Abstract
Hint1 is a haploinsufficient tumor suppressor gene and the underlying molecular mechanisms for its tumor suppressor function are unknown. In this study we demonstrate that HINT1 participates in ionizing radiation (IR)–induced DNA damage responses. In response to IR, HINT1 is recruited to IR-induced foci (IRIF) and associates with γ-H2AX and ATM. HINT1 deficiency does not affect the formation of γ-H2AX foci; however, it impairs the removal of γ-H2AX foci after DNA damage and this is associated with impaired acetylation of γ-H2AX. HINT1 deficiency also impairs acetylation of ATM and activation of ATM and its downstream effectors, and retards DNA repair, in response to IR. HINT1-deficient cells exhibit resistance to IR-induced apoptosis and several types of chromosomal abnormalities. Our findings suggest that the tumor suppressor function of HINT1 is caused by, at least in part, its normal role in enhancing cellular responses to DNA damage by regulating the functions of both γ-H2AX and ATM.
Introduction
DNA double strand breaks (DSB) resulting from exposure to ionizing radiation (IR), various chemical agents, or even from normal DNA replication are the most lethal type of DNA lesion because the damage to both DNA strands prevents use of the complimentary DNA strand as a template for DNA repair. Therefore, the proper removal of DSB requires the coordinated action of diverse signaling pathways involved in cell cycle checkpoints and DNA repair (van Gent et al., 2001). Inefficient or inaccurate repair of DSB can cause genomic instability and enhance carcinogenesis (van Gent et al., 2001).
Although the precise sequence of events that occurs in the process of repairing DSB is not clear, changes in chromatin structure are believed to play an important role in this process (Downs et al., 2007). Thus, phosphorylation of a histone variant, H2AX, is one of the earliest events after DSB (Rogakou et al., 1998). Within seconds after exposure to IR, H2AX is phosphorylated at a highly conserved Serine 139 residue in the C-terminal tail of this histone. This phosphorylated protein is called γ-H2AX. The γ-H2AX protein forms distinctive foci in the nucleus that are observable by immunofluorescent staining (Rogakou et al., 1999). γ-H2AX provides a scaffold for other checkpoint signaling and DNA repair proteins to accumulate in the vicinity of DSB, forming IR-induced foci (IRIF; Paull et al., 2000). Because DNA is wrapped around histone octamers and packaged further into higher order structures, during the subsequent process of DNA repair, chromatin remodeling activity is required to relax this tight packaging and allow access of repair proteins to the damaged sites in DNA. After DNA repair is complete, chromatin remodeling complexes also play an important role in restoring normal cell cycle progression (Papamichos-Chronakis et al., 2006). Therefore, chromatin remodeling functions in all stages of DNA damage responses (DDR) in eukaryotic cells.
DDR also involve activation of checkpoints that delay progression of the cell cycle, and thus allow time for DNA repair. Activation of the ATM protein is also one of the earliest cellular responses to DSB (Bakkenist and Kastan, 2003). In undamaged cells, ATM resides as a catalytically inactive dimer or multimer. Upon DNA damage, it is first acetylated by a specific histone acetyltransferase called TIP60 (Sun et al., 2005). This is followed by autophosphorylation at Ser1981, which causes the activation of ATM kinase activity (Bakkenist and Kastan, 2003; Sun et al., 2005). The active ATM monomers activate via phosphorylation several downstream effectors including Chk1, Chk2, and p53, all of which are involved in cell cycle checkpoints or apoptosis.
In this study we examined the role of a novel tumor suppressor gene, Hint1, in DDR elicited by DSB. The HINT1 protein belongs to the histidine triad superfamily of nucleotide-binding proteins and is evolutionarily highly conserved (Lima et al., 1996). In previous studies we demonstrated that Hint1 deficiency in mice resulted in increased susceptibility to both spontaneous and carcinogen-induced tumor formation (Su et al., 2003; Li et al., 2006). We also discovered that Hint1 −/− mouse embryonic fibroblasts (MEFs) are more resistant to the cytotoxicity of IR than Hint1 +/+ MEFs (Su et al., 2003). These findings suggested that the HINT1 protein might play a role in cellular responses to DNA damage. Because of the importance of γ-H2AX in the chromatin remodeling pathway and ATM in the checkpoint and apoptosis pathway in maintaining genome integrity, we investigated the role of HINT1 in these two pathways of DDR after treating control or HINT1-deficient cells with IR or the radiomimetic drug bleomycin.
Results
HINT1-deficient cells display prolonged γ-H2AX foci and impaired acetylation of γ-H2AX after γ radiation
To study the role of HINT1 in DDR, we used MEFs derived from the Hint1 +/+, +/−, and −/− mice, which were previously generated in our laboratory (Su et al., 2003; Li et al., 2006). We first examined the expression of HINT1 in these cells by real-time RT-PCR and immunoblot analysis. We found that the mRNA level in Hint1 +/− cells is ∼50% of the wild-type cells and undetectable in Hint1 −/− cells (Fig. 1 A). The expression of HINT1 protein is also high in the Hint1 +/+ MEFs, decreased by half in the +/− MEFs, and undetectable in the −/− cells (Fig. 1 B). Because of the importance of the formation of γ-H2AX foci in response to DSB, we compared the kinetics of γ-H2AX foci formation and removal in the three different genotypes of Hint1 MEFs after exposing the cells to 4 Gy of γ radiation. Colocalization of γ-H2AX foci with p-ATM foci was observed in these cells (unpublished data). Cells were counted as positive if they contained five or more γ-H2AX foci per nucleus. In untreated Hint1 +/+ MEFs, <10% of the cells were positive for γ-H2AX foci. However, in untreated Hint1 +/− or −/− MEFs, 30–40% of the cells were γ-H2AX positive, and this increase was statistically significant (Fig. 1 C). This suggests that HINT1-deficient cells accumulate spontaneous DNA damage at a higher rate than the corresponding wild-type cells. After γ radiation, the cells in all three genotypes showed increases in γ-H2AX foci, suggesting that HINT1 is not required for the formation of IR-induced γ-H2AX foci. The percentage of cells positive for γ-H2AX foci as a function of postirradiation time is shown also in Fig. 1 C. γ-H2AX–positive cells gradually declined at 3 h after IR treatment in Hint1 +/+ cells, and by 24 h the percentage of γ-H2AX–positive cells observed was similar to sham-treated control. However, 90% of the Hint1 −/− and ∼75% of the Hint1 +/− cells remained positive for γ-H2AX staining for up to 24 h (Fig. 1 C). We further examined whether the prolonged γ-H2AX foci colocalize with the MRN repair complex. Double fluorescent staining was performed with γ-H2AX and Rad50 antibodies, and DAPI was used as a counterstain of the nucleus. In the Hint1 +/+ cells, Rad50 foci colocalized with γ-H2AX foci at 1 h after IR, and both foci diminished at 24 h after IR (Fig. 1 D). However, in the Hint1 −/− cells, even at 24–48 h after IR, Rad50 foci were observable and these foci colocalized with γ-H2AX (Fig. 1 D). Because Rad50 is a component of the MRN complex, which is recruited to γ-H2AX foci in response to DNA DSB (Maser et al., 1997; Paull and Lee, 2005), prolonged Rad50 foci further indicate that there are defects of γ-H2AX dynamics in HINT1-deficient cells. The significant differences of γ-H2AX and Rad50 foci removal between wild-type and HINT1-deficient MEFs suggest that HINT1 is required for the normal removal of IRIF that occurs within a few hours after IR.
Figure 1.
