Abstract
Survivin is a key cellular protein thought to function in apoptotic regulation, mitotic progression, or possibly both. In this study, we describe the isolation of two conditional knockouts of the survivin gene in chicken DT40 cells. DT40 cells lacking Survivin die in interphase after failing to complete cytokinesis. However, these cells show normal sensitivity to the chemotherapeutic agent etoposide. Expression of Survivin mutants against a null background to reassess the role of several key residues reveals that DT40 cells can grow normally if their sole Survivin is missing a widely studied cyclin-dependent kinase phosphorylation site or sites reportedly essential for binding to Smac or aurora B. Mutations in the nuclear export sequence or dimerization interface render cells temperature sensitive for growth. As an important caveat for other studies in which protein function is studied by transient transfection, three of the Survivin mutants fail to localize in the presence of the wild-type protein but do localize and indeed support life in its absence.
Introduction
The chromosomal passenger protein complex (CPC), a key regulator of mitosis consisting of aurora B kinase, inner centromere protein (INCENP), Survivin, and Borealin/Dasra B (Cooke et al., 1987; Adams et al., 2000; Gassmann et al., 2004; Ruchaud et al., 2007), is essential for correction of kinetochore attachment errors, completion of cytokinesis, and numerous other mitotic functions (Ruchaud et al., 2007).
Survivin is a cell cycle–regulated protein whose expression peaks in mitosis (Li et al., 1998; for reviews see Wheatley and McNeish, 2005; Lens et al., 2006). Survivin forms both a dimer (Chantalat et al., 2000; Muchmore et al., 2000) and a three-helix bundle with the N terminus of INCENP and the N terminus of Borealin/Dasra B (Bourhis et al., 2007; Jeyaprakash et al., 2007). In the bundle, Survivin is a monomer, with Borealin docking to the surface that forms the interface in Survivin homodimers. The three-helix bundle is essential for CPC targeting and function in mitosis.
Survivin helps mediate the mitotic localization of the CPC (Carvalho et al., 2003; Klein et al., 2006; Knauer et al., 2006; Vader et al., 2006) and may contribute to aurora B activity in Xenopus laevis and fission yeast (Bolton et al., 2002; Petersen and Hagan, 2003), although this has been challenged (Honda et al., 2003). Survivin and its budding yeast homologue Bir-1 are required for spindle checkpoint function (Carvalho et al., 2003; Lens et al., 2003; Petersen and Hagan, 2003). However, the exact role of Survivin in mitosis remains controversial.
Survivin is an inhibitor of apoptosis protein (IAP) with a single baculovirus IAP repeat (BIR) domain and has been proposed to link cell proliferation and cell death (Li et al., 1998; for reviews see Wheatley and McNeish, 2005; Altieri, 2006). Unlike IAPs involved in apoptosis control, Survivin lacks a C-terminal RING finger and contains only one BIR domain (residues 18–88; Crook et al., 1993; Ambrosini et al., 1997).
Survivin is overexpressed in many tumors (Ambrosini et al., 1997; Li, 2003), and cells overexpressing the protein are resistant to many apoptotic stimuli. Conversely, loss of Survivin expression or function can cause spontaneous apoptosis or sensitize cancer cells to apoptotic stimuli (Li et al., 1998; Mahotka et al., 1999; Jiang et al., 2001; Mirza et al., 2002; Carvalho et al., 2003; Temme et al., 2003; Beltrami et al., 2004; Song et al., 2004). Survivin may regulate caspase-3 activity (Tamm et al., 1998; Li et al., 1999; Conway et al., 2000; Shin et al., 2001), but it does not inhibit caspase-3 directly (Banks et al., 2000). Survivin homologues in Schizosaccharomyces pombe (Uren et al., 1999; Rajagopalan and Balasubramanian, 2002), Caenorhabditis elegans (Fraser et al., 1999; Speliotes et al., 2000), Xenopus laevis (Bolton et al., 2002), and mice (Uren et al., 2000) lack obvious antiapoptotic functions (but see Walter et al., 2006). However, Drosophila melanogaster deterin can exhibit antiapoptotic activity in transfected cells (Jones et al., 2000), and murine Survivin is essential for thymocyte development (Okada et al., 2004).
The role of Survivin in mitosis and apoptosis remains unclear, possibly because Survivin is studied in numerous cell types under a wide range of experimental conditions and usually in the presence of the wild-type protein. In this study, we describe a conditional knockout of Survivin in DT40 cells. Our results support some of the published conclusions about Survivin function; however, several structural features previously reported to be essential for Survivin function turn out to be nonessential for cell viability when examined against a null background.
Results
Isolation of Survivin conditional knockout cells
We deleted the entire 725-bp ORF encoding survivin in chicken DT40 B lymphocytes (Fig. 1 A; Buerstedde and Takeda, 2006). Two knockouts were isolated. The first wild-type allele was replaced with a neomycin (KO1) or histidinol (KO2) selectable marker. Heterozygotes were cotransfected with two constructs, one encoding the tetracycline transactivator (tTA) plus a second with a survivin cDNA under control of a tTA-responsive promoter (tetO). The remaining allele was replaced with a histidinol (KO1) or puromycin (KO2) selectable marker (Fig. 1 C). The two knockouts differed in the control of the tTA transcription factor, which was under the control of the strong cytomegalovirus promoter in KO1 and the much weaker cellular chicken KIF4 promoter in KO2 (Fig. 1 B). Addition of doxycycline blocks tTA binding to the promoter driving the rescue cDNA, resulting in the shutoff of wild-type Survivin expression.
Figure 1.
Generation of Survivin conditional knockout in DT40 cells. (A) Diagram of the survivin locus and targeting vectors used. Arrowheads, EcoRI cleavage sites. (B) Strategy for rescue and shutoff of Survivin expression. (C) Table showing constructs used for the two knockout cell lines. (D) Southern blot of wild-type, heterozygote, and Survivin-null clones. EcoRI-digested genomic DNA was hybridized with the 5′ external probe (red bar) shown in A. Superscripts 1 and 2 refer to heterozygotes from knockouts 1 and 2, respectively. (E) Repression of rescue Survivin mRNA confirmed by RT-PCR of total RNA from heterozygote and KO1 cells incubated with doxycycline. (F) Real-time PCR confirms survivin repression by doxycycline. Values were normalized relative to actin mRNA. Values for cells grown in doxycycline are shown as striped bars. Error bars indicate SD. (G) Immunoblotting analysis of Survivin repression for KO1. 20 μg of whole cell lysate from DT40 (wild type [WT]) and SurvivinOFF cells was subjected to 12.5% SDS-PAGE and probed with affinity-purified polyclonal anti-Survivin antibody. Loading control, anti–α-tubulin. (H) Immunoblotting analysis of Survivin repression for KO2 performed as for G.
Specific targeting events were confirmed by Southern blotting using EcoRI digestion and a 5′ external probe (Fig. 1, A and D). For KO1, the probe recognized a 4.1-kb band corresponding to the wild-type survivin allele and a 4.9-kb or 6-kb band after targeted integration of the histidinolres or neomycinres constructs, respectively. For KO2, the correctly targeted alleles gave bands of 4.9 (histidinolres) and 4.8 kb (puromycinres).
Conventional and quantitative RT-PCR confirmed that doxycycline addition caused a rapid and dramatic decrease in expression of the survivin rescue cDNA, which could be monitored by RT-PCR, as it carries a 36-bp deletion in the 3′ untranslated region. survivin mRNA levels fell by 75% at 4 h after doxycycline addition and by 99% at 24 h (Fig. 1, E and F).
The level of Survivin protein in KO2 cultures growing without doxycycline was similar to or slightly below that in wild-type cells (Fig. 1 H, 0 h), whereas in KO1, Survivin was substantially overexpressed (Fig. 1 G). Remarkably, the growth of KO1 and KO2 cells appeared to be identical; thus, an excess of the canonical isoform of Survivin did not appear to confer a growth advantage on KO1 cells (Fig. 2 C). Doxycycline addition caused a rapid drop in Survivin levels, which became essentially undetectable by 36 h in KO2. Loss of Survivin was more variable for KO1, and in some experiments cells died before the protein was completely lost from cultures. Indirect immunofluorescence staining of KO2 cells using antibody against chicken Survivin showed no detectable Survivin signal during mitosis after 60 h in doxycycline (Fig. 2, A″ and A‴).
Figure 2.
Phenotype of cells after Survivin shutoff. (A) Distribution of Survivin (red), α-tubulin (green), and DNA (blue) in SurvivinON (A and A′) and SurvivinOFF (A″ and A‴) mitotic cells. Bar, 5 μm. (B) Phase-contrast images of SurvivinON and SurvivinOFF cultures (the latter after 60-h growth in doxycycline). Bar, 10 μm. (C) Cells from both survivin knockouts KO1 and KO2 die after exposure to doxycycline. (D) Annexin V–positive (apoptotic) cells appear 36 h after shutoff of survivin transcription in KO1. (E) INCENP (red) is diffuse, but kinetochores (CENP-H–GFP, green) appear normal in SurvivinOFF cells (bottom). Top, SurvivinON control. Bar, 5 μm. (F) INCENP and H3S10ph levels decrease shortly after Survivin levels drop after addition of doxycycline to KO1 cells. Loading control, anti–α-tubulin. WT, wild type. (G) Micrograph showing decreased H3S10ph staining (green) in SurvivinOFF cells (bottom). Top, SurvivinON control. Bar, 5 μm.
In this study, we use the term “SurvivinON” to refer to cells with the genotype Survivin−/−:tetSurvivin, which was grown without doxycycline. We use “SurvivinOFF” to refer to Survivin−/−:tetSurvivin cells exposed to doxycycline.
Survivin is essential for life in DT40 cells
SurvivinON cell lines grew with a doubling time (12 h) similar to that of wild-type DT40 cells (Fig. 2 C). In contrast, SurvivinOFF cells ceased proliferating after 36 h in doxycycline. By 60 h, most cells had died by apoptosis, with the few survivors being much larger than their SurvivinON counterparts (Fig. 2, B and C). Annexin V–positive apoptotic cells began to accumulate in SurvivinOFF cultures at 36 h, and by 48 h, 80% of the cells were annexin V positive (Fig. 2 D).
Previous RNAi studies showed that CPC members are codependent for protein stability and localization (Adams et al., 2001; Carvalho et al., 2003; Honda et al., 2003; Gassmann et al., 2004). Indeed, when Survivin protein levels dropped in SurvivinOFF cells, INCENP levels also decreased (Fig. 2 F). INCENP failed to localize to prophase and metaphase centromeres in SurvivinOFF cells, whereas CENP-H–GFP appeared to target normally at kinetochores (Fig. 2 E). A decrease in H3S10ph levels after depletion of Survivin suggested a partial loss of aurora B kinase activity (Fig. 2, F and G). However, aurora B levels did not change (unpublished data).
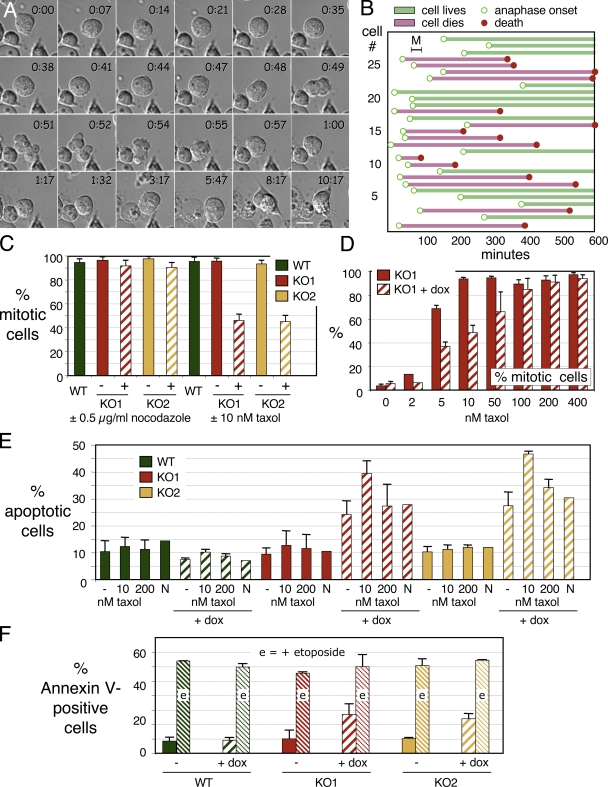
Cell death phenotype after loss of Survivin
Despite an earlier report that Survivin promotes entry of CD34+ cells into the cell cycle (Fukuda et al., 2002), our time-lapse imaging confirmed that cells lacking Survivin after 36 h in doxycycline continued to cycle through mitosis (see Figs. 2–4). The mitotic index of SurvivinOFF cultures was slightly elevated compared with wild-type DT40 or SurvivinON cells after 60 h in doxycycline (Fig. 3 A).
Figure 3.
Survivin-depleted cells fail to complete cytokinesis. (A) The mitotic index of SurvivinOFF cells is increased for both KO1 and KO2 relative to wild-type (WT) DT40. (B) Shutoff of Survivin expression leads to an increase in multinucleated (primarily binucleated) cells. Values for cells grown in doxycycline are shown as striped bars. Error bars indicate SD. (C) Selected frames from videos of SurvivinON and SurvivinOFF cells. Merged images show differential interference contrast (red) and histone H2B-mRFP (green), with H2B-mRFP also shown in grayscale. Time is given in hours/minutes. A control SurvivinON cell (top) completes mitosis normally. The SurvivinOFF cell achieves a metaphase alignment (0:25) but exhibits lagging chromosomes in anaphase, and cytokinesis ultimately fails. Bar, 5 μm. (D) Synchronization of DT40 cells by centrifugal elutriation. (E) Cells harvested later in the cell cycle die before those harvested earlier in the cell cycle. (F) Cells harvested later in the cell cycle fail cytokinesis before those harvested earlier in the cell cycle.
Figure 4.
Cells lacking Survivin die in interphase after failing to complete cytokinesis. (A) Selected frames from a video in which a SurvivinOFF cell fails to complete cytokinesis and dies by apoptosis in the subsequent interphase. Time is given in hours/minutes. Bar, 10 μm. (B) Analysis of cell death in SurvivinOFF cells from videos like those shown in A. Bars begin at anaphase onset (open circles) and terminate either at cell death (closed circles) or at the end of the video. Bar M shows the mean length of mitosis in DT40 (30 min). (C) SurvivinOFF cells exhibit a normal mitotic arrest in nocodazole but not in 10 nM taxol. (D) SurvivinOFF cells arrest in mitosis at taxol concentrations ≥100 nM. (E) The death of SurvivinOFF cells is exacerbated by 10 nM but not by 200 nM taxol. (F) SurvivinOFF and SurvivinON control cultures exhibit similar levels of apoptotic death after exposure to 10 μM etoposide. Values for cells grown in doxycycline are shown as striped bars. Error bars indicate SD. WT, wild type.
As in previous studies in which Survivin protein was knocked down by antisense nucleotides, mouse knockout, or RNAi (Fraser et al., 1999; Li et al., 1999; Speliotes et al., 2000; Uren et al., 2000; Carvalho et al., 2003; Lens et al., 2003), the most dramatic phenotype observed in mitotic SurvivinOFF cells was a highly penetrant failure in cytokinesis, as observed by scoring the percentage of multinucleate cells (Fig. 3, B and F) or by direct visualization in time-lapse live cell imaging (Fig. 3 C, Fig. 4 A, Fig. S1 A, and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1). By 60 h in doxycycline, >50% of the surviving cells were binucleated compared with <5% of SurvivinON cells and wild-type DT40 cells (Fig. 3 B). Rarely, we also observed cells with four or more nuclei (Fig. S1 B).
Live cell imaging revealed that in many SurvivinOFF mitoses, the chromosomes appeared to separate but cytokinesis failed, giving rise to binucleated progeny (Fig. 3 C, and Videos 3 and 4, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1). Other SurvivinOFF cells achieved a normal metaphase chromosome alignment and anaphase onset, but sister chromatid separation failed and the chromatids collapsed back on one another, forming larger nuclei (Fig. S1 A and Video 5). Such cells formed single large nuclei. This may explain why not all cells appear multinucleated after Survivin depletion. Thus, Survivin is required during both chromosome segregation and cytokinesis.
SurvivinOFF cells die after completing mitosis and failing cytokinesis
The death of SurvivinOFF cells was linked to the cell cycle, as shown in experiments using centrifugal elutriation, a noninvasive selection synchrony procedure that does not perturb cell cycle progression (Gillespie and Henriques, 2006). SurvivinOFF cells were elutriated after incubation in doxycycline for 36 h, at which time Survivin protein had fallen to undetectable levels. Fractions of cells synchronized in G1 (fraction 2), S (fraction 3), and G2/M phases (fractions 4 and 5; Fig. 3 D) were harvested and cultured, and apoptotic cells were scored every 2 h by visual inspection (Fig. 3 E) and annexin V and propidium iodide staining (not depicted).
G1- and S-phase cells (fractions 2 and 3) remained healthy for 10–15 h after synchronization, by which time many had passed through a subsequent G2/M. In contrast, G2/M cells (fractions 4 and 5) died much more quickly. Most cells had become binucleated before they died (Fig. 3 F), suggesting that Survivin-depleted cells died in interphase after failing in cytokinesis.
Live cell analysis confirmed this hypothesis (Fig. 4, A and B; and Videos 6 and 7, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1). In one study, 46% (19/41) of elutriated cells from fraction 2 failed in cytokinesis and 12% died in interphase. A further 27% were binucleate at the start of the video and died before reentering mitosis. In another study, 29 cells entered anaphase during filming. Of these, 15 died in interphase after exiting mitosis (Fig. 4 B and Video 7). The mean interval between anaphase onset and death was 276 ± 114 min (range of 40–450 min). Given the 12-h cell cycle time for DT40, death of SurvivinOFF cells likely occurs in late G1 or S phase. This cell death was not delayed by the pancaspase inhibitor Z-VAD (unpublished data) and therefore was not caused by disregulation of caspases.
Spindle checkpoint function in SurvivinOFF cells
As expected from published work (Carvalho et al., 2003; Lens et al., 2003), SurvivinOFF DT40 cells exhibited a normal mitotic arrest in 0.5 μg/ml nocodazole (Fig. 4 C). Their response to taxol was more complex, however. At taxol concentrations between 5 and 50 nM, SurvivinOFF cells showed a significantly decreased mitotic index, indicative of a defective checkpoint response (Fig. 4, C and D). However, at taxol concentrations of ≥100 nM, SurvivinOFF cells experienced a normal mitotic arrest.
FACS analysis showed a 15% increase in TUNEL-positive apoptotic SurvivinOFF cells at 10 nM taxol compared with untreated SurvivinOFF controls (Fig. 4 E). This increase was not seen at high doses of taxol (200 nM) or with nocodazole. This could be explained if SurvivinOFF cells override the checkpoint at low doses of taxol, exit mitosis, and are more susceptible to die in interphase. SurvivinOFF cells exposed to high doses of taxol and blocked in mitosis might be more protected from dying during the time course of the experiment. These data further support our hypothesis that exit from an aberrant mitosis leads to death of SurvivinOFF cells during interphase.
SurvivinOFF cells exhibit normal apoptosis in response to etoposide and staurosporine
Many published studies report that down-regulation of Survivin by RNAi or antisense oligonucleotides can enhance spontaneous or drug-induced apoptosis (Ambrosini et al., 1998; Li et al., 1999; Jiang et al., 2001; Pennati et al., 2002; Ling and Li, 2004; Pennati et al., 2004; Wang et al., 2005; Zaffaroni et al., 2007). Furthermore, a wide range of studies showed that Survivin overexpression can have a cytoprotective effect (for reviews see Wheatley and McNeish, 2005; Altieri, 2006).
These effects on apoptosis are not universal, as loss of Survivin from DT40 cells had no effect on their response to a well-defined proapoptotic stimulus. SurvivinOFF cells were incubated with etoposide for 3 h after exposure to doxycycline for 30 or 36 h to repress Survivin. When cells were collected and analyzed by TUNEL staining, we observed no significant difference in levels of apoptosis between cultures with or without Survivin (Fig. 4 F). Likewise, Survivin overexpression did not appear to provide a protective effect against etoposide-induced cell death. KO1-SurvivinON cells significantly overexpress Survivin, whereas KO2-SurvivinON cells express Survivin at endogenous levels (Fig. 1, G and H, compare 0 h with wild type), yet both cell lines undergo a similar response to etoposide.
Exposure to staurosporine can significantly increase the fraction of mouse embryos positive for activated caspase-3 after exposure to antisense survivin oligonucleotides (Kawamura et al., 2003). However, SurvivinOFF DT40 cells pretreated with doxycycline for 30 h were not more sensitive to staurosporine than SurvivinON or wild-type DT40 cells (unpublished data). Thus, with the exception of low doses of taxol, the loss of Survivin does not change the sensitivity of DT40 cells to drugs that induce the intrinsic apoptotic response.
Functional assessment of various Survivin structural motifs and interactions
Most prior mutagenesis studies of Survivin were performed with the human protein, which is ∼60% identical to chicken Survivin. Because Survivin is well conserved, we could identify many of the corresponding residues in the chicken protein (Fig. 5 A). In subsequent figures (Fig. 5 F), we show the position of the mutated residues in the Survivin dimer or CPC structure after modeling the structure of chicken Survivin using the human Survivin (hSurvivin) coordinates (Chantalat et al., 2000; Jeyaprakash et al., 2007).
Figure 5.
Functional analysis of Survivin protein in SurvivinON/OFF cells. (A) Alignment of human and chicken Survivin sequences showing conserved residues mutated in this study. (B and C) Chicken (g) and human (h) Survivin-GFP rescues the life of SurvivinOFF cells. Growth curves for cells expressing GFP fusion proteins are green. (D and E) Chicken and human Survivin-GFP colocalize normally with INCENP (red) in SurvivinOFF cells. (F) The location of residue T36 in the CPC and dimer structures of Survivin (Chantalat et al., 2000; Jeyaprakash et al., 2007). (G) Expression of SurvivinT36A/E-GFP and loss of wild-type (WT) Survivin expression from cultures grown in doxycycline. (H) Chicken SurvivinT36A-GFP rescues the life of SurvivinOFF cells. (I) gSurvivinT36A-GFP colocalizes normally with INCENP in SurvivinON/OFF cells. Bars, 5 μm.
SurvivinOFF cells were fully rescued by expression of wild-type chicken or human Survivin fused at its C terminus to GFP (Fig. 5, B and C; and Fig. S1, C and D). SurvivinOFF:Survivin-GFP cells grew normally in the presence and absence of doxycycline. Furthermore, Survivin-GFP localized to centromeres in prometaphase and to the spindle midzone in anaphase (Fig. 5, D and E). Importantly, in knockout cells expressing exogenous forms of Survivin, the wild-type doxycycline-regulated rescue Survivin became undetectable by 36 h in doxycycline. These observations enabled us to test a range of Survivin mutants for their ability to rescue Survivin function.
The BIR domain is essential for Survivin function
In human Survivin, four conserved residues, Cys57, Cys60, His77, and Cys84, form a zinc finger that stabilizes the structure of the BIR domain (Chantalat et al., 2000; Verdecia et al., 2000). To confirm the role of the BIR domain in Survivin function in vivo, SurvivinC86A (equivalent to hSurvivinC84A) and SurvivinC59A (equivalent to hSurvivinC57A) mutants were stably transfected into KO1, where they were expressed at levels comparable with the rescue Survivin (Fig. 6, B and F).
Figure 6.
The BIR domain is crucial for Survivin function. (A) Location of C86 in the CPC form of Survivin. (B) Expression of SurvivinC86A-GFP and loss of wild-type (WT) Survivin expression from cultures grown in doxycycline. (C) SurvivinC86A-GFP fails to rescue the life of SurvivinOFF cells; cells expressing this protein as their sole form of Survivin die with kinetics essentially identical to those of SurvivinOFF cells. (D) SurvivinC86A-GFP fails to localize in SurvivinON/OFF cells. INCENP localization is normal in SurvivinON cells, so SurvivinC86A-GFP fails to act as a dominant-negative mutant. Panel a shows the normal localization of Survivin-GFP in a control experiment. (E) Location of C59 in the Survivin dimer. (F) Expression of SurvivinC59A-GFP and loss of wild-type Survivin expression from cultures grown in doxycycline. (G) Survivin C59A-GFP fails to rescue the life of SurvivinOFF cells. (H) SurvivinC59A-GFP fails to localize in SurvivinON/OFF cells. INCENP localization is normal in SurvivinON cells, so SurvivinC59A-GFP also fails to act as a dominant-negative mutant. Bars, 5 μm.
Despite a previous report that hSurvivinC84A was dominant negative (Li et al., 1999), SurvivinON cells expressing both mutants grew with normal kinetics and showed normal INCENP localization (Fig. 6). Neither mutant Survivin localized normally in mitosis, and after doxycycline addition, cells expressing only the Survivin mutants died with kinetics resembling SurvivinOFF cells. Consistent with this loss of function, neither SurvivinC86A nor SurvivinC59A could target INCENP to centromeres or the spindle midzone. Thus, the zinc-binding residues of the BIR domain are crucial for Survivin functions in mitosis.
Cdk phosphorylation on T34 is dispensable for Survivin function
Survivin can be phosphorylated on T34 by Cdk1–cyclin B1 (O'Connor et al., 2000). Expression of the nonphosphorylated SurvivinT34A mutant produced a dominant-negative phenotype in human cancer cells, resulting in caspase-9–dependent apoptosis (Grossman et al., 2001; Yan et al., 2006). Treatment of cells with the CDK inhibitor flavopiridol resulted in a decrease in Survivin levels, and nonphosphorylated SurvivinT34A reportedly exhibited decreased stability relative to wild-type Survivin (Wall et al., 2003).
Chicken mutants SurvivinT36A-GFP and SurvivinT36E-GFP (equivalent to hSurvivin T34A and T34E) were stably expressed in KO1 cells (Fig. 5, F and G). Both mutant Survivins localize correctly on centromeres at metaphase and the spindle midzone in anaphase (Fig. 5 I and Fig. S3 B, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1). Surprisingly, cells expressing both mutants grew normally in the presence of doxycycline (Fig. 5 H) even though the tet-regulated wild-type rescue Survivin was entirely undetectable at any blot exposure (Fig. 5 G). Thus, contrary to prediction, CDK phosphorylation of T36 is dispensable for Survivin function in DT40 cells.
Asp residues required for interaction with Smac are dispensable for Survivin function
Several studies claim that Survivin binds Smac/Diablo and antagonizes its proapoptotic function (Muchmore et al., 2000; McNeish et al., 2005; Sun et al., 2005; Kim et al., 2006). These studies implicated hSurvivin residues D53 and D71 in the regulation of apoptosis (Muchmore et al., 2000; Song et al., 2004), and transfection of HeLa cells with hSurvivinD53A and hSurvivinD71A mutants caused spontaneous apoptosis (Song et al., 2003, 2004).
To test the role of D53 in Survivin function, we isolated clones of Survivin conditional knockout (KO1) cells stably expressing chicken SurvivinD55A (equivalent to hSurvivinD53A) at levels comparable with the wild-type rescue Survivin (Fig. 7 B). These cells grew normally in the absence of doxycycline and continued to grow, after a lag, in its presence with SurvivinD55A-GFP as the only form of Survivin detectable in the cells (Fig. 7 C). SurvivinD55A-GFP localized properly to centromeres, the central spindle and the midbody in mitotic cells (Fig. 7 D). Thus, residue D55 (and presumably interaction with Smac) is not essential for Survivin function in DT40 cells.
Figure 7.
Smac and aurora B binding are not essential for Survivin function. (A) Location of D55 in the Survivin dimer. (B) Expression of SurvivinD55A-GFP and loss of wild-type (WT) Survivin expression from cultures grown in doxycycline. (C) SurvivinD55A-GFP rescues the life of SurvivinOFF cells. (D) SurvivinD55A-GFP colocalizes normally with INCENP in SurvivinON/OFF cells. (E) Location of D72 and D73 in the Survivin dimer. (F) Expression of SurvivinD72A/D73A-GFP and loss of wild-type Survivin expression from cultures grown in doxycycline. (G) SurvivinD72A/D73A-GFP fails to localize in SurvivinON cells; INCENP localization is normal (a and b). SurvivinD72A/D73A-GFP fails to localize in SurvivinOFF cells at prometaphase and metaphase; INCENP localization at centromeres is also compromised (c). SurvivinD72A/D73A-GFP and INCENP localize normally in SurvivinOFF cells at anaphase/telophase (d). (H) SurvivinD72A/D73A-GFP rescues the life of SurvivinOFF cells. Bars, 5 μm.
Asp residues required for interaction with aurora B are required for Survivin targeting to centromeres
In addition to interfering with putative antiapoptotic functions, hSurvivin mutant D70A/D71A was reported to no longer interact with aurora B and to cause multinucleation of HeLa cells (Cao et al., 2006). Therefore, we introduced the analogous mutation (SurvivinD72A/D73A; Fig. 7 E) into SurvivinON/OFF cells. This Survivin mutant revealed unexpected aspects of CPC behavior in early mitosis.
Immunoblotting confirmed that SurvivinD72A/D73A-GFP was expressed at relatively high levels in these cell lines (Fig. 7 F); however, it was unable to localize to centromeres or the spindle midzone in the presence of wild-type Survivin (Fig. 7 G, a and b). These cells grew with normal kinetics, so this mutant did not exhibit a dominant-negative phenotype. Strikingly, SurvivinOFF cells expressing SurvivinD72A/D73A-GFP continued to grow with normal kinetics in the presence of doxycycline, even though the wild-type rescue Survivin was now undetectable (Fig. 7, F and H). Therefore, the antiapoptotic and aurora B–binding role attributed to these residues in human Survivin is dispensable for Survivin function in DT40 cells.
An unexpected result was obtained when the localization of SurvivinD72A/D73A-GFP was examined in detail in SurvivinOFF cells. Remarkably, SurvivinD72A/D73A-GFP failed to localize to centromeres during prophase through metaphase in these cells (Fig. 7 G, c). However, the protein localized to the anaphase spindle midzone and subsequently concentrated at the midbody in telophase (Fig. 7 G, d). In SurvivinOFF cells, this protein induced an identical behavior in INCENP and aurora B, which were diffuse during prophase through metaphase but localized to the spindle midzone and midbody in late mitosis (Fig. 7 G and Fig. S3).
Because SurvivinD72A/D73A-GFP was defective in centromere localization during prometaphase, we hypothesized that these cells might exhibit a defective spindle checkpoint response to taxol. Indeed, SurvivinOFF cells expressing SurvivinD72A/D73A-GFP showed a slight but significant checkpoint defect in the presence of low-dose taxol (Fig. S2 C, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1).
Thus, analysis of the SurvivinD72A/D73A-GFP mutant reveals the surprising fact that INCENP and Survivin concentration at centromeres is not essential for mitotic progression, at least when the proteins subsequently target to the spindle midzone and midbody during anaphase.
Temperature-sensitive phenotype of mutants of the linker region between the BIR domain and C-terminal α helix
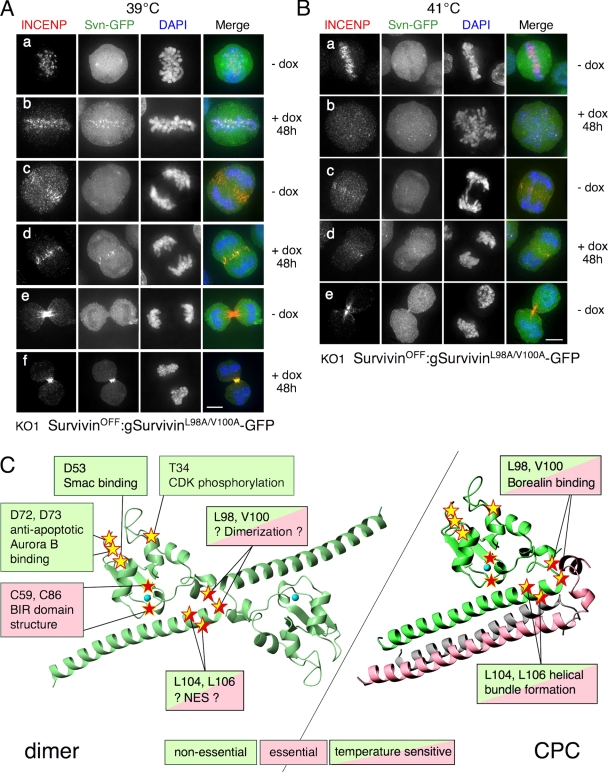
The linker region (residues 82–102) between the BIR domain and C-terminal α helix is a site of molecular contact with Borealin in the CPC (Bourhis et al., 2007; Jeyaprakash et al., 2007) and with Survivin itself in the homodimer (Chantalat et al., 2000; Sun et al., 2005). Human Survivin also contains a nuclear export sequence (NES) that overlaps this region (Colnaghi et al., 2006; Stauber et al., 2006; Engelsma et al., 2007; Knauer et al., 2007). We analyzed the role of this region in Survivin function by mutating four highly conserved residues: L98, V100, L104, and L106. V100, which is conserved in mouse, zebrafish, and Drosophila, corresponds to L98 in hSurvivin.
We isolated SurvivinON/OFF cells expressing either SurvivinL98A/V100A (equivalent to hSurvivinL96A/L98A) or SurvivinL104A/L106A (equivalent to hSurvivinL102A/L104A) at levels similar to the wild-type Survivin (Fig. 8 B, 39°C). These cells continued to grow at 39°C and 37°C, respectively, in doxycycline long after SurvivinOFF cells had died (Fig. 8, C and D) so both mutants could rescue Survivin's essential functions. Strikingly, neither linker mutant could rescue SurvivinOFF cells at 41°C, a temperature at which DT40 cells grow normally. At 41°C, SurvivinOFF cells expressing either mutant started dying after 48-h growth in doxycycline, ∼24 h later than SurvivinOFF cells (Fig. 8, E and F). Indeed, if SurvivinOFF:SurvivinL98A/V100A-GFP cells grown at permissive temperature with doxycycline for 1 wk were shifted to 43°C, they failed to grow, whereas wild-type DT40 or SurvivinOFF:Survivin-GFP cells grew normally (Fig. S4 C, available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1). We conclude that both Survivin linker mutants are temperature sensitive for function.
Figure 8.
Survivin linker mutants are temperature sensitive. (A) Location of L98, V100, L104, and L106 in the CPC. (B) Expression of SurvivinL98A/V100A-GFP and SurvivinL104A/L106A-GFP and loss of wild-type (WT) Survivin expression from cultures grown in doxycycline at 39 or 41°C. (C) SurvivinL98A/V100A-GFP rescues the life of SurvivinOFF cells at 39°C. (D) SurvivinL104A/L106A-GFP rescues the life of SurvivinOFF cells at 37°C. (E and F) Cells expressing solely SurvivinL98A/V100A-GFP and SurvivinL104A/L106A-GFP die at 41°C, albeit with slightly delayed kinetics.
Interestingly, neither Survivin mutant could compete with the wild-type protein for its localization in mitosis. In SurvivinON cells growing at 39°C, SurvivinL98A/V100A-GFP localization was diffuse in all stages of mitosis (Fig. 9 A, a, c, and e). These data are consistent with the analysis of a comparable human Survivin mutant (Knauer et al., 2006). Remarkably, SurvivinL98A/V100A-GFP did target to centromeres, the spindle midzone, and the midbody in SurvivinOFF cells grown with doxycycline at 39°C (Fig. 9 A, b, d, and f). Similar phenotypes were observed for SurvivinL104A/L106A-GFP at 37°C (Fig. S4 A).
Figure 9.
A Survivin linker mutant localizes aberrantly. (A) SurvivinL98A/V100A-GFP fails to localize at 39°C in the presence of wild-type rescue Survivin (a, c, and e). In the absence of wild-type rescue Survivin, a subset of SurvivinL98A/V100A-GFP localizes normally (b, d, and f). INCENP localizes normally in all cells. (B) SurvivinL98A/V100A-GFP fails to localize at 41°C in the presence or absence of wild-type rescue Survivin. INCENP localizes normally only in the presence of wild-type rescue Survivin (a, c, and e). (C) Summary of the mutational analysis of Survivin. Bars, 5 μm.
INCENP localized correctly in SurvivinON cells express either mutant at 39 or 41°C, so the mutants did not act as dominant negatives (Fig. 9 B, a, c, and e; and Fig. S4 B, a, c, and e). Under SurvivinOFF conditions, cells expressing these mutants had a Survivin-null phenotype at 41°C, with diffuse Survivin and INCENP localization, multinucleation, and multipolar spindles (Fig. 9 B, b and d; and Fig. S4 B, b and d). In conclusion, the ability of two Survivints mutants to confer viability correlates perfectly with their ability to localize to mitotic structures, providing further support for the notion that it is the mitotic functions of Survivin that are essential for the life of DT40 cells.
Discussion
Survivin's role in mitosis and apoptosis remains poorly understood. A bias (>1,500/2,000 PubMed listings) toward studies of apoptosis belies the fact that the phenotypes of survivin mutants in yeasts (Rajagopalan and Balasubramanian, 1999; Uren et al., 1999) and C. elegans (Fraser et al., 1999; Speliotes et al., 2000) indicate a role in mitosis rather than cell death regulation. Indeed, even though DT40 cells have widely been used to study apoptosis (Ruchaud et al., 2006), our experiments show that it is the mitotic and not an antiapoptotic role for Survivin that is essential for life in these cells.
Studies of Survivin in vertebrates have used RNAi, antisense oligonucleotides, dominant-negative mutants, and mouse knockouts (Fraser et al., 1999; Li et al., 1999; Speliotes et al., 2000; Carvalho et al., 2003; Lens et al., 2003). Of these, the first three are subject to the caveat that the elimination of Survivin is never complete, and because aurora B is a kinase, even low levels of Survivin could in principle support significant amounts of kinase activity. Phenotypic analysis of the mouse knockouts is challenging, as Survivin is essential for embryonic life, with death occurring by 4.5 d after coitum (Uren et al., 2000). As yet, no conditional mouse mutant for Survivin has been made.
In this study, we characterize two conditional knockouts of Survivin in chicken DT40 cells. A principal advantage of this system is that after shutoff of the regulated Survivin cDNA responsible for keeping cells alive, levels of the wild-type protein and its mRNA become essentially undetectable in the culture. This is significant, as a previous RNAi study of aurora kinase localization produced very different results depending on whether or not the endogenous protein was present when various mutants were expressed (Scrittori et al., 2005). We obtained similar results for three mutants, SurvivinL98A/V100A, SurvivinL104A/L106A, and SurvivinD72A/D73A, which fail to localize in the presence of wild-type Survivin but do so, at least partially (and are functional), in its absence. Thus, the null background was essential for study of these features of the Survivin molecule.
Our experiments confirm several widely held beliefs about Survivin but contradict others. For example, Survivin is essential for the completion of cytokinesis. Survivin function is apparently linked with that of the CPC, and the cytokinesis phenotype could reflect aberrant regulation of MKLP-1 (Yang et al., 2004; Guse et al., 2005; Zhu et al., 2005; Klein et al., 2006), taxins (Delaval et al., 2004), Ect2 (Chalamalasetty et al., 2006), PLK1 (Goto et al., 2006; Burkard et al., 2007), or other aurora B substrates.
Cells lacking Survivin execute the earlier stages of mitosis through metaphase chromosome alignment without obvious difficulties. This contrasts with previous RNAi analyses of Survivin and other CPC members (Adams et al., 2001; Carvalho et al., 2003; Lens et al., 2003; Gassmann et al., 2004). A recent RNAi study also found that Survivin was not required for chromosome movements in early mitosis (Rosa et al., 2006); however, that and other studies described abnormal spindles in Survivin-depleted cells (Giodini et al., 2002; Okada et al., 2004; Rosa et al., 2006). In contrast, we observed no gross spindle abnormalities in mitosis of SurvivinOFF cells.
Although cell death after loss of Survivin is clearly linked to the cell cycle, this death occurs in interphase, not during mitosis or mitotic exit. Using centrifugal elutriation, a nonperturbing method for selecting synchronized cell populations, we found that cells harvested later in the cell cycle die significantly before those harvested earlier. Indeed, our live cell analysis indicates that cells die during midinterphase on mean ∼5 h after mitotic exit. Considering the 12-h division cycle of DT40 cells, this may correspond to a point in G1/S analogous to that at which some normal human cells arrest in a p53-dependent manner after Survivin RNAi (Yang et al., 2004). DT40 cells lack functional p53, and the absence of a checkpoint could explain why they die. At present, no CPC-dependent functions have been described during interphase, so this forms an intriguing point for future study.
Although Survivin is essential for a spindle checkpoint response to low-dose taxol, surprisingly, higher doses of taxol did cause a tight mitotic arrest in cells lacking Survivin. The effects of taxol on microtubule plus end dynamics are not simple. As the taxol concentration increases, the plus ends become progressively less dynamic (Derry et al., 1995; Kelling et al., 2003). One possible explanation for our results, suggested to us by M.A. Jordan, is that Survivin might be required to impose a spindle checkpoint when microtubule plus ends are minimally dynamic but may no longer be required when those dynamics are completely suppressed (unpublished data; Jordan, M.A., personal communication). Alternatively, it may be that in low-dose taxol, the spindle checkpoint can eventually be silenced, perhaps transiently, even though microtubule dynamics are perturbed (Brito and Rieder, 2008). Survivin could promote checkpoint function by somehow decreasing the probability of transient checkpoint inactivation. Whatever the explanation, it is clear that one cannot simply generalize that cells lacking Survivin are unresponsive to taxol.
Either chicken or human Survivin fused to GFP can support the life of SurvivinON/OFF cells for extended periods in culture after shutoff of the wild-type rescue cDNA. Thus, this system is well suited to test the functionality and phenotype of various mutant forms of Survivin in the absence of detectable levels of wild-type protein (mutants summarized in Fig. 9 C).
Although some Survivin mutants behaved as expected, others clearly did not. For example, perturbation of the Survivin BIR domain renders the protein completely nonfunctional during mitosis. However, contrary to expectations (Li et al., 1999), two Survivin BIR domain mutants failed to exhibit dominant-negative activity. Because the BIR domain probably promotes protein–protein interactions (Srinivasula and Ashwell, 2008) but does not participate either in dimer formation (Chantalat et al., 2000) or in formation of the CPC three-helix bundle (Bourhis et al., 2007; Jeyaprakash et al., 2007), its role in Survivin function remains to be determined.
CDK phosphorylation of Survivin is reportedly essential for cell life (O'Connor et al., 2000; Grossman et al., 2001; Yan et al., 2006). However, DT40 cells whose sole Survivin had its conserved CDK site mutated to either Ala or Glu grew normally and exhibited normal CPC behavior in mitosis. Thus, regulation by CDKs is not essential for Survivin function.
Several studies implicate Smac/Diablo, the mitochondrial antagonist of IAPs, as a binding partner of Survivin in apoptosis regulation (Muchmore et al., 2000; Song et al., 2003, 2004; McNeish et al., 2005; Sun et al., 2005; Kim et al., 2006). The SurvivinD55A mutant was reported to abolish this interaction and to have a significant proapoptotic phenotype (Song et al., 2004). However, contrary to expectations, we find that DT40 cells can live, albeit growing more slowly, with this mutant as the sole Survivin.
Survivin residues 80 and 106 contain an NES enabling hSurvivin to shuttle between the nucleus and cytoplasm (Rodriguez et al., 2002, 2006; Colnaghi et al., 2006) and required for CPC targeting to centromeres (Knauer et al., 2006). This region forms a docking surface for Survivin itself in the homodimer or for Borealin in the CPC (Bourhis et al., 2007; Jeyaprakash et al., 2007). Survivin dimer formation impedes Crm1 binding by the NES (Engelsma et al., 2007), and Survivin may be preferentially exported from the nucleus as a monomer.
We isolated two mutants of the docking surface that render DT40 cells temperature sensitive for growth. Cells dependent on SurvivinL98A/V100A or SurvivinL104A/L106A grow at 39°C or 37°C, respectively, but die at 41°C. DT40 cells normally grow at 41°C, and indeed, the diurnal deep muscle temperature of chickens is 45°C during periods of activity (Simpson, 1912). Because cells dependent on SurvivinL98A/V100A or SurvivinL104A/L106A are viable at permissive temperature, these mutants must support CPC formation in vivo. In contrast, hSurvivinF101A/L102A (F103A/L104A in chicken Survivin) is a monomer in vitro (Engelsma et al., 2007). We suggest that at nonpermissive temperature, SurvivinL98A/V100A or SurvivinL104A/L106A mutants may be unable to form either the CPC or homodimer interactions.
Survivin may direct the localization of the CPC to centromeres (Vader et al., 2006), but the significance of this localization is unclear. Is centromere localization required for CPC activities in correcting microtubule attachment errors or in the spindle checkpoint response? Remarkably, cells expressing solely SurvivinD72A/D73A, which was reported to block the binding of both Smac and aurora B (Cao et al., 2006), grow normally even though the mutant Survivin is unable to target to centromeres under all conditions tested. In the absence of wild-type Survivin, INCENP also fails to localize in early mitosis, suggesting that SurvivinD72A/D73A cannot target it to centromeres. Surprisingly, when these cells enter anaphase, INCENP exhibits a robust localization to the central spindle and midbody. This may explain why these cells complete mitosis, execute cytokinesis, and proliferate with SurvivinD72A/D73A as their sole Survivin. These observations suggest that Survivin is not responsible for INCENP targeting during anaphase/telophase, or that if it is, targeting must occur by a different mechanism from that in early mitosis.
The SurvivinD72A/D73A mutant reveals that INCENP and Survivin concentration at centromeres is not essential for mitotic progression, at least when the proteins subsequently target to the spindle midzone and midbody during anaphase. Interestingly, cells whose growth depends on SurvivinD72A/D73A exhibit a compromised spindle checkpoint in response to taxol but do not show a dramatic increase in the fraction of kinetochore attachment errors. It could be that lower amounts of transient CPC at centromeres serve to correct attachment errors but that higher levels or more stably associated CPC is required for the checkpoint response to low-dose taxol. This is the first evidence that CPC function at centromeres is required for a normal spindle checkpoint response to low-dose taxol. It will be important in the future to identify the ligands that interact with D72 and D73 on the BIR domain to target Survivin to centromeres.
Survivin continues to guard its secrets closely, and the fact that three different Survivin mutants localize differently in the presence and absence of wild-type Survivin points to the importance of a genetically clean system such as that described here for the definitive functional dissection of the protein. DT40 cells have proven to be a good system for the study of apoptotic cell death (Ruchaud et al., 2006). Thus, even though our studies show that cell death after loss of Survivin is linked to the cell cycle rather than apoptotic disregulation, this system may in the future be able to answer remaining questions concerning the roles of Survivin in mitosis and apoptosis.
Materials and methods
Cell culture and targeting constructs
DT40 cells were grown in Roswell Park Memorial Institute culture media supplemented with 10% FBS and 1% chicken serum in 5% CO2 at 39°C (Buerstedde and Takeda, 1991). Doxycycline at a final concentration of 0.5 μg/ml was added to the culture medium to repress transcription of the survivin rescue transgene. The survivin gene locus was isolated from the λ Fix II DT40 genomic library. To disrupt the survivin gene, targeting vectors (vector pUHG10.3 backbone) containing a selectable marker that confers resistance to neomycin, puromycin, or histidinol were constructed (Fig. 1 A).The resistance cassettes were flanked by a 5′ genomic arm situated upstream of the initiation codon of the survivin ORF and a 3′ genomic arm situated downstream of its stop codon. For the survivin rescue construct, primers AAGAGCTCAAATGGCGGCCTATGCTGAAATGCTGCCC and CAGTTATTGAGACAGCGTGGCCTAAGGGCCCATGTTCTCTATC were used to amplify the chicken survivin cDNA. The PCR product consisting of the ORF of chicken survivin and a smaller 3′ untranslated region was cloned into pUHG10.3. After linearization, all constructs were transfected by electroporation as previously described (Samejima et al., 2001). After drug selection, DNA from resistant clones was extracted and analyzed by Southern blotting after digestion with EcoRI, PflMI, or AflII and probed with a 5′ external probe (Fig. 1).
Immunoblotting and antibodies
Whole cell lysates were prepared, and the equivalent to one million cells was loaded onto a polyacrylamide gel. SDS-PAGE and immunoblotting were performed according to standard procedures. Anti–α-tubulin antibody (clone B512) and anti-H3 phospho-Ser10 were purchased from Sigma-Aldrich and Millipore, respectively. Rabbit polyclonal (WCE1186) and mouse monoclonal anti-INCENP (3D3) were previously described (Cooke et al., 1987; Earnshaw and Cooke, 1991). Rabbit polyclonal anti–chicken Survivin (WCE43D) was raised against 6xHis-GgSurvivin and affinity purified. Rabbit polyclonal anti–chicken aurora B (WCE56A) was raised against GST-Ggaurora B residues 80–210.
Indirect immunofluorescence microscopy
Cells were incubated at 39°C on polylysine-coated slides (Polysine; VWR International) for 15 min before fixation in 4% PFA /cytoskeletal buffer (137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 0.4 mM KH2PO4, 2 mM MgCl2, 2 mM EGTA, 5 mM Pipes, and 5.5 mM glucose) at 37°C and permeabilization in 0.15% Triton X-100 in cytoskeletal buffer. After blocking in 1% BSA/PBS, cells were probed with the aforementioned antibodies, and slides were mounted using Vectashield (Vector Laboratories). All image stacks were taken using a microscope (IX-70; Olympus) with a charge-coupled device camera (CH350; Photometrics) controlled by DeltaVision SoftWorx (Applied Precision, LLC) and a 100× S Plan Apocromat NA 1.4 objective using a Sedat filter set (Chroma Technology Corp.) and running at RT. Image stacks were deconvolved, and maximum projections were generated using SoftWorx. All files were saved as TIFF files and exported to Photoshop (Adobe) for final presentation. Levels were adjusted similarly for each experimental dataset to lower nonspecific background haze using the standard Photoshop adjust levels tool.
Live cell imaging
All videos (except Fig. 3 C and Video 4) were taken using a Perfect Focus microscope (TE-2000E; Nikon) with a camera (CoolSnap HQ2; Photometrics) controlled by Metamorph (MDS Analytical Technologies) using a 100× NA 1.4 Plan Apochromat objective. Chicken DT40 Survivin knockout cells stably expressing H2B-RFP were elutriated, and fraction 2 cells were transferred onto 40-mm coverslips coated with concanavalin A (EMD). These coverslips were placed into a Nikon chamber (Bioptechs) and kept at 39°C in the presence of Roswell Park Memorial Institute culture media without phenol red. Three-dimensional datasets were collected every 2 or 10 min, and video frames were processed with ImagePro Plus software (version 6.0; Media Cybernetics, Inc.).
The videos shown in Fig. 3 C and Video 4 were taken using a microscope (IX-70; Olympus) with a charge-coupled device camera (CH350; Photometrics) controlled by DeltaVision SoftWorx and a 100× NA 1.4 S Plan Apocromat objective using a Sedat filter set. The temperature was controlled using a Weather Station (Precision Control) and set for 39°C. DT40 cells were placed on concanavalin A–coated coverslips and imaged every minute as z stacks. Image stacks were deconvolved, and maximum projections were generated using SoftWorx. Video files were constructed from selected stills and saved as .mov files.
Cell elutriation
108 cells resuspended in 5 ml of culture media supplemented with 1 mM EDTA were elutriated using an elutriator (JE-5 rotor; Beckman Coulter) at a steady flow rate of 40 ml/s and a starting speed of 3,750 rpm. After equilibration, the speed was adjusted to collect the different fractions (fraction 2, 3,250 rpm; fraction 3, 3,000 rpm; fraction 4, 2,750 rpm; and fraction 5, 2,500 rpm). Fractions were spun down, immediately placed in fresh warm medium, and kept at 39°C.
Site-directed mutagenesis
Survivin point mutants were generated by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis kit; Stratagene) using the plasmid WTGg-Survivin-pEGFPN1 and transfected into the knockout cells by electroporation. Stable knockout lines homogeneously expressing the GFP fusion protein were isolated and kept at 39°C.
Growth curves and multinucleation/mitosis index measurements
Growth curves were calculated by seeding the various cell lines at 2 × 105 cells/ml at 39°C (unless indicated otherwise) and counting the cell number every 12 h using a hemocytometer. To avoid the effects of overgrowth, cells were diluted to 2 × 105 cells/ml whenever the number exceeded 106 cells/ml. The cell number at each time point was multiplied by the appropriate dilution factor to get a true count. This protocol gives linear cell growth profiles regardless of the length of the experiment.
For the assessment of the multinucleation/mitosis indexes, a total of 1,000 cells were scored for each time point. The multinucleation index was calculated by dividing the number of multinucleated interphase cells by the total number of interphase cells. The mitotic index was calculated by dividing the number of mitotic cells by the number of interphase and mitotic cells.
Apoptosis assays
Annexin V staining was performed according to the manufacturer's instructions (BioVision, Inc.). Cells were analyzed by flow cytometry using a FACScalibur machine (Becton Dickinson), and results were quantified using CellQuest software (Becton Dickinson).
Online supplemental material
Fig. S1 presents further characterization of the phenotype of SurvivinOFF cells, focusing on their failure to complete cytokinesis. It also shows further evidence for their rescue by Survivin-GFP. Fig. S2 shows that a T36E phosphomimetic mutant does not disturb Survivin localization and that the SurvivinD72A/D73A-GFP mutant, which is defective in localizing to centromeres in early mitosis, exhibits a defective checkpoint response to 10 nm taxol. Fig. S3 shows that aurora B kinase fails to localize to centromeres in early mitosis but localizes normally in anaphase/telophase in cells expressing SurvivinD72A/D73A. Fig. S4 presents further localization data that Survivin linker mutant SurvivinL104A/L106A-GFP fails at 37 and 41°C. It shows that SurvivinL98A/V100A-GFP is temperature sensitive for growth. Video 1 shows the growth of wild-type DT40 cells in culture. Video 2 shows that DT40 cells lacking Survivin fail to complete cytokinesis and subsequently die. Video 3 shows that a SurvivinON DT40 cell expressing wild-type Survivin completes mitosis normally. Video 4 shows a cell lacking Survivin that reaches anaphase, during which sister chromatid separation occurs but then fails to complete cytokinesis. Video 5 shows a cell lacking Survivin that fails to complete anaphase as sister chromatids collapse back together and cytokinesis fails. Videos 6 and 7 show that cells lacking Survivin fail to complete cytokinesis and subsequently die during interphase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806118/DC1.
Supplementary Material
Acknowledgments
We thank G. Zachos for advice on elutriation, M.A. Jordan for helpful suggestions, and C. Rieder for communication of results before publication.
This work was supported by a grant from the Wellcome Trust, of which W.C. Earnshaw is a Principal Research Fellow. A. Carvalho was supported in part by the Fundacao para a Ciencia e Tecnologia of Portugal.
Note added in proof. While this paper was in the proof stage, it came to our attention that there is a second distantly related Survivin gene in chicken (ENSGALP0000038612). We refer to this as Survivin-2. This encodes for a putative larger (173 aa) protein that shows 33% identity with GgSurvivin-1 and 34% identity with HsSurvivin. GgSurvivin-1 and HsSurvivin share 58% identity. We do not know if Survivin-2 protein is expressed in DT40 cells. Importantly, even if it is expressed, Survivin-2 is not sufficient to rescue cell life, localize the CPC to centromeres, stabilize INCENP, or promote full aurora B activity (detected by phosphorylation of H3 on S10) in cells where Survivin-1 is depleted
Z. Yue and A. Carvalho contributed equally to this paper.
Z. Yue's present address is Dept. of Cell and Developmental Biology, University of Dundee, Dundee DD1 5EH, Scotland, UK.
A. Carvalho and R. Gassmann's present address is Dept. of Cellular and Molecular Medicine, Ludwig Institute for Cancer Research, La Jolla, CA 92093.
F. Lai's present address is Centre for Genomic Regulation, Barcelona 08003, Spain.
E. Gudmundsdottir's present address is NimbleGen Systems of Iceland LLC, 113 Reykjavik, Iceland.
C.G. Morrison's present address is Dept. of Biochemistry, National University of Ireland Galway, Galway, Ireland.
Abbreviations used in this paper: BIR, baculovirus IAP repeat; CPC, chromosomal passenger protein complex; hSurvivin, human Survivin; IAP, inhibitor of apoptosis protein; INCENP, inner centromere protein; NES, nuclear export sequence; tTA, tetracycline transactivator.
References
- Adams, R.R., S.P. Wheatley, A.M. Gouldsworthy, S.E. Kandels-Lewis, M. Carmena, C. Smythe, D.L. Gerloff, and W.C. Earnshaw. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., H. Maiato, W.C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and Aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri, D.C. 2006. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 18:609–615. [DOI] [PubMed] [Google Scholar]
- Ambrosini, G., C. Adida, and D.C. Altieri. 1997. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 3:917–921. [DOI] [PubMed] [Google Scholar]
- Ambrosini, G., C. Adida, G. Sirugo, and D.C. Altieri. 1998. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J. Biol. Chem. 273:11177–11182. [DