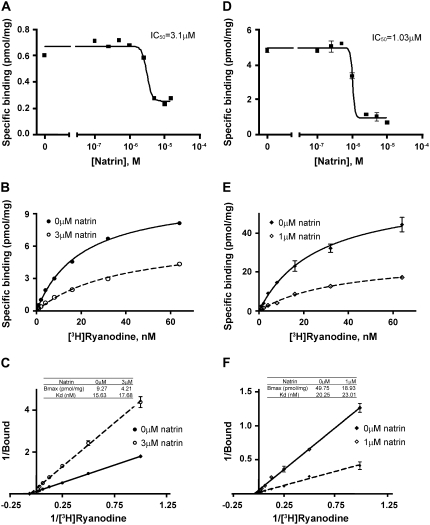
After the co-immunoprecipitation experiments, we examined the functional effects of natrin on the RyR1 calcium- release channel, by carrying out [3H]ryanodine binding assays. Ryanodine binding reflects the open probability of the calcium channel, because ryanodine is known to bind specifically to RyR in its open conformation (29). As shown in Fig. 2, A and D, natrin toxin produced a strong inhibition of [3H]ryanodine binding, both to skeletal muscle SR vesicles and to purified RyR1 molecules. The half-maximal inhibitory concentration of natrin was found to be 3.1 μM for SR vesicles, and 1.03 μM for purified RyR1.
FIGURE 2.
Natrin toxin reduces [3H]ryanodine binding to RyR1. Concentration dependence for blockage of [3H]ryanodine binding to skeletal SR vesicles (A) and to purified RyR1 (D). The SR vesicles (0.25 mg/mL) or purified RyR1 (25 μg/mL) were incubated (see Materials and Methods) with various concentrations of natrin (0–10 μM) in binding buffer containing 10 μM [Ca2+] and 2 nM [3H]ryanodine. We also preformed equilibrium saturation assays of [3H]ryanodine binding to skeletal SR vesicles (B and C) and to purified RyR1 (E and F). Experiments were performed in binding buffer containing 10 μM [Ca2+], 2 nM [3H]ryanodine, and various concentrations of unlabeled ryanodine (1–64 nM). (B) Saturation curve of [3H]ryanodine binding to skeletal SR vesicles in the absence or presence of 3 μM natrin. (C) Data-fitting according to Lineweaver-Burk equation. (E) Saturation curve of [3H]ryanodine binding to purified RyR1 in absence or presence of 1 μM natrin. (F) Data-fitting.
Next, we performed equilibrium saturation assays of [3H]-ryanodine binding to skeletal SR vesicles (Fig. 2 B) and to purified RyR1s (Fig. 2 E). Fig. 2, B and E, shows that natrin reduced [3H]ryanodine's capability to bind both SR vesicles and purified RyR1s. For SR vesicles, a Lineweaver-Burk plot (Fig. 2 C) visualized that 3 μM natrin reduced the total binding capacity (Bmax) of [3H]ryanodine from 9.27 to 4.21 pmol/mg, whereas the binding affinity was virtually unchanged, as revealed by the dissociation constant (Kd) that only changed from 15.63 to 17.68 nM. For purified RyR1s, 1 μM natrin reduced the Bmax of [3H]ryanodine from 49.75 to 18.93 pmol/mg, whereas the Kd changed slightly, from 20.25 to 23.01 nM (Fig. 2 F). These kinetic data suggest that natrin toxin behaves as a typical noncompetitive antagonist of RyR1: it binds to a site distinct from the ryanodine-binding site. We think that the reduction of our Bmax is attributable to the binding of natrin inhibiting the conformational changes of RyR1 (see Discussion), blocking the access and reducing the binding of ryanodine to the binding sites, because ryanodine is known to bind specifically to RyR1 in its open conformation (29). However, other explanations are also possible, e.g., the effects of natrin toxin, just as of ruthenium red, could involve blocking the low-affinity binding site while noncompetitively altering high-affinity binding (30). Additional experiments will be necessary to understand the reduced (apparent) Bmax.
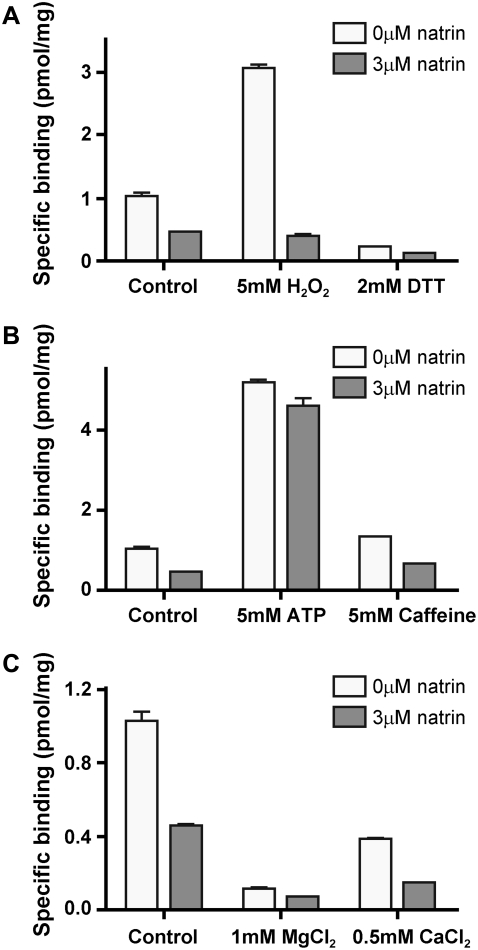
In addition, we investigated the effects of natrin and various RyR1 channel modulators on [3H]ryanodine binding to skeletal SR vesicles. The modulators tested were the oxidizing and reducing reagents H2O2 and dithiothreitol (DTT), the RyR1 channel activators ATP and caffeine, and the RyR1 channel inhibitors Mg2+ and Ca2+ (at mM concentrations). As shown in Fig. 3 A, in the absence of natrin, 5 mM H2O2 increased [3H]ryanodine binding to SR vesicles from 1.13 ± 0.06 (mean ± SD, n = 2) to 3.07 ± 0.07 pmol/mg (n = 2), and 2 mM DTT decreased binding to 0.23 ± 0.007 pmol/mg (n = 2). When SR vesicles were treated additionally with 3 μM natrin, [3H]ryanodine binding was reduced to 0.46 ± 0.01 (n = 2, control), 0.37 ± 0.06 (n = 2, 5 mM H2O2), and 0.13 ± 0.01 pmol/mg (n = 2, 2 mM DTT). The RyR1 activators 5 mM caffeine and 5 mM ATP both increased [3H]ryanodine binding from 1.13 ± 0.06 pmol/mg (n = 2) to 5.18 ± 0.11 for ATP (n = 2), and from 1.13 ± 0.06 pmol/mg to 1.33 ± 0.03 pmol/mg for caffeine (n = 2, Fig. 3 B). When SR vesicles were treated additionally with of 3 μM natrin, the corresponding ryanodine binding decreased to 4.59 ± 0.27 (n = 2) and 0.67 ± 0.01 (n = 2) pmol/mg, respectively (Fig. 3 B). Under inhibitory conditions, 1 mM MgCl2 and 0.5 mM CaCl2 decreased [3H]ryanodine binding to 0.11 ± 0.02 (n = 2) and 0.39 ± 0.01 (n = 2) pmol/mg (without natrin), respectively, and the addition of 3 μM natrin decreased binding further to 0.07 ± 0.005 (n = 2) or 0.15 ± 0.006 (n = 2) pmol/mg (Fig. 3 C). In summary, the [3H]ryanodine binding data demonstrate that natrin toxin inhibits [3H]ryanodine binding for RyR1 channels in skeletal SR in both its activated and inactivated states, as well as under a variety of redox modifications.
FIGURE 3.
Effects of RyR1 channel modulators on natrin inhibition of [3H]ryanodine binding to skeletal SR vesicles. Equilibrium [3H]ryanodine binding experiments, in different redox states (A), or in presence of channel activators (B) or channel inhibitors (C), were performed both with and without 3 μM natrin. Experimental conditions were the same as in Fig. 2. Data are presented as mean ± SD.
Natrin toxin blocks RyR1 calcium-release channel activity
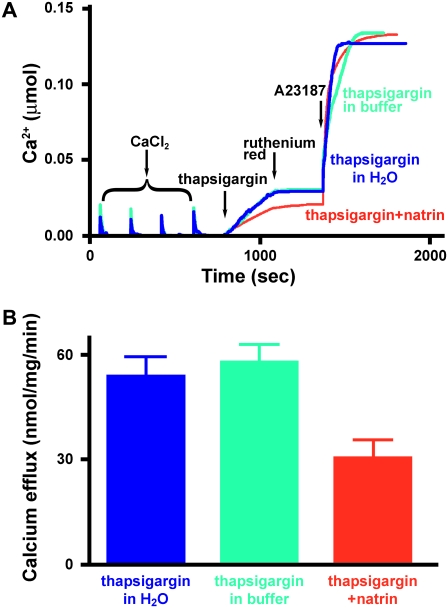
To examine further the effects of that natrin on RyR1 channel activity, we visualized Ca2+ efflux events by RyR1 from native SR vesicles in real time. Extravesicular [Ca2+] was monitored by a Ca2+ indicator, antipyrylazo III (24). Skeletal SR vesicles were partially loaded with Ca2+ by four sequential additions of CaCl2, each initially increasing the extravesicular [Ca2+] by ∼7.5 μM from the preceding level (Fig. 4 A). After suppression of Ca2+-ATPase activity by the addition of 200 nM thapsigargin, the rates of Ca2+ efflux were recorded. Changes in fluorescence intensity, corresponding to calcium efflux events, were recorded for 10 min after the addition of thapsigargin, and while extravesicular [Ca2+] was still increasing, we added ruthenium red (5 μM) to stop the Ca2+ efflux, confirming that RyR1 channels were mediating this calcium efflux event. Finally, the Ca2+ ionophore A23187 (3 μg/mL) was added to release all Ca2+ remaining in the SR vesicles.
FIGURE 4.
Natrin toxin blocks RyR1 calcium efflux from skeletal SR vesicles. (A) Extravesicular [Ca2+] was monitored by the Ca2+ indicator, antipyrylazo III. (Blue) 200 nM thapsigargin in H2O; (green) 200 nM thapsigargin in buffer (30 mM KCl, 25 mM Na-HEPES, pH 7.0); (red) 200 nM thapsigargin + 0.5 μM natrin in buffer (30 mM KCl, 25 mM Na-HEPES, pH 7.0). (B) Averaged maximal rates of calcium efflux with or without natrin. Data are presented as mean ± SE.
As shown in Fig. 4 B, in the absence of natrin toxin, the maximal rate of calcium efflux was 52.82 ± 4.62 nmol/mg/min (200 nM thapsigargin in H2O; mean ± SE, n = 12) and 59.28 ± 4.01 nmol/mg/min (200 nM thapsigargin in buffer; n = 8). When 0.5 μM natrin was added with thapsigargin, the maximal release rate was reduced to 31.69 ± 4.38 nmol/mg/min (200 nM thapsigargin + 0.5 μM natrin; n = 11, p < 0.05). These data demonstrate that natrin inhibits calcium release from SR through the RyR1 channel.
Two-dimensional image-averaging and classification
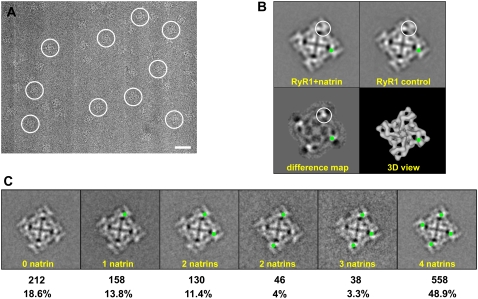
The purified natrin toxin was incubated with RyR1 to promote complex formation. The samples were applied to carbon-coated electron microscopy grids, which were then rapidly frozen in liquid ethane. Both the RyR1 + natrin complexes and the control RyR1 (without added natrin toxin) were imaged by cryo-EM. Fig. 5 A shows a typical electron micrograph in which the individual RyR1 + natrin complexes are visible.
FIGURE 5.
Cryo-electron microscopy and two-dimensional analysis. (A) Portion of cryo-EM micrograph of RyR1 + natrin complexes, showing individual complexes embedded in a thin layer of vitreous ice. The tetrameric structure of RyR1 is well-preserved. Several individual particles are marked with white circles. Scale bar, 500 Å. (B) Two-dimensional averages of top views of RyR1 control and RyR1 + natrin. Two-dimensional top view of RyR1 + natrin was averaged from 245 particle images, and two-dimensional top view of RyR1 control was averaged from 266 particle images. The difference map was obtained by subtracting the RyR1 control average from the RyR1 + natrin average. Top view represents the projection of RyR1 as seen from the cytoplasmic side, as depicted in a reference image of the three-dimensional structure. The largest differences shown in the difference map, corresponding to additional masses contributed by the binding of natrin molecules, are seen as density-maxima bright white areas, and are encircled. The corresponding location of major difference in the difference map is also highlighted by green dots. The width of each frame is 544 Å. (C). Partial occupancy of natrin-binding sites on RyR1 particles. Green dots represent binding sites occupied with natrin molecules (see text for details).
Ryanodine receptor-1 is a homo-tetramer that consists of four identical subunits and possesses fourfold symmetry in its three-dimensional structure (31). By selecting particles with their fourfold-symmetry axes that oriented perpendicular to the carbon support film, we computed two-dimensional averages (Fig. 5 B). This view represents a projection of the three-dimensional structure of RyR as seen from the cytoplasmic side (i.e., three-dimensional view in Fig. 5 B). Overall the two-dimensional average for RyR1 + natrin agreed closely with that for the RyR1 control, apart from the presence of small regions of additional density. To identify the subtle differences between RyR1 + natrin and the RyR1 control, we created a difference map through subtraction of the image of the RyR1 control average from that of RyR1 + natrin. The brightest areas shown in the difference map in Fig. 5 B represent the protein mass present in the RyR1 + natrin complex, but absent in the RyR1 control (one corner is highlighted by a white circle). The location sites of increased mass density were located in regions that correspond to the “clamp” (i.e., assemblages of domains that form the corners of the square-shaped cytoplasmic part of the receptor) in the three-dimensional structure of RyR1 (green dots were used to highlight the relative position in two-dimensional averages and in the three-dimensional view). The four maximum differences almost certainly correspond to the additional masses attributable to the binding of four natrin molecules to one RyR1 tetramer, an interpretation confirmed by a more precise mapping of natrin-binding sites within the three-dimensional structure (see below).
Before computing the three-dimensional reconstruction of the RyR1 + natrin complex, we had to address a problem of heterogeneity of the data set, caused by partial occupancy of the four natrin-binding sites on one RyR1 homo-tetramer. Because the binding affinity between natrin and purified RyR1 is ∼1 μM, we estimated that, under the conditions used for cryo-EM, ∼90% of the natrin-binding sites on RyR1 should be occupied. However, this estimate is for optimal binding conditions, and the actual binding on electron microscopy grids is likely to be lower. For RyR1, the case is even more complicated, because each RyR1 tetramer has four natrin-binding sites. Besides the RyR1 molecules showing full occupancy, bound with four natrin molecules (Fig. 5 C, right), there are five other categories of RyR1 molecules that show less than the full complement of four bound natrin molecules (i.e., with 0, 1, 2, or 3 natrin molecules; furthermore, there are two categories of RyR1 + natrin complexes with two natrin molecules bound, as depicted in Fig. 5 C, middle). To estimate the actual percentages of RyR1 with the various complements of bound natrin molecules, we performed a two-dimensional classification analysis. Top images of RyR1 + natrin complexes (N = 1142) were subjected to a multireference supervised classification (32). As shown in Fig. 5 C, ∼48.9% of the particles had four natrin molecules bound, whereas 51.1% had fewer. This classification scheme allowed us to eliminate most of the incompletely occupied RyR1 particles in the subsequent image-processing. We only used particles with natrin-binding sites fully occupied to compute the final three-dimensional reconstruction of the RyR1 + natrin complex.
Determination of natrin-binding locations in three-dimensional structure of RyR1
Fig. 6, A and B, shows surface representations of three-dimensional reconstructions of the control RyR1 and RyR1 + natrin complex, in three views. The final resolution was estimated to be 21 Å by Fourier shell correlation, with a cutoff of 0.5 (27). The reconstructions show the distinctive mushroom shape of RyR, consisting of a large cytoplasmic assembly with at least 11 domains, and the smaller transmembrane domain assembly (31,33). As expected from the two-dimensional data, the two reconstructions are highly comparable, but an obvious difference appears on the top surface of domains 5 and 6, as shown in the side views in Fig. 6, A and B (red circles). We generated a three-dimensional difference map by subtracting the volume of the control RyR1 from the volume of the RyR1 + natrin complex. The main positive difference regions are superimposed on the three-dimensional reconstruction of control RyR1 (Fig. 6 C, orange). This difference map clearly indicates that there is one bound natrin per RyR1 subunit, located at the top surface of domains 5 and 6. The three-dimensional locations of the natrin-binding site are also consistent with the two-dimensional difference map.
FIGURE 6.
Three-dimensional surface representations of RyR1 control and RyR1 + natrin complex, and three-dimensional difference map. Three-dimensional reconstruction of RyR1 control is shown in yellow (A), and RyR1 + natrin complex is shown in orange (B). (C) Difference map (RyR1 + natrin minus RyR1, orange) is superimposed on three-dimensional reconstruction of RyR1 control (yellow). Three-dimensional reconstructions are shown in three views: (left) top view from cytoplasmic surface, which in situ would face the transverse tubule; (middle) side view; (right) bottom view showing surface that would face the SR lumen. Numerals 1–11 on the cytoplasmic assembly indicate distinguishable domains, according to our earlier nomenclature (31,33).
Docking the crystal structure of natrin into the cryo-EM density map of the RyR1 + natrin complex
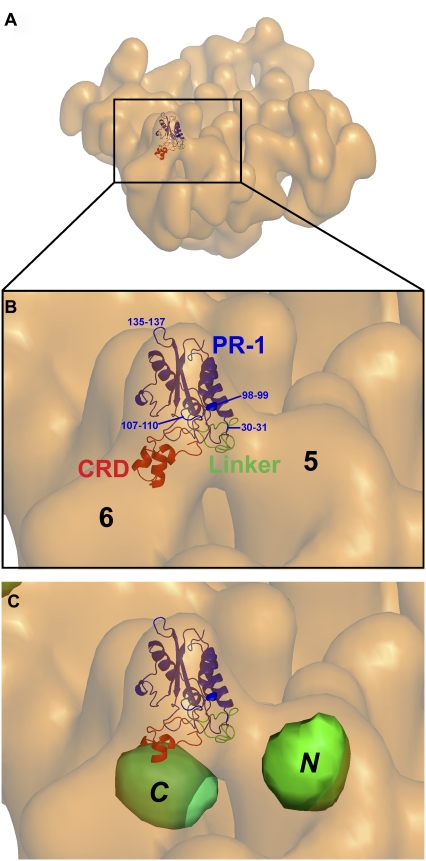
The atomic structure of natrin toxin was solved by x-ray crystallography (18). It consists of three domains: a pathogenesis-related protein of the group 1 (PR-1)-like domain (residues 6–160), a short linker (residues 161–182), and the CRD (residues 183–221). A docking of the atomic structure of natrin was performed interactively by fitting the atomic coordinates (Protein Data Bank code 2GIZ) into the cryo-EM surface envelope of the RyR1 + natrin map. Because the three-dimensional structure of the RyR1 + natrin complex was reconstructed from RyR1 with natrin fully bound particles, the crystal structure of natrin fitted quite well into the cryo-EM density map that assigned to the natrin molecule, the cross-correlation coefficient value between the difference map and the fitted atomic coordinates is 0.76, as determined by the SPIDER program (28). Other docking orientations, e.g., a 180° rotation around the vertical axis (switching the CRD and linker domain) or a 180° rotation around the horizontal axis (switching the linker and PR-1 domain), gave lower cross-correlation coefficient values. Additional studies, e.g., improving the resolution of the RyR1 + natrin complex and flexible docking, are needed to refine our docking model further. Fig. 7 illustrates the fitting of natrin in a view where the cytoplasmic-facing side is tilted from the fourfold symmetry axis. Only small portions of three loops (residues 30–31, 107–110, and 135–137), and of two α-helices (residues 11–13, not visible in this view, and 98–99) from the PR-1 domain of natrin protrude beyond the electron microscopy density. Details of docking results are shown in Supplementary Material, Movie S1. Our docking model clearly indicates that the linker domain and the CRD of natrin directly interact with domains 5 and 6 of RyR1, respectively (Fig. 7 B), whereas the PR-1 domain points away from RyR1, and thus does not appear to participate in binding.
FIGURE 7.
Docking of x-ray crystal structure of natrin into cryo-EM density map of RyR1 + natrin complex. Atomic coordinates of natrin molecule (Protein Data Bank code 2GIZ) were manually fitted into our cryo-EM density map of RyR1 + natrin complex, using the program O. (A) Tilted view of RyR1 + natrin complex with one natrin molecule docked in natrin-binding site of one subunit in RyR1 homo-tetramer. (B) A zoom view of clamp region from A, showing that natrin binds to domains 5 and 6 of RyR1. Domain rendering of natrin molecule: PR-1 domain (amino-acid residues 1–160, blue), linker (residues 161–182, green), and CRD (residues 183-221, red). Numbers indicate residues that protrude slightly out of the density map envelope. (C) Detailed view of clamp-shaped structure, showing relative positions of previously mapped amino-terminal (N) and central (C) mutation hotspots.
DISCUSSION
Natrin blocks the RyR1 calcium-release channel by stabilizing a domain-domain interaction within RyR1
Domains 5 and 6 in RyR1, with which the natrin toxin directly interacts, contain the two mutation hotspots that we previously mapped (34,35). Over 100 mutations in RyR1 were identified in families with malignant hyperthermia and central-core disease, and these mutations are largely clustered in three regions of the ∼5,000-amino-acid sequence of RyR1: the amino-terminal region (amino-acid residues 35–614), central region (amino-acid residues 1728–2728), and carboxyl-terminal region (amino-acid residues 3348–4973) (36). The two mutation hotspots that lie in the cytoplasmic region, the amino-terminal region, and the central region are well-separated in the primary sequence (>1100 amino acids). According to a hypothesis proposed by Ikemoto and Yamamoto (37), the two mutation hotspots occur in structural domains that physically interact in the three-dimensional structure, and changes in the strength of their interaction affect channel gating. In that hypothesis, the amino-terminal domain and central domain interact so as to serve as a regulatory switch for channel-gating activity; a tight “zipping” of the interacting domains stabilizes the channel in the closed state. A mutation in either domain weakens the domain-domain interaction, thus increasing the tendency toward “unzipping.” Such an unzipping causes activation and leakiness of the Ca2+ release channel. In support of their hypothesis, the authors found that addition of the synthetic peptides DP4 (corresponding to the Leu2442-Pro2477 region in RyR1's central mutation domain) or DP1 (corresponding to Leu590-Cys609 in RyR1's N-terminal mutation domain) enhanced [3H]ryanodine binding and induced Ca2+ release from the SR (38,39). The synthetic peptide DP4 also induced contraction in skinned muscle fibers at an inhibitory Mg2+ concentration (40), and increased the frequency of Ca2+ sparks in saponin-permeabilized fibers because of an increased open probability of the channels (41).
Previous studies pertaining to interdomain interactions relied mainly on indirect biochemical assays to detect conformational changes in RyR isoforms, either RyR1 or RyR2 (38,39), and little information describing local or global changes in RyR1's three-dimensional architecture has been reported. We used three-dimensional cryo-EM to test the domain-switch hypothesis, i.e., to determine the three-dimensional localization of the two mutation hotspots. We previously mapped one central mutation hotspot (Ser2367) in the three-dimensional structure of RyR2 by green fluorescent protein (GFP) labeling (34). Another site in the amino-terminal mutation hotspot (Ser437) was located by the same method (35). The GFP inserted in the amino-terminal region sequence (Ser437) was mapped to a region near domains 5 and 9, whereas the GFP in the central region (Ser2367) mapped to the bridge between domains 5 and 6 (labeled N and C in Fig. 7 C). Although those results were obtained for the structure of RyR2, they can be extrapolated to RyR1 because of the high degree of sequence identity (42,43) and structural similarity of the two isoforms (31,44). Fig. 7 C shows that the two GFP-mapped locations are on opposite sides of domain 5 in the clamp region of the RyR cytoplasmic assembly, a spatial relationship consistent with the interdomain zipping-unzipping hypothesis of Ikemoto and Yamamoto (37).
Our structural information, together with the domain-switch hypothesis, leads us to propose a molecular mechanism for natrin's inhibition of the RyR1 calcium-release channel. As illustrated in Fig. 7 C, the CRD of natrin interacts with the central mutation domain, and the linker interacts with the N-terminal domain. It is likely that the binding of natrin to RyR1 strengthens the interaction between the two domains, thereby minimizing domain-switch unzipping and stabilizing the closed state of the RyR1 channel. As revealed in three-dimensional reconstruction, the binding of natrin caused domains 5 and 6 to approach closer to one another. These movements also appear prominently in the two-dimensional difference map as two dark (negative) density areas (corresponding to the reduced mass attributable to the movement of domains) next to the bright white (positive) density maxima (attributable mainly to the additional mass of natrin and secondarily to domain movements).
Our findings are pertinent to the development of novel therapeutics for muscular dystrophies. Dantrolene is a drug that suppresses RyR1 Ca2+ release from SR in skeletal muscle, and is used as a therapeutic agent in individuals susceptible to malignant hyperthermia (45). In addition, JTV519 is a 1,4-benzothiazepine derivative, found to improve contractility and to prevent the development of calcium leak through the inhibition of the mutant cardiac RyR2 and skeletal RyR1 (46). Both drugs stabilize RyR by restoring the domain-domain interaction that is disordered by disease-causing mutations (46,47). Moreover, fatigue of skeletal muscles is directly linked to the channel leakage of RyR1, and an RyR1 stabilizer can block the leakage, reduce muscle damage, and boost stamina in mice (48).
CRD is the binding domain that directly interacts with various ion channels
Our docking model reveals that the CRD and the linker are the major parts of the natrin molecule that directly interact with RyR1. In an earlier study, the CRD of Tpx-1, a CRISP-2 subclass protein, was demonstrated to inhibit RyR2, whereas it activated RyR1 (12). Previous studies also attributed the interaction between other ion channels and CRISPs to the CRD. Stecrisp is another snake-venom toxin that belongs to the CRISP-3 family, and the CRD of stecrisp has structural features similar to those of two ion channel blockers, Bgk and Shk (19). Isolated from the sea anemone, Bgk and Shk are 37-residue and 35-residue peptides that function as voltage-sensitive K+ channel blockers (49,50). Natrin and stecrisp show a high degree of sequence identity, and can also block the BKCa channel (17) and the Kv1.3 channel (18). Natrin also shows high sequence homology with other two CRISPs, pseudechetoxin and pseudecin. These are two ion channel blockers that target the cyclic nucleotide-gated ion channel (19). For both of these latter CRISPs, the CRD contains the site of interaction with the target cyclic nucleotide-gated ion channels (51,52). Another study demonstrated that the CRD of natrin plays a crucial role in the interaction between this toxin and the Kv1.3 potassium channel (18). Taken together, these findings suggest the existence of a familial mode of interaction of the CRISPs with their various targeted ion channels. In each case, the CRD is the domain of the CRISP that binds to the channel, and thereby modulates its functioning.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Joshua Strauss for technical assistance, the Wadsworth Center's Electron Microscopy Core Facility, and the Resource for Visualization of Biological Complexity (funded by National Institutes of Health Biotechnological Resource Grant RR01219).
This work was supported by American Heart Association grant 0430076N to Z.L., by National Institutes of Health grant AR40615 to T.W., by the Natural Science Foundation of China (grant 30330160 to S.-F.S and C.-C.Y, and grant 30370379 to C.-C.Y), by the Trans-Century Talent-Awarding Program, Ministry of Education, China (to C.-C.Y.), and by the National Basic Research Program of China (grant 2004CB720005 to S.-F.S).
Qiang Zhou, Qiong-Ling Wang, and Xing Meng contributed equally to this work.
Editor: Kenton J. Swartz.
References
- 1.Kierszenbaum, A. L., O. Lea, P. Petrusz, F. S. French, and L. L. Tres. 1981. Isolation, culture, and immunocytochemical characterization of epididymal epithelial cells from pubertal and adult rats. Proc. Natl. Acad. Sci. USA. 78:1675–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellerman, D. A., V. G. Da Ros, D. J. Cohen, D. Busso, M. M. Morgenfeld, and P. S. Cuasnicu. 2002. Expression and structure-function analysis of DE, a sperm cysteine-rich secretory protein that mediates gamete fusion. Biol. Reprod. 67:1225–1231. [DOI] [PubMed] [Google Scholar]
- 3.Kasahara, M., J. Gutknecht, K. Brew, N. Spurr, and P. N. Goodfellow. 1989. Cloning and mapping of a testis-specific gene with sequence similarity to a sperm-coating glycoprotein gene. Genomics. 5:527–534. [DOI] [PubMed] [Google Scholar]
- 4.Maeda, T., J. Nishida, and Y. Nakanishi. 1999. Expression pattern, subcellular localization and structure-function relationship of rat Tpx-1, a spermatogenic cell adhesion molecule responsible for association with Sertoli cells. Dev. Growth Differ. 41:715–722. [DOI] [PubMed] [Google Scholar]
- 5.Kratzschmar, J., B. Haendler, U. Eberspaecher, D. Roosterman, P. Donner, and W. D. Schleuning. 1996. The human cysteine-rich secretory protein (CRISP) family. Primary structure and tissue distribution of CRISP-1, CRISP-2 and CRISP-3. Eur. J. Biochem. 236:827–836. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki, Y., and T. Morita. 2004. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 44:227–231. [DOI] [PubMed] [Google Scholar]
- 7.Jalkanen, J., I. Huhtaniemi, and M. Poutanen. 2005. Mouse cysteine-rich secretory protein 4 (CRISP4): a member of the CRISP family exclusively expressed in the epididymis in an androgen-dependent manner. Biol. Reprod. 72:1268–1274. [DOI] [PubMed] [Google Scholar]
- 8.Franzini-Armstrong, C., and F. Protasi. 1997. Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiol. Rev. 77:699–729. [DOI] [PubMed] [Google Scholar]
- 9.Morrissette, J., J. Kratzschmar, B. Haendler, R. el-Hayek, J. Mochca-Morales, B. M. Martin, J. R. Patel, R. L. Moss, W. D. Schleuning, R. Coronado, and L. D. Possani. 1995. Primary structure and properties of helothermine, a peptide toxin that blocks ryanodine receptors. Biophys. J. 68:2280–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nobile, M., V. Magnelli, L. Lagostena, J. Mochca-Morales, L. D. Possani, and G. Prestipino. 1994. The toxin helothermine affects potassium currents in newborn rat cerebellar granule cells. J. Membr. Biol. 139:49–55. [DOI] [PubMed] [Google Scholar]
- 11.Nobile, M., F. Noceti, G. Prestipino, and L. D. Possani. 1996. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 110:15–20. [DOI] [PubMed] [Google Scholar]
- 12.Gibbs, G. M., M. J. Scanlon, J. Swarbrick, S. Curtis, E. Gallant, A. F. Dulhunty, and M. K. O'Bryan. 2006. The cysteine-rich secretory protein domain of Tpx-1 is related to ion channel toxins and regulates ryanodine receptor Ca2+ signaling. J. Biol. Chem. 281:4156–4163. [DOI] [PubMed] [Google Scholar]
- 13.Yamazaki, Y., F. Hyodo, and T. Morita. 2003. Wide distribution of cysteine-rich secretory proteins in snake venoms: isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch. Biochem. Biophys. 412:133–141. [DOI] [PubMed] [Google Scholar]
- 14.Chang, T.-Y., S.-H. Mao, and Y.-W. Guo. 1997. Cloning and expression of a cysteine-rich venom protein from Trimeresurus mucrosquamatus (Taiwan habu). Toxicon. 35:879–888. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Y., Q. Lu, X. Zhou, S. Zhu, R. Li, W. Wang, and Y. Xiong. 2003. Purification and cloning of cysteine-rich proteins from Trimeresurus jerdonii and Naja atra venoms. Toxicon. 42:539–547. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki, Y., H. Koike, Y. Sugiyama, K. Motoyoshi, T. Wada, S. Hishinuma, M. Mita, and T. Morita. 2002. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 269:2708–2715. [DOI] [PubMed] [Google Scholar]
- 17.Wang, J., B. Shen, M. Guo, X. Lou, Y. Duan, X. P. Cheng, M. Teng, L. Niu, Q. Liu, Q. Huang, and Q. Hao. 2005. Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel. Biochemistry. 44:10145–10152. [DOI] [PubMed] [Google Scholar]
- 18.Wang, F., H. Li, M. Liu, H. Song, H. Han, Q. Wang, C. Yin, Y. Zhou, Z. Qi, Y. Shu, Z. Lin, and T. Jiang. 2006. Structural and functional analysis of natrin, a venom protein that targets various ion channels. Biochem. Biophys. Res. Commun. 351:443–448. [DOI] [PubMed] [Google Scholar]
- 19.Guo, M., M. Teng, L. Niu, Q. Liu, Q. Huang, and Q. Hao. 2005. Crystal structure of the cysteine-rich secretory protein Stecrisp reveals that the CRD has a K+ channel inhibitor-like fold. J. Biol. Chem. 280:12405–12412. [DOI] [PubMed] [Google Scholar]
- 20.Shikamoto, Y., K. Suto, Y. Yamazaki, T. Morita, and H. Mizuno. 2005. Crystal structure of a CRISP family Ca2+-channel blocker derived from snake venom. J. Mol. Biol. 350:735–743. [DOI] [PubMed] [Google Scholar]
- 21.Saito, A., S. Seiler, A. Chu, and S. Fleischer. 1984. Preparation and morphology of sarcoplasmic reticulum terminal cisternae from rabbit skeletal muscle. J. Cell Biol. 99:875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inui, M., A. Saito, and S. Fleischer. 1987. Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. J. Biol. Chem. 262:1740–1747. [PubMed] [Google Scholar]
- 23.Yin, C. C., and F. A. Lai. 2000. Intrinsic lattice formation by the ryanodine receptor calcium-release channel. Nat. Cell Biol. 2:669–671. [DOI] [PubMed] [Google Scholar]
- 24.Dulhunty, A. F., D. R. Laver, E. M. Gallant, M. G. Casarotto, S. M. Pace, and S. Curtis. 1999. Activation and inhibition of skeletal RyR channels by a part of the skeletal DHPR II–III loop: effects of DHPR Ser687 and FKBP12. Biophys. J. 77:189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Z., J. Zhang, M. Sharma, P. Li, S. R. W. Chen, and T. Wagenknecht. 2001. Three-dimensional reconstruction of the recombinant type 3 ryanodine receptor and localization of its amino terminus. Proc. Natl. Acad. Sci. USA. 98:6104–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra, A., P. Penczek, R. K. Agrawal, I. S. Gabashvilli, R. A. Grassucci, R. Jünemann, M. Burkhardt, K. H. Nierhaus, and J. Frank. 1998. Escherichia coli 70S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280:103–116. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal, R. K., P. Penczek, R. A. Grassucci, and J. Frank. 1998. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA. 95:6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du, G. G., J. P. Imredy, and D. H. MacLennan. 1998. Characterization of recombinant rabbit cardiac and skeletal muscle Ca2+ release channels (ryanodine receptors) with a novel [3H]ryanodine binding assay. J. Biol. Chem. 273:33259–33266. [DOI] [PubMed] [Google Scholar]
- 30.Chu, A., M. Diaz-Munoz, M. J. Hawkes, K. Brush, and S. L. Hamilton. 1990. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum calcium release channel. Mol. Pharmacol. 37:735–741. [PubMed] [Google Scholar]
- 31.Radermacher, M., V. Rao, R. Grassucci, J. Frank, A. P. Timerman, S. Fleischer, and T. Wagenknecht. 1994. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J. Cell Biol. 127:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank, J. 2006. Multivariate data analysis and classification of images. In Three-Dimensional Electron Microscopy of Macromolecular Assemblies. J. Frank, editor. Oxford University Press, New York. 145–192.
- 33.Samso, M., T. Wagenknecht, and P. D. Allen. 2005. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Z., R. Wang, J. Zhang, S. R. W. Chen, and T. Wagenknecht. 2005. Localization of a disease-associated mutation site on the three-dimensional structure of cardiac ryanodine receptor. J. Biol. Chem. 280:37941–37947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, R., W. Chen, S. Cai, J. Zhang, J. Bolstad, T. Wagenknecht, Z. Liu, and S. R. W. Chen. 2007. Localization of an NH2-terminal disease-causing mutation hotspot to the “clamp” region in the three-dimensional structure of the cardiac ryanodine receptor. J. Biol. Chem. 282:17785–17793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarty, T. V., K. A. Quane, and P. J. Lynch. 2000. Ryanodine receptor mutations in malignant hyperthermia and central core disease. Hum. Mutat. 15:410–417. [DOI] [PubMed] [Google Scholar]
- 37.Ikemoto, N., and T. Yamamoto. 2002. Regulation of calcium release by interdomain interaction within ryanodine receptors. Front. Biosci. 7:d671–d683. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, T., R. El-Hayek, and N. Ikemoto. 2000. Postulated role of interdomain interaction within the ryanodine receptor in Ca2+ channel regulation. J. Biol. Chem. 275:11618–11625. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, T., and N. Ikemoto. 2002. T-tubule depolarization-induced local events in the ryanodine receptor, as monitored with the fluorescent conformational probe incorporated by mediation of peptide A. J. Biol. Chem. 277:984–992. [DOI] [PubMed] [Google Scholar]
- 40.Lamb, G. D., G. S. Posterino, T. Yamamoto, and N. Ikemoto. 2001. Effects of a domain peptide of the ryanodine receptor on Ca2+ release in skinned skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 281:C207–C214. [DOI] [PubMed] [Google Scholar]
- 41.Shtifman, A., C. W. Ward, T. Yamamoto, J. Wang, B. Olbinski, H. H. Valdivia, N. Ikemoto, and M. F. Schneider. 2002. Interdomain interactions within ryanodine receptors regulate Ca2+ spark frequency in skeletal muscle. J. Gen. Physiol. 119:15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zorzato, F., J. Fujii, K. Otsu, M. Phillips, N. M. Green, F. A. Lai, G. Meissner, and D. H. MacLennan. 1990. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 265:2244–2256. [PubMed] [Google Scholar]
- 43.Otsu, K., H. F. Willard, V. K. Khanna, F. Zorzato, N. M. Green, and D. H. MacLennan. 1990. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 265:13472–13483. [PubMed] [Google Scholar]
- 44.Sharma, M., P. Penczek, R. Grassucci, H. B. Xin, S. Fleischer, and T. Wagenknecht. 1998. Cryoelectron microscopy and image analysis of the cardiac ryanodine receptor. J. Biol. Chem. 273:18429–18434. [DOI] [PubMed] [Google Scholar]
- 45.Ward, A., M. O. Chaffman, and E. M. Sorkin. 1986. Dantrolene: a review of its pharmacodynamic and pharmacokinetic properties and therapeutic use in malignant hyperthermia, the neurolept malignant syndrome, and an update of its use in muscle spasticity. Drugs. 32:130–168. [DOI] [PubMed] [Google Scholar]
- 46.Oda, T., M. Yano, T. Yamamoto, T. Tokuhisa, S. Okuda, M. Doi, T. Ohkusa, Y. Ikeda, S. Kobayashi, N. Ikemoto, and M. Matsuzaki. 2005. Defective regulation of interdomain interactions within the ryanodine receptor plays a key role in the pathogenesis of heart failure. Circulation. 111:3400–3410. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi, S., M. L. Bannister, J. P. Gangopadhyay, T. Hamada, J. Parness, and N. Ikemoto. 2005. Dantrolene stabilizes domain interactions within the ryanodine receptor. J. Biol. Chem. 280:6580–6587. [DOI] [PubMed] [Google Scholar]
- 48.Bellinger, A. M., S. Reiken, M. Dura, P. W. Murphy, S. X. Deng, D. W. Landry, D. Nieman, S. E. Lehnart, M. Samaru, A. LaCampagne, and A. R. Marks. 2008. Remodeling of ryanodine receptor complex causes “leaky” channels: a molecular mechanism for decreased exercise capacity. Proc. Natl. Acad. Sci. USA. 105:2198–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tudor, J. E., P. K. Pallaghy, M. W. Pennington, and R. S. Norton. 1996. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat. Struct. Biol. 3:317–320. [DOI] [PubMed] [Google Scholar]
- 50.Dauplais, M., A. Lecoq, J. Song, J. Cotton, N. Jamin, B. Gilquin, C. Roumestand, C. Vita, C. L. C. de Medeiros, E. G. Rowan, A. L. Harvey, and A. Menez. 1997. On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 272:4302–4309. [DOI] [PubMed] [Google Scholar]
- 51.Brown, R. L., T. L. Haley, K. A. West, and J. W. Crabb. 1999. Pseudechetoxin: a peptide blocker of cyclic nucleotide-gated ion channels. Proc. Natl. Acad. Sci. USA. 96:754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamazaki, Y., R. L. Brown, and T. Morita. 2002. Purification and cloning of toxins from elapid venoms that target cyclic nucleotide-gated ion channels. Biochemistry. 41:11331–11337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.