HINT1-deficient MEFs display prolonged γ-H2AX staining and impaired acetylation of γ-H2AX. (A) Real-time RT-PCR quantification of mRNA levels in exponential growing Hint1 MEFs. †, P < 0.0001 compared with Hint1 +/+ MEFs. (B) Immunoblot analysis of HINT1 expression in exponential growing Hint1 MEFs. Densitometry analysis was performed on four independent blots. (C) Hint1 MEFs were untreated (0 min) or treated with 4 Gy of IR, and were then fixed after the indicated times. Cells were stained with an anti–γ-H2AX antibody and counterstained for the nucleus with DAPI. Cells with five or more γ-H2AX foci were considered positive. The percentage of positive cells was determined in 500 cells per slide and plotted against the time after treatment. Three repeat experiments were performed and mean and standard deviations were determined. *, P < 0.05 compared with Hint1 +/+ MEFs; **, P < 0.05 compared with both Hint1 +/+ and +/− MEFs. (D) Hint1 MEFs were untreated (control) or treated with 4 Gy of IR, and were fixed after the indicated times. Cells were stained with Rad50 (green) and γ-H2AX antibody (red), and counterstained for the nucleus with DAPI (blue). Images were merged using the ImageJ software. Representative micrographs of Hint1 MEFs are shown. Bars, 10 μm. (E) Hint1 MEFs were untreated (0) or treated with 4 Gy of IR, and whole cell lysates were collected after the indicated times. Immunoprecipitates were obtained using a γ-H2AX antibody or mock mouse IgG (lane M) and resolved by SDS-PAGE. Immunoblots were then performed using a γ-H2AX antibody, an acetyllysine antibody, or an ac-H2A(K5) antibody. Error bars represent the standard deviation of data from triplicate experiments. Repeat studies gave similar results.
The precise mechanisms by which γ-H2AX foci are removed are not known. γ-H2AX may be dephosphorylated in chromatin by protein phosphatases acting in situ or it may be first replaced by unmodified H2AX, and then dephosphorylated in the nucleoplasm. Current data supports the latter mechanism because thus far direct dephosphorylation of γ-H2AX has been seen only in vitro (Nazarov et al., 2003; Chowdhury et al., 2005; Keogh et al., 2006). Moreover, it was recently discovered that during DNA repair in Drosophila melanogaster the removal of phospho-H2Av, the Drosophila homologue of human γ-H2AX, requires the histone acetylase activity of the TIP60 protein complex (Kusch et al., 2004), and in HeLa cells TIP60 also acetylates H2AX in response to IR (Ikura et al., 2007). There is increasing evidence that the TIP60 complex plays an important role in the regulation of DDR because it is recruited to the chromatin surrounding DSB and contributes to the dynamics of DNA repair through a series of chromatin modifications (Bird et al., 2002; Downs et al., 2004; Kusch et al., 2004; Murr et al., 2006).
A recent paper showed that the HINT1 protein associates with the TIP60 complex in SW480 human colon cancer cells (Weiske and Huber, 2006). This finding prompted us to examine whether the persistence of γ-H2AX foci in HINT1-deficient MEFs is caused by impaired acetylation of γ-H2AX because of absence of HINT1 from the TIP60 complex. To examine whether γ-H2AX is acetylated in response to IR, we immunoprecipitated the γ-H2AX protein from lysates of Hint1 MEFs at 0, 30 min, 1 h, or 3 h after a 4-Gy dose of γ radiation, using conditions that dissociate histones from nucleosomes in the preparation procedure (see Materials and methods). Using an anti–γ-H2AX antibody in an immunoblot analysis of these immunoprecipitates, we found that even in unirradiated Hint1 +/+ cells, there was a basal level of γ-H2AX, and that this level increased at 30 min, and then declined at 3 h after IR (Fig. 1 E). The levels of γ-H2AX in the Hint1 +/− and −/− cells at time 0 and at 3 h were greater than that in the +/+ cells. These results are consistent with those described in Fig. 1 C. We then used an antibody to acetyllysine in immunoblots to examine whether in these cells γ-H2AX is acetylated in response to IR, as is the case with its Drosophila orthologue (Kusch et al., 2004). There was a low level of acetylation of γ-H2AX in the untreated Hint1 +/+ cells. This increased at 30 min, and then declined at 1 h after IR. However, in both Hint1 +/− and −/− MEFs, the acetylation of γ-H2AX was completely abolished, with or without IR (Fig. 1 E). We also used the ac-H2A(K5) antibody to examine whether this acetylated γ-H2AX protein is specifically acetylated at the conserved lysine site because several other lysines are also reported to be acetylated after IR (Yamamoto and Horikoshi, 1997). In the Hint1 +/+ MEFs there was a basal level of acetylation of γ-H2AX at K5, and at 30 min after IR this level increased markedly, and then declined at 1 h. However, no evidence of acetylation of K5 was detected in the γ-H2AX protein in Hint1 +/− or −/− cells, before and after IR (Fig. 1 E). These findings suggest that HINT1 is haploinsufficient with respect to its role in mediating the acetylation of γ-H2AX because this acetylation is impaired in Hint1 +/− MEFs. The acetylation of γ-H2AX appears to be associated with the capability of removing γ-H2AX foci in the later stages of DDR because only the Hint1 +/+ MEFs, in which γ-H2AX is acetylated in response to IR, are able to remove γ-H2AX foci at 3 h. The Hint1 +/− and −/− MEFs, which are impaired in γ-H2AX acetylation, fail to remove γ-H2AX foci even 24 h after IR. As a result, these cells display prolonged γ-H2AX nuclear foci staining (Figs. 1 C). These findings are consistent with previous studies on the role of γ-H2AX acetylation in the removal of γ-H2AX foci in Drosophila cells (Kusch et al., 2004).
HINT1 is recruited to IRIF in response to γ radiation and interacts with γ-H2AX
The results described in the previous section clearly demonstrate that HINT1 plays a role in DDR and is involved in the chromatin remodeling pathway after IR. Because IR triggers the recruitment to discrete nuclear foci of several proteins that play a role in signaling DNA damage and repair (Paull et al., 2000), it was of interest to determine whether the HINT1 protein also forms IRIF. For this purpose a FLAG-tagged Hint1 expression plasmid was constructed and transiently expressed in Hint1 −/− MEFs. These cells were either untreated or treated with a 4-Gy dose of γ radiation and fixed at the indicated times. IRIF formation was analyzed by immunofluorescent staining with specific antibodies to the FLAG tag or γ-H2AX. We found that in untreated cells, HINT1 is localized predominantly in the cytoplasm, although diffuse staining of HINT1 was also observed in the nucleus (Fig. 2 A). The cytoplasmic patterns of FLAG-Hint1 staining did not change in response to IR. However, we observed remarkable changes of FLAG-Hint1 localization in the nucleus in response to IR. There were very few foci in untreated cells, and these foci did not colocalize with γ-H2AX. At 30 min after IR, HINT1 formed distinct nuclear foci that colocalized with γ-H2AX foci (Fig. 2 A). These foci remained detectable at 3 h and disappeared at 18 h after IR. Interestingly, the kinetics of HINT1 IRIF formation correlates well with that of γ-H2AX foci (Fig. 2 B). The fact that HINT1 is recruited to IRIF provides strong evidence that HINT1 plays an important role in DDR.
Figure 2.
The HINT1 protein is recruited to IRIF and associates with γ-H2AX. (A) Hint1 −/− MEFs were transiently transfected with FLAG-Hint1, grown on coverslips, and treated with 4 Gy of IR. Cells were then fixed after the indicated times and stained with an anti-FLAG antibody (green) together with an anti–γ-H2AX antibody (red) and counterstained for the nucleus with DAPI. Adjacent cells not expressing FLAG-Hint1 were used as an internal negative control. Repeat studies gave similar results. (B) The kinetics of HINT1 and γ-H2AX nuclear foci formation before and after IR treatment. For each genotype and time point, the number of foci per cell was analyzed in 20 images (error bars represent standard deviation). (C) Hint1 −/− MEFs were transiently transfected with FLAG-Hint1 or the control vector (p3xFLAG). 48 h later cells were treated with or without 4 Gy of IR, and whole cell lysates were prepared for immunoprecipitation using the FLAG antibody. Immunoblots were performed using either a FLAG antibody, a γ-H2AX antibody, or an ATM antibody. Repeat studies gave similar results. *, P < 0.05 compared with untreated cells.
To examine whether HINT1 binds to γ-H2AX, we immunoprecipitated the HINT1 protein from lysates of FLAG-Hint1–expressing MEFs using a FLAG antibody with or without prior radiation of these cells. We found that after, but not before, radiation, γ-H2AX coprecipitated with HINT1 (Fig. 2 C). Although the γ-H2AX band was weak this result was reproducible.
HINT1 is required for the acetylation of ATM and activation of the ATM checkpoint pathway
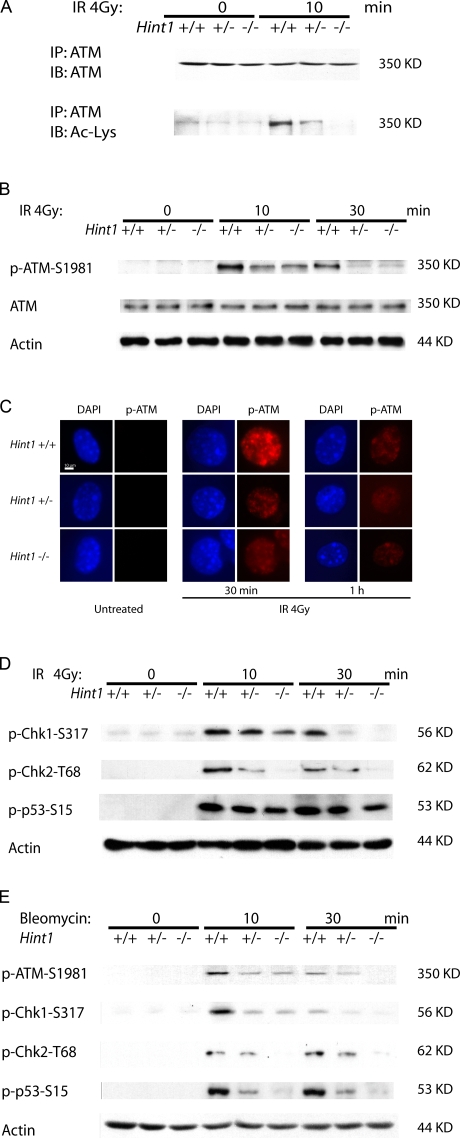
Our previous study showed that Hint1 −/− MEFs are more resistant to the cytotoxic effects of high doses of IR (Su et al., 2003). Because cells with extensive DNA damage are normally eliminated by apoptosis (van Gent et al., 2001), this resistance to IR-induced cytotoxicity of Hint1 −/− cells suggests that these cells are defective in this response. Therefore, the ATM checkpoint pathway of DDR was examined in HINT1-deficient cells. Because ATM is recruited to IRIF (Bakkenist and Kastan, 2003; Carson et al., 2003), we first examined whether HINT1 is associated with ATM. We immunoprecipitated the HINT1 protein from lysates of FLAG-Hint1–expressing MEFs, with or without prior radiation of these cells. We found that HINT1 is associated with ATM even before radiation, and after radiation this association is increased (Fig. 2 C). The impaired acetylation of γ-H2AX in HINT1-deficient cells prompted us to further examine whether HINT1 is also required for ATM acetylation. For this purpose the ATM protein was immunoprecipitated from cell lysates of Hint1 MEFs before and after treating the cells with 4 Gy of γ radiation, and then an acetyllysine antibody was used in immunoblots to detect acetylation of ATM. We found that IR caused a marked increase in ATM acetylation in the Hint1 +/+ cells. However, in Hint1 +/− MEFs the acetylation of ATM was reduced, and in the Hint1 −/− MEFs there was no detectable acetylation of ATM (Fig. 3 A). Therefore, HINT1 is required for the acetylation of ATM induced by IR.
Figure 3.
Loss of HINT1 impairs the acetylation and activation of ATM. (A) Hint1 MEFs were untreated or treated with 4 Gy of IR, and whole cell lysates were collected after 10 min. Immunoprecipitates were obtained using an anti-ATM antibody and resolved on SDS-PAGE. Immunoblots were then performed using either an ATM antibody or an acetyllysine antibody. (B) Hint1 +/+, +/−, and −/− MEFs were untreated (0) or treated with 4 Gy of IR. After the indicted times, cell lysates were collected and immunoblot analyses done for the levels of p-ATM-S1981 and ATM. Actin was used as an internal loading control. (C) Hint1 +/+, +/−, and −/− MEFs were untreated or treated with 4 Gy of IR and fixed after the indicated times. Cells were then stained with an antibody to p-ATM-S1981 (red) and counterstained for the nucleus with DAPI (blue). Bar, 10 μm. (D and E) Cell lysates from Hint1 MEFs untreated (0) or treated with 4 Gy of IR (D) or 100 nM bleomycin (E) were collected at the indicated times, and immunoblot analyses were done for the levels of p-Chk1-S317, p-Chk2-T68, and p-p53-S15. A repeated experiment yielded similar results.
One of the major roles of ATM in DDR is the phosphorylation of downstream targets that mediate activation of cell cycle checkpoints and apoptosis (Kitagawa and Kastan, 2005). Therefore, activation of this pathway was investigated in Hint1 +/+, +/−, and −/− MEFs after treatment with 4 Gy of γ radiation. Cell extracts were prepared and immunoblots were performed using a p-ATM-S1981 antibody to detect the active form of ATM. We found by immunoblots that ATM is activated in Hint1 +/+ MEFs at 10–30 min after IR; however, this activation was markedly reduced in both the Hint1 +/− and −/− cells (Fig. 3 B). At the same time, neither HINT1 deficiency nor IR altered the protein levels of ATM (Fig. 3 B). We also used an immunofluorescent staining method to examine the activation of ATM in intact cells, and the results were consistent with immunoblot results. Thus, treatment with 4 Gy of γ radiation caused a marked increase in the levels of p-ATM-S1981 at 30 min after IR in the Hint1 +/+ cells. The p-ATM protein formed distinct foci in these cells (Fig. 3 C). In the Hint1 +/+ cells there was a decrease in p-ATM at 1 h after IR, and at this time point the Hint1 −/− MEFs exhibited a further significant decrease in the level of p-ATM. At 1 h after IR, the level of p-ATM was also decreased in the irradiated Hint1 +/− MEFs when compared with the Hint1 +/+ cells, but this decrease was not statistically significant. This may be because of the increased signal/noise ratio at low fluorescence intensity.
We next examined whether the reduced activation of ATM also affects the activation of its downstream targets such as Chk1, Chk2, and p53. For this purpose phosphospecific antibodies against Chk1, Chk2, and p53 were used in immunoblots. As expected, the levels of p-Chk1-Ser317, p-Chk2-Thr68, and p-p53-Ser15 were markedly increased at 10 min after exposure of the cells to 4 Gy of IR. The levels of all three phosphoproteins were reduced in HINT1-deficient cells at this time point and also at 30 min after IR, when compared with the Hint1 +/+ cells (Fig. 3 D). This decrease was greater in the Hint1 −/− than the Hint1 +/− cells (Fig. 3 D). Similar effects were observed in a parallel study using 100 nM of the radiomimetic agent bleomycin as the DNA-damaging agent (Fig. 3 E).
The activation of p53 and other downstream effectors of ATM is reduced in HINT1-deficient MEFs
To identify genes that are differentially regulated in response to DSB in Hint1 MEFs we used Affymetrix GeneChip technology to analyze the gene expression profiles of these cells after IR. We found that the up-regulation of several DNA damage-inducible genes in response to IR was reduced in Hint1 +/− and −/− MEFs in response to IR (unpublished data). These genes include p21WAF1/CIP1, GADD45α, GADD153, and p53-inducible protein 1 (TP53inp1). To confirm these results, the mRNA levels of these genes were measured by real-time RT-PCR, using GAPDH as an internal control. We found that when compared with the Hint1 +/+ cells, there was a statistically significant (P < 0.05) decrease in activation of all four genes in Hint1 −/− MEFs at 48 h after treatment with 4 Gy of γ radiation (Fig. 4 A). In Hint1 +/− MEFs, there was also a significant decrease in induction of the expression of p21, GADD45α, and GADD153, but not TP53inp1 (Fig. 4 A). We also confirmed the real-time RT-PCR results on p21 by immunoblots of protein levels of p21 in untreated and 4-Gy γ radiation–treated Hint1 MEFs. The expression levels of p21 were similar in the three genotypes of untreated cells. 48 h after IR there was a marked increase in the protein level of p21 in Hint1 +/+ MEFs. However, induction of the p21 protein was reduced in Hint1 +/− cells and further reduced in Hint1 −/− cells (Fig. 4 B).
Figure 4.
Loss of HINT1 is associated with reduced activation of IR-inducible genes. (A) Hint1 MEFs were untreated or treated with 4 Gy of IR, and total RNA was collected after the indicated time points. The mRNA levels of p21, GADD45α, GADD153, and TP53inp1 were determined by real-time RT-PCR. (B) Whole cell lysates of control or IR (4 Gy)-treated Hint1 MEFs were collected for immunoblot analysis of the level of the p21 protein. Actin was used as a loading control. Error bars represent standard deviation from triplicate experiments. *, P < 0.05 when compared with Hint1 +/+ MEFs at each time point; §, P < 0.05 when compared with Hint1 +/− MEFs.
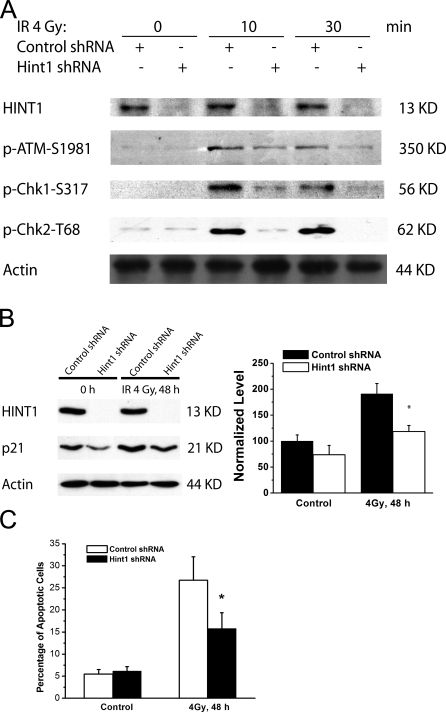
Because deficient expression of HINT1 causes decreased induction of the stress-inducible genes described in Fig. 4 A, it was of interest to determine whether HINT1-deficient cells are also impaired in induction of apoptosis. However, we were not able to detect a significant increase of apoptosis even in the Hint1 +/+ MEFs after treatment with 4 Gy of γ radiation (unpublished data). Therefore, we developed stable derivatives of SW480 colon cancer cells in which HINT1 is suppressed by transfection with a Hint1–short hairpin RNA (shRNA) vector (Weiske and Huber, 2005). For comparison we also developed a control SW480 derivative that had been transfected with a control shRNA vector. Expression of the HINT1 protein was nearly undetectable in Hint1-shRNA SW480 cells. Both cell lines displayed similar growth rate. These cells also exhibited reduced activation of ATM and downstream targets such as Chk1 and Chk2 in response to IR (Fig. 5 A). The level of the p21 protein was also reduced (Fig. 5 B). After treatment with 4 Gy of γ radiation, the level of p21 was induced in the control shRNA-transfected cells, whereas the induction of p21 was markedly reduced in the Hint1-shRNA–transfected cells (Fig. 5 B). Both the control and HINT1-deficient SW480 cells displayed activation of γ-H2AX with or without IR, perhaps reflecting the combined effects of loss of HINT1 expression and their transformed state (unpublished data). Using FACS flow cytometry we quantitated the sub-G1 population as an index of apoptosis. Without IR the percentage of apoptosis in both cell types was ∼5%. At 48 h after IR this rose to ∼25% in the control cells, but only to ∼15% in the Hint1-shRNA knockdown cells (Fig. 5 C). Therefore, decreased expression of HINT1 significantly impairs the induction of apoptosis by IR.
Figure 5.
Loss of HINT1 causes resistance to IR-induced apoptosis. SW480 cells were transfected with control shRNA or Hint1-shRNA and selected with G418, and then stable clones were pooled. (A) Whole cell lysates collected at the indicated times from untreated or IR (4 Gy)-treated pools were used for immunoblot analysis of HINT1, p-ATM-S1981, p-Chk1-S317, and p-Chk2-T68. Actin was used as a loading control. (B) Whole cell lysates of control or IR (4 Gy)-treated pools were collected for immunoblot analysis of the level of the p21 protein. Actin was used as a loading control. (C) Control SW480 pools and SW480 pools in which HINT1 was stably knocked down by shRNA were either untreated (0 h) or treated with 4 Gy of IR. Apoptotic cells were analyzed by sub-G1 content using a flow cytometry method. A repeat experiment yielded similar results. Error bars represent the standard deviation from triplicate experiments. *, P < 0.05 when compared with the control shRNA-transfected cells at each time point.
HINT1-deficient MEFs also display retarded DNA rejoining after γ radiation, chromosome aberrations, and genomic instability
To further address whether the spontaneous and prolonged γ-H2AX foci staining in the HINT1-deficient MEFs was caused by an inability to remove H2AX after DNA breaks had been repaired or by the presence of unrepaired DNA breaks, we compared DNA-rejoining capabilities of Hint1 +/+ and −/− MEFs. We first performed alkaline comet assays because this method is sensitive enough to detect low levels of DNA breaks. Hint1 MEFs were either mock treated or irradiated with a 4-Gy dose of γ radiation on ice, and then allowed to recover at 37°C for 0 min, 1 h, or 8 h. The tail moment was analyzed in 100 randomly chosen comets of the three genotypes of cells. Consistent with the spontaneous γ-H2AX foci staining, the unirradiated Hint1 −/− MEFs exhibited an increased tail moment compared with the Hint1 +/+ MEFs (Fig. 6, A and B). The Hint1 +/+ MEFs rejoined most of the DNA breaks after 1 h. However, the DNA breaks in the Hint1 −/− MEFs remained unrepaired at 8 h after exposure to IR (Fig. 6, A and B). Therefore, HINT1 deficiency also causes retarded DNA repair of spontaneous and IR-induced DNA strand breaks.
Figure 6.
HINT1-deficient MEFs display retarded DNA rejoining. (A and B) Analysis of time course kinetics of DNA rejoining by alkaline comet assay. Hint1 MEFs were irradiated with 4 Gy of γ radiation on ice and postincubated for the indicated recovery times. Comet slides were prepared, lysed in an alkaline buffer, and electrophoresed. Comet images were captured using a fluorescent microscope, and the tail moment was analyzed in 100 randomly chosen comets using comet analysis software. Representative comet images and the histogram of cell number versus tail moment (A) and the mean of tail moments observed at different times are shown (B). *, P < 0.005 compared with Hint1 +/+ MEFs (nonparametric Mann-Whitney test). A repeat experiment gave similar results. (C) PFGE analysis of DNA DSB repair in Hint1 MEFs. Hint1 MEFs were irradiated with 40 Gy of γ radiation on ice and postincubated for the indicated recovery times. Agarose plugs were prepared, cells were lysed by proteinase K, and genomic DNA was electrophoresed. The percentage of DSB unrepaired were calculated and DNA-PKcs −/− MEFs were used for comparison. Error bars represent the standard deviation from triplicate experiments. *, P < 0.05 compared with Hint1 +/+ MEFs (Student's t test).
To further address whether the HINT1-deficient cells exhibit defects specifically in rejoining DNA DSB, pulse-field gel electrophoresis (PFGE)–based DNA repair assays were performed to examine the DNA-rejoining ability of MEFs after exposure to IR. Hint1 MEFs were either mock treated or irradiated with a 40-Gy dose of γ radiation on ice, and then allowed to recover at 37°C for 0, 1 , 2, or 48 h. Chromosomal DNA was prepared for PFGE and unrepaired DNA DSB was determined following the procedure described previously (Block et al., 2004). For comparison, we also included DNA-dependent protein kinase catalytic subunit (DNA-PKcs)–deficient MEFs as a negative control because these cells are defective in nonhomologous end joining repair of DSB (Kurimasa et al., 1999a). We found that wild-type MEFs are able to repair most DNA DSB within 1 h, whereas both Hint1 −/− and DNA-PKcs −/− MEFs were not able to repair DNA DSB at 1 or 2 h after radiation (Fig. 6 C). Surprisingly, both the Hint1 −/− and DNA-PKcs −/− MEFs rejoined most of their DNA DSB at late time points such as 48 h after radiation (Fig. 6 C). It is possible that in DNA-PKcs −/− MEFs a back-up DNA repair pathway is involved in rejoining DNA DSB at a slower rate than DNA-PK–dependent nonhomologous end joining (Wu et al., 2008). Our results do not exclude the possibility that a low level of DSB is still present in both the Hint1 −/− and DNA-PKcs −/− MEFs at late time points, which cannot be detected by PFGE. In addition, PFGE cannot distinguish correctly rejoined DSB from misrejoined DSB. Nevertheless, the markedly slower repair of DNA DSB after IR in Hint1 −/− MEFs correlates with the persistent γ-H2AX foci, which reflect, at least in part, the persistence of DNA DSB, and not simply immobilized H2AX, at the repaired sites.
Collectively, our findings suggest that HINT1-deficient cells display defects in both DNA repair and ATM checkpoint pathways. To examine whether this is associated with genomic instability, micronuclei formation and chromosomal abnormalities were analyzed in the three genotypes of Hint1 MEFs. We first examined the frequency of abnormalities in unirradiated cells and found that the frequencies of micronuclei, unequal nuclear division, and nuclear blebbing were significantly higher in +/− and −/− cells than in +/+ cells (Fig. 7, A–C). No chromatin bridges or multinucleation were observed in the +/+ cells; however, these abnormalities were observed in both the +/− and −/− cells (Fig. 7 C). After treatment of these cells with 4 Gy of IR, there was a significant increase in the frequency of micronuclei formation in all three genotypes (Fig. 7 B). A substantial increase in micronuclei number per binucleated cell was also observed after IR treatment in the HINT1-deficient cells (Fig. 7 B).
Figure 7.
HINT1-deficient cells display increased chromosome aberrations. Hint1 MEFS were treated with or without a 4-Gy dose of IR and 18 h later were treated with cytochalasin B for 22 h. Cells were then fixed and stained with Acridine orange for the analysis of chromosomal abnormalities. (A) Representative images showing normal binuclear morphology of untreated Hint1 +/+ MEF and chromosome aberrations in untreated Hint1 −/− MEFs, including micronuclei (top right), unequal division (middle left), nuclear blebbing (middle right), chromatin bridge (bottom left), and multinucleation (bottom right). (B) Micronuclei frequency per binucleated cell was compared in Hint1 MEFs untreated or treated with 4 Gy of IR. (C) Chromosomal abnormality frequencies of the indicated type per binucleated cell were compared in Hint1 MEFs untreated or treated with 4 Gy of IR. (D) Telomere labeling of Hint1 +/+ and −/− MEFs. CF, centromeric fragment; CB, chromosome break; arrows, chromatid breaks. (E) Chromosomal analysis of Hint1 +/+ and −/− MEFs. *, P < 0.05 when compared with the +/+ MEFs (Fisher's exact test).
We next determined whether HINT1 deficiency is associated with chromosomal abnormalities. For this purpose, metaphase chromosomes of the three genotypes of MEFs were subject to telomere fluorescence in situ hybridization. A fluorescent Cy3-labeled peptide nucleic acid probe that is specific for telomeric sequences was used to localize telomeres (T-FISH). In addition, the chromosomes were counterstained with DAPI, which binds with high affinity to ataxia telangiectasia–rich centromeric heterochromatin regions of mouse chromosomes. A remarkable feature of the Hint1 −/− MEF metaphases was the increased frequency of spontaneous chromatid/chromosome breaks (Fig. 7, D and E, CB), as well as increased centromeric instability manifested as centromere fragments (Fig. 7, D and E, CF). Representative metaphase spreads obtained from Hint1 +/+ and −/− MEFs hybridized with the telomere-specific probe are shown (Fig. 7 D). Two prominent telomeric signals were observed in both arms of all of the chromosomes, and the centromeric fragments observed in Hint1 −/− cells frequently showed the telomeric signal.
Different types of spontaneous chromosomal anomalies were observed in Hint1 −/− MEFs, including breaks (chromosome and chromatid types), fragments (centric and acentric), and exchange type aberrations. The types and frequencies of spontaneous chromosomal aberrations observed per cell using T-FISH are presented in Fig. 7 E. The frequency of chromosome breaks per cell detected in Hint1 −/− cells was fourfold higher than in Hint1 +/+ MEFs (1.63 per metaphase for Hint1 −/− cells vs. 0.41 per metaphase for Hint1 +/+ cells). No exchange type aberrations were detected in Hint1 +/+ MEFs, but chromosomal exchanges and an increase in centromere fragments were observed in Hint1 −/− MEFs (Fig. 7 E). These results suggest that HINT1 deficiency causes a marked increase in spontaneous chromosomal abnormalities, presumably as a consequence of the impairments in DDR described in Fig. 7.
Discussion
HINT1 is a highly conserved protein in almost all known living species and it is ubiquitously expressed in all mammalian tissues (Klein et al., 1998; Li et al., 2006). Two proteins in the same HIT family as HINT1, namely FHIT (Ishii et al., 2006) and aprataxin (Ahel et al., 2006), have been implicated in DDR. The present study provides the first direct evidence that the tumor suppressor HINT1 plays an important role in modulating the appropriate responses of mammalian cells to DNA damage. Indeed, HINT1-deficient MEFs display defects in both the γ-H2AX chromatin remodeling pathway involved in the repair of DNA strand breaks and the ATM checkpoint pathway that can arrest cell cycle progression and induce apoptosis in response to DNA damage. Thus, we found that in response to IR, the HINT1 protein translocates to IRIF, where it associates with γ-H2AX foci (Fig. 2). The formation of γ-H2AX foci is normally a very early and transient response of cells to DSB (van Gent et al., 2001). Spontaneous formation of γ-H2AX foci was seen in several DNA repair–deficient cells, such as cells derived from ataxia telangiectasia or Nijmegen breakage syndrome patients (Kuhne et al., 2004). Interestingly, we found that HINT1-deficient MEFs also display γ-H2AX foci in the absence of irradiation (Fig. 1), which suggests that these cells have increased levels of spontaneous DNA damage. We found that HINT1-deficient MEFs display a marked persistence of γ-H2AX foci after exposure to IR (Fig. 1), which suggests that these cells are defective in DNA repair. Comet assay and PFGE-based DNA repair assays (Fig. 6) further confirmed the reduced DNA repair capability of HINT1-deficient MEFs compared with wild-type cells. These findings are of interest because another member of the HIT protein family, aprataxin, has recently been shown to play an important role in resolving abortive DNA ligation intermediates and in DNA repair (Ahel et al., 2006).
HINT1-deficient cells are also defective in activation of ATM in response to IR. Thus, we found that both acetylation and activation of ATM are significantly reduced in HINT1-deficient MEFs and in Hint1-shRNA knockdown derivatives of SW480 colon cancer cells, as manifested by decreased phosphorylation of the S1981 residue in ATM and decreased phosphorylation of its downstream effectors Chk1(S317), Chk2(T68), and p53(S15) (Fig. 3 and Fig. 5 A). The induction by IR of several DNA damage–inducible genes, including p21, GADD45α, GADD153, and TP53inp1, is also defective in HINT1-deficient MEFs (Fig. 4 A). Cellular levels of the p21 protein are also decreased in HINT1-deficient MEFs and in Hint1-shRNA SW480 cells (Fig. 4 B and Fig. 5 B). Thus, it appears that HINT1 normally plays an important role in the ATM checkpoint pathway that is activated in response to DNA damage. Despite these findings, HINT1-deficient MEFs are more resistant to the cytotoxicity of IR (Su et al., 2003) and HINT1-deficient SW480 derivatives are more resistant to the induction of apoptosis after IR than control SW480 cells (Fig. 5 C). These results appear to be paradoxical because loss of ATM activity is often associated with increased cellular sensitivity to DNA damage. However, reduced activation of ATM-mediated signaling pathways may also result in reduced apoptotic potential. Barlow et al. (1997) have demonstrated that ATM selectively regulates the p53-dependent cell cycle checkpoint and apoptotic pathways. Another study has demonstrated the requirement for both ATM and Bax gene products in IR-induced apoptosis (Chong et al., 2000). It has been recently found that inactivation of ATM reduces apoptosis in Myc-transgenic mice (Pusapati et al., 2006), and ATM loss impairs Myc apoptotic responses (Maclean et al., 2007). Cells with reduced apoptotic potential may be able to survive with a considerable amount of genomic instability. In a clonogenic survival assay, decreased sensitivity to increasing doses of γ radiation was found in Hint1 −/− MEFs (Su et al., 2003). Because this assay is based on the number of surviving colonies, the “decreased radiation sensitivity” observed with Hint1 −/− cells (Su et al., 2003) does not indicate that HINT1-deficient cells are also devoid of genomic instability. Because Hint1 +/− and −/− MEFs display a substantial amount of spontaneous and IR-induced genomic instability associated with micronuclei and nucleoplasmic bridges (Fig. 7), these findings are consistent with the observation that the apoptotic potential is impaired in HINT1-deficient cells. HINT1 may also directly affect targets downstream of ATM, including p53 and p21, thereby directly affecting the process of apoptosis. Indeed, overexpression of HINT1 induces PARP cleavage, activates caspase-3, and enhances expression of p53 (Weiske and Huber, 2006). Therefore, the combined effects of loss of HINT1 expression on cellular responses to DNA DSB could be complex, including transient responses such as reduced activation of ATM and downstream responses such as reduced apoptosis. The method used to assess the effects of HINT1 deficiency on sensitivity to the cytotoxic effects of IR (colony formation assay at 2–3 wk) in MEFs (Su et al., 2003) involves complex cellular processes, including clonal selection of resistant cells, and therefore may not directly address the sensitivity of HINT1-deficient cells to DNA DSB.
Although ATM acetylation is markedly decreased in Hint1 −/− cells, ATM activation (phosphorylation of downstream targets) is not completely inhibited (Fig. 3), perhaps because ATM can be activated by mechanisms other than acetylation but at a lower efficiency. Furthermore, p-ATM foci still colocalize with γ-H2AX in Hint1 −/− cells (unpublished data), and this may be sufficient for activating γ-H2AX because in HINT1-deficient MEFs the reduced activation of ATM does not affect the phosphorylation of H2AX (Fig. 1). Thus, the function of ATM may not be completely lost in these cells. Alternatively, activation of γ-H2AX in HINT1-deficient cells may be mediated by the DNA-PKcs because DNA-PKcs and ATM function redundantly in the phosphorylation of H2AX in response to IR, and γ-H2AX foci are formed even in ATM −/− cells (Stiff et al., 2004). Whether deficiency of DNA DSB repair in Hint1 −/− MEFs was caused by insufficient activation of ATM is not clear, and it will be of interest to study whether in Hint1 −/− MEFs the retarded DNA DSB repair is caused by inefficient nonhomologous end joining or compromised mechanism of other repair pathways.
It was of interest that, despite the fact that the Hint1 −/− MEFs have defects in the DSB repair pathway, including loss of ATM activation and the presence of unrepaired DSB, Hint1 −/− cells are more resistant to cytotoxicity by IR than Hint +/+ cells (Su et al., 2003). It is possible that in HINT1-deficient cells the functions of ATM are fine tuned so that proapoptotic effects induced by IR are inhibited whereas prosurvival effects are not. It is also possible that HINT1 has other activities that are required for inducing apoptosis after IR. Nevertheless, genetic or epigenetic inactivation of HINT1 might contribute to the development of radioresistant tumors, and, on the other hand, transient inactivation of HINT activity might be a strategy for developing compounds that protect normal cells from the cytotoxicity caused by IR.
There is increasing evidence that protein acetylation plays an important role in regulating DDR (Squatrito et al., 2006). Perhaps the most novel aspect of the present study is our finding that HINT1-deficient cells are impaired in the acetylation of both γ-H2AX and ATM in response to DNA damage (Fig. 1 D and Fig, 3 A) and that HINT1 associates with ATM and γ-H2AX (Fig. 2 C). These findings suggest that HINT1 may be important for mediating and coordinating the acetylation events necessary to induce both the early (ATM activation) and late (removal of γ-H2AX) steps of DDR. Additional studies are required to determine the mechanism by which HINT1 influences the state of acetylation of ATM and γ-H2AX. HINT1 has intrinsic hydrolase activity for phosphoramidate and acyladenylate (Bieganowski et al., 2002; Chou et al., 2007), but further studies are required to determine if these activities are relevant to its functions in DDR pathways.
Collectively, the defects in the γ-H2AX chromatin remodeling pathway in HINT1-deficient cells, combined with the defects in the ATM checkpoint pathway, may explain why HINT1-deficient MEFs display an increase in various types of chromosomal abnormalities and genomic instability (Fig. 7). These findings confirm the haploinsufficient function of HINT1 at a mechanistic level. It was of interest that the biological and cellular effects caused by decreased levels of HINT1 are not entirely dose dependent. Thus, Hint1 +/− MEFs exhibit decreased acetylation of γ-H2AX to an extent similar to Hint1 −/− cells, and they also display prolonged activation of γ-H2AX similar to Hint1 −/− cells (Fig. 1). In contrast, we did observe dose dependency for several other cellular functions of HINT1, such as decreased activation of ATM, p21, and other p53 target genes (Figs. 4 and 5). Recent studies indicate that the effects of gene dosage can be complex and cannot be linearly correlated to expression levels (Kondrashov and Koonin, 2004). This is particularly relevant to genes encoding subunits of protein complexes because stoichiometry imbalance can cause alterations of interaction with the target element, which leads to an abnormal phenotype (Birchler et al., 2005). The HINT1 protein forms a stable homodimer (Lima et al., 1996) and possibly functions in various protein complexes (Korsisaari and Makela, 2000; Guang et al., 2004; Weiske and Huber, 2006; Ajit et al., 2007; Wang et al., 2007) and, therefore, stoichiometry balance may be important for its functions. Because of the strong haploinsufficient phenotype of Hint1 +/− cells, loss of heterozygosity in the Hint1 gene or decreased HINT1 expression of human tissues may increase the risk of tumor development in these tissues. Recent promoter methylation studies indicate that the Hint1 gene promoter is partially methylated in several lung cancer and colon cancer cell lines (Yuan et al., 2004; Wang et al., 2007), and that this is associated with decreased expression of HINT1 RNA and protein. Because HINT1 plays an important role in DDR, this decreased expression of HINT1 may increase the likelihood of genome instability and thereby enhance tumor development.
Materials and methods
Cell lines
Hint1 +/− mice were mated to generate Hint1 +/+, +/−, and −/− embryos and each genotyped as previously described (Li et al., 2006). Preparation of MEFs was also described previously (Su et al., 2003). For each experiment cells were pooled from at least four embryos. Repeat experiments were performed using different pools. Passage 4 MEFs were used in all experiments, except where otherwise indicated. SW480 cells (American Type Culture Collection) were transfected with a plasmid encoding a control shRNA or a Hint1-shRNA, using the Lipofectin reagent (Invitrogen) following the manufacturer's protocol, and selected under G418 treatment for 3 wk. Resistant clones were pooled and used for subsequent studies. DNA-PKcs −/− MEFs (PK33N) were provided by D. Chen (The University of Texas Southwestern Medical Center at Dallas, Dallas, TX; Kurimasa et al., 1999b).
Chemicals and antibodies
Bleomycin, colchicine, and cytoclasin B were purchased from Sigma-Aldrich. The HINT1 antibody was previously described (Klein et al., 1998). The ATM and TIP60 antibodies were purchased from Santa Cruz Biotechnology, Inc.; histone H2A and γ-H2AX antibodies from Millipore; ac-H2A(K5) antibody from Abcam; p-ATM-S1981, p-Chk1-S317, p-Chk2-T68, p-p53-S15, and p21 antibodies from Cell Signaling Technology; acetyllysine and Rad50 antibodies from Millipore; actin, FLAG, and HA antibodies from Sigma-Aldrich; HRP-conjugated anti–mouse IgG or anti–rabbit IgG secondary antibodies from GE Healthcare; and FITC-conjugated anti–rabbit IgG, Texas red–conjugated anti–rabbit IgG, and Texas red–conjugated anti–mouse IgG secondary antibodies from Vector Laboratories. Immunoblot analysis procedures were described previously (Li et al., 2006).
Plasmids
To construct FLAG-Hint1 expression constructs, the full-length Hint1 cDNA was amplified by PCR from pcDNA3-Hint1 (Klein et al., 1998) and subcloned into the p3xFLAG-CMV-7.1 vector (Sigma-Aldrich). The plasmids encoding a control shRNA or a Hint1-shRNA were provided by O. Huber (Institute of Clinical Chemistry and Pathobiochemistry, Berlin, Germany; Weiske and Huber, 2005).
Immunofluorescent microscopy
Cells were grown on coverslips and fixed in acetone-methanol at a ratio of 1:1, or in 4% paraformaldehyde, and prepermeabilized with 0.5% Triton X-100. Coverslips were blocked with 5% skim milk and incubated with the indicated primary antibodies for 1 h at 37°C, followed by incubation with a FITC-conjugated anti–rabbit IgG and/or a Texas red–conjugated anti–mouse IgG. Translocation of FLAG-Hint1 was detected by double staining using a polyclonal FLAG antibody and a monoclonal γ-H2AX antibody, after the Hint1 −/− MEFs were transfected with FLAG-Hint1 using PlasFect reagent (Bioline) following the manufacturer's protocol, and treated with 4 Gy of γ radiation. Images were analyzed using a fluorescent microscope (Nikon) with a Nikon PlanFluor 40× oil objective lens (NA 1.30).
Acid extraction of histones
Exponentially growing cells were treated as described and at indicated time points were collected in lysis buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and 1.5 mM PMSF), and 1 N HCl was used to adjust final concentration to 0.1 N. Samples were incubated on ice for 30 min and centrifuged for 10 min at 14,000 g at 4°C. The supernatant was dialyzed at 4°C against 0.1 M acetate for 1 h twice and three times in ddH2O for 1 h, 3 h, and overnight, respectively. The dialyzed samples were then subject to immunoblot analysis.
Immunoprecipitation
For immunoprecipitation of ATM, whole cell lysates were prepared from Hint1 MEFs after IR and incubated with an ATM antibody. For immunoprecipitation of H2AX, cells were lysed in modified radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2.5 mM MgCl2, 0.5 mM CaCl2, 0.1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM DTT, 25% glycerol, and 1 μM MS-275) with EDTA-free Halt protease and phosphatase inhibitor cocktails (Thermo Fisher Scientific) and sonicated on ice for 10 s twice. DNase was added to the extract and then centrifuged, and the supernatant was diluted to a protein concentration of 0.1 mg/ml for immunoprecipitation with a γ-H2AX antibody. The precipitates were subjected to immunoblotting with the indicated antibodies. For the association of HINT1 and γ-H2AX or ATM, whole cell lysates from FLAG-Hint1 expressing MEFs, with or without IR, were incubated with an anti-FLAG antibody, and precipitates were subject to immunoblotting with a FLAG or specific antibodies to γ-H2AX or ATM.
Real-time RT-PCR
Real-time quantitative RT-PCR was performed on two technical replicates of at least two biological sample replicates, using the MX3000 system (Stratagene). Primer sequences used are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200711150/DC1).
Alkaline comet assay
Passage 2 Hint1 MEFs were mock treated or irradiated on ice (4 Gy). Cells were allowed to recover at 37°C for the indicated times, and alkaline comet assay was performed using the CometAssay kit (Trevigen) following the manufacturer's protocol. Comet images stained with 1 μg/ml SYBRGreen were captured using a fluorescent microscope (Nikon) with a Nikon PlanFluor 20× objective lens (NA 0.5). The tail moment was analyzed using Euclid comet analysis software.
PFGE-based DSB repair assay
Cells undergoing exponential growth were irradiated with 40 Gy and then allowed to recover at 37°C for the indicated times. PFGE assays were performed using a CHEF DRII system (Bio-Rad Laboratories) with a 120° reorientation angle, following the manufacturer's protocol. In brief, cells were trypsinized and washed with PBS twice, resuspended in 50 μl of cell suspension buffer (10 mM Tris-HCl, pH 7.2, 20 mM NaCl, and 50 mM EDTA) at a concentration of 108 cells/ml, and mixed with equal volumes of 2% agarose to form plugs. The plugs were incubated with lysis buffer (100 mM EDTA, pH 8.0, 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, and 1 mg/ml proteinase K) for 15 h at 50°C, and then washed in wash buffer (20 mM Tris-HCl, pH 8.0, and 50 mM EDTA) four times. Plugs were inserted into wells of a 1% agarose gel and subjected to PFGE in 0.5× TBE buffer at 3 V/cm for 48 h with linearly increasing switch times of 50–5,000 s at 12°C. Gels were stained with ethidium bromide and visualized under UV illumination. Images were captured using a digital camera (Canon) and ethidium bromide staining was quantitated using ImageJ. The percentage of unrepaired DSB was estimated as follows: [A/(A + B)]/[Ao/(Ao + Bo)] × 100, where A is DNA migrating into the gel, B is DNA retained within the plug, Ao is DNA migrating into the gel when no repair was allowed, and Bo is DNA retained within the plug when no repair was allowed, as previously described (Block et al., 2004).
Micronuclei analysis and chromosome aberration analysis
Passage 2 Hint1 MEFs were grown on coverslips with or without treatment of a 4-Gy dose of IR. 3–6 μg/ml of cytochalasin B was added to the cultures at 18 h after treatment. At 22 h after adding cytochalasin B, cells were collected and micronuclei analysis was performed as described previously (Balajee and Geard, 2004). Cell were stained with Acridine orange and observed for micronuclei and other chromosome aberrations under a fluorescent microscope using a DAPI/FITC/TRITC triple filter (Nikon) with a PlanFluor 20× objective lens (NA 0.5; Nikon). Results from multiple experiments of micronuclei analysis or chromosome aberration analysis were pooled together to obtain higher power of statistical analysis.
Telomere labeling
Exponential growing passage 2 Hint1 +/+ and −/− MEFs were treated with 0.3 μg/ml colchicine for 12 h and then fixed and stained as described previously (Shen et al., 2007). The images were captured using an imaging microscope (EZ.C1; Axioplan 2; Carl Zeiss, Inc.), with a 63× oil objective lens (NA 1.40; Apochromat; Carl Zeiss, Inc.).
Statistical analysis
Data from real-time RT-PCR, immunoblot, foci analysis, apoptosis analysis, and PFGE-based DSB repair assays were analyzed using Student's t test. Standard deviations are shown in the corresponding figures. In comet assays, because the distribution of tail moments was not Gaussian, the nonparametric Mann-Whitney test was used for statistical analysis. Fisher's exact test was used in micronuclei analysis and metaphase analysis by telomere labeling. The p-values are indicated in the text and/or figures, and a difference between groups of P < 0.05 was considered statistically significant.
Online supplemental material
Table S1 shows the primer sequences used in real-time RT-PCR experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200711150/DC1.
Supplementary Material
Acknowledgments
We thank R. Baer and W. Gu for critical comments on the manuscript; D. Chen for providing the DNA-PKcs −/− MEFs; O. Huber for the Hint1-shRNA plasmid; B.M. Sutherland, R. Rothstein, and R.J. Reid for assistance with the PFGE assays; and X. Wang, W.Q. Xing, Y. Jin, and J.A. Meador for technical assistance.
This work was supported by the Environmental Protection Agency (X97279801-0 to H. Li); the Office of Science, Office of Biological and Environmental Research, US Department of Energy (grant DE-FG02-05ER64055 to A.S. Balajee); the National Institutes of Health (grants ES 12888 and ES 09089 to T.K. Hei); and the T.J. Martell Foundation, the National Foundation for Cancer Research, and the Entertainment Industry Foundation (I.B. Weinstein).
Abbreviations used in this paper: DDR, DNA damage responses; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSB, double strand breaks; IR, ionizing radiation; IRIF, Ionizing radiation-induced foci; MEF, mouse embryonic fibroblast; PFGE, pulse-field gel electrophoresis; shRNA, short hairpin RNA.
References
- Ahel, I., U. Rass, S.F. El-Khamisy, S. Katyal, P.M. Clements, P.J. McKinnon, K.W. Caldecott, and S.C. West. 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 443:713–716. [DOI] [PubMed] [Google Scholar]
- Ajit, S.K., S. Ramineni, W. Edris, R.A. Hunt, W.T. Hum, J.R. Hepler, and K.H. Young. 2007. RGSZ1 interacts with protein kinase C interacting protein PKCI-1 and modulates mu opioid receptor signaling. Cell. Signal. 19:723–730. [DOI] [PubMed] [Google Scholar]
- Bakkenist, C.J., and M.B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506. [DOI] [PubMed] [Google Scholar]
- Balajee, A.S., and C.R. Geard. 2004. Replication protein A and gamma-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Exp. Cell Res. 300:320–334. [DOI] [PubMed] [Google Scholar]
- Barlow, C., K.D. Brown, C.X. Deng, D.A. Tagle, and A. Wynshaw-Boris. 1997. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 17:453–456. [DOI] [PubMed] [Google Scholar]
- Bieganowski, P., P.N. Garrison, S.C. Hodawadekar, G. Faye, L.D. Barnes, and C. Brenner. 2002. Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3. J. Biol. Chem. 277:10852–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J.A., N.C. Riddle, D.L. Auger, and R.A. Veitia. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21:219–226. [DOI] [PubMed] [Google Scholar]
- Bird, A.W., D.Y. Yu, M.G. Pray-Grant, Q. Qiu, K.E. Harmon, P.C. Megee, P.A. Grant, M.M. Smith, and M.F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 419:411–415. [DOI] [PubMed] [Google Scholar]
- Block, W.D., D. Merkle, K. Meek, and S.P. Lees-Miller. 2004. Selective inhibition of the DNA-dependent protein kinase (DNA-PK) by the radiosensitizing agent caffeine. Nucleic Acids Res. 32:1967–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, C.T., R.A. Schwartz, T.H. Stracker, C.E. Lilley, D.V. Lee, and M.D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, M.J., M.R. Murray, E.C. Gosink, H.R. Russell, A. Srinivasan, M. Kapsetaki, S.J. Korsmeyer, and P.J. McKinnon. 2000. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc. Natl. Acad. Sci. USA. 97:889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T.F., I.B. Tikh, B.A. Horta, B. Ghosh, R.B. De Alencastro, and C.R. Wagner. 2007. Engineered monomeric human histidine triad nucleotide-binding protein 1 hydrolyzes fluorogenic acyl-adenylate and lysyl-tRNA synthetase-generated lysyl-adenylate. J. Biol. Chem. 282:15137–15147. [DOI] [PubMed] [Google Scholar]
- Chowdhury, D., M.C. Keogh, H. Ishii, C.L. Peterson, S. Buratowski, and J. Lieberman. 2005. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 20:801–809. [DOI] [PubMed] [Google Scholar]
- Downs, J.A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S.J. Kron, S.P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 16:979–990. [DOI] [PubMed] [Google Scholar]
- Downs, J.A., M.C. Nussenzweig, and A. Nussenzweig. 2007. Chromatin dynamics and the preservation of genetic information. Nature. 447:951–958. [DOI] [PubMed] [Google Scholar]
- Guang, W., H. Wang, T. Su, I.B. Weinstein, and J.B. Wang. 2004. Role of mPKCI, a novel mu-opioid receptor interactive protein, in receptor desensitization, phosphorylation, and morphine-induced analgesia. Mol. Pharmacol. 66:1285–1292. [DOI] [PubMed] [Google Scholar]
- Ikura, T., S. Tashiro, A. Kakino, H. Shima, N. Jacob, R. Amunugama, K. Yoder, S. Izumi, I. Kuraoka, K. Tanaka, et al. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27:7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, H., K. Mimori, H. Inoue, T. Inageta, K. Ishikawa, S. Semba, T. Druck, F. Trapasso, K. Tani, A. Vecchione, et al. 2006. Fhit modulates the DNA damage checkpoint response. Cancer Res. 66:11287–11292. [DOI] [PubMed] [Google Scholar]
- Keogh, M.C., J.A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J.C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, et al. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 439:497–501. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and M.B. Kastan. 2005. The ATM-dependent DNA damage signaling pathway. Cold Spring Harb. Symp. Quant. Biol. 70:99–109. [DOI] [PubMed] [Google Scholar]
- Klein, M.G., Y. Yao, E.D. Slosberg, C.D. Lima, Y. Doki, and I.B. Weinstein. 1998. Characterization of PKCI and comparative studies with FHIT, related members of the HIT protein family. Exp. Cell Res. 244:26–32. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., and E.V. Koonin. 2004. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20:287–290. [DOI] [PubMed] [Google Scholar]
- Korsisaari, N., and T.P. Makela. 2000. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J. Biol. Chem. 275:34837–34840. [DOI] [PubMed] [Google Scholar]
- Kuhne, M., E. Riballo, N. Rief, K. Rothkamm, P.A. Jeggo, and M. Lobrich. 2004. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 64:500–508. [DOI] [PubMed] [Google Scholar]
- Kurimasa, A., S. Kumano, N.V. Boubnov, M.D. Story, C.S. Tung, S.R. Peterson, and D.J. Chen. 1999. a. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimasa, A., H. Ouyang, L.J. Dong, S. Wang, X. Li, C. Cordon-Cardo, D.J. Chen, and G.C. Li. 1999. b. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc. Natl. Acad. Sci. USA. 96:1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W.H. Macdonald, S.K. Swanson, R.L. Glaser, J.R. Yates III, S.M. Abmayr, M.P. Washburn, and J.L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 306:2084–2087. [DOI] [PubMed] [Google Scholar]
- Li, H., Y. Zhang, T. Su, R.M. Santella, and I.B. Weinstein. 2006. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene. 25:713–721. [DOI] [PubMed] [Google Scholar]
- Lima, C.D., M.G. Klein, I.B. Weinstein, and W.A. Hendrickson. 1996. Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins. Proc. Natl. Acad. Sci. USA. 93:5357–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean, K.H., M.B. Kastan, and J.L. Cleveland. 2007. Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Mol. Cancer Res. 5:705–711. [DOI] [PubMed] [Google Scholar]
- Maser, R.S., K.J. Monsen, B.E. Nelms, and J.H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr, R., J.I. Loizou, Y.G. Yang, C. Cuenin, H. Li, Z.Q. Wang, and Z. Herceg. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8:91–99. [DOI] [PubMed] [Google Scholar]
- Nazarov, I.B., A.N. Smirnova, R.I. Krutilina, M.P. Svetlova, L.V. Solovjeva, A.A. Nikiforov, S.L. Oei, I.A. Zalenskaya, P.M. Yau, E.M. Bradbury, and N.V. Tomilin. 2003. Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat. Res. 160:309–317. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., J.E. Krebs, and C.L. Peterson. 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20:2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T.T., and J.H. Lee. 2005. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 4:737–740. [DOI] [PubMed] [Google Scholar]
- Paull, T.T., E.P. Rogakou, V. Yamazaki, C.U. Kirchgessner, M. Gellert, and W.M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895. [DOI] [PubMed] [Google Scholar]
- Pusapati, R.V., R.J. Rounbehler, S. Hong, J.T. Powers, M. Yan, K. Kiguchi, M.J. McArthur, P.K. Wong, and D.G. Johnson. 2006. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc. Natl. Acad. Sci. USA. 103:1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou, E.P., D.R. Pilch, A.H. Orr, V.S. Ivanova, and W.M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., C. Boon, C. Redon, and W.M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H., A.S. Balajee, J. Wang, H. Wu, C. Eng, P.P. Pandolfi, and Y. Yin. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 128:157–170. [DOI] [PubMed] [Google Scholar]
- Squatrito, M., C. Gorrini, and B. Amati. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16:433–442. [DOI] [PubMed] [Google Scholar]
- Stiff, T., M. O'Driscoll, N. Rief, K. Iwabuchi, M. Lobrich, and P.A. Jeggo. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- Su, T., M. Suzui, L. Wang, C.S. Lin, W.Q. Xing, and I.B. Weinstein. 2003. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc. Natl. Acad. Sci. USA. 100:7824–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., X. Jiang, S. Chen, N. Fernandes, and B.D. Price. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA. 102:13182–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent, D.C., J.H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196–206. [DOI] [PubMed] [Google Scholar]
- Wang, L., Y. Zhang, H. Li, Z. Xu, R.M. Santella, and I.B. Weinstein. 2007. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 67:4700–4708. [DOI] [PubMed] [Google Scholar]
- Weiske, J., and O. Huber. 2005. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J. Cell Sci. 118:3117–3129. [DOI] [PubMed] [Google Scholar]
- Weiske, J., and O. Huber. 2006. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J. Biol. Chem. 281:27356–27366. [DOI] [PubMed] [Google Scholar]
- Wu, W., M. Wang, S.K. Singh, T. Mussfeldt, and G. Iliakis. 2008. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair (Amst.). 7:329–338. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., and M. Horikoshi. 1997. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 272:30595–30598. [DOI] [PubMed] [Google Scholar]
- Yuan, B.Z., A.M. Jefferson, N.C. Popescu, and S.H. Reynolds. 2004. Aberrant gene expression in human non small cell lung carcinoma cells exposed to demethylating agent 5-aza-2'-deoxycytidine. Neoplasia. 6:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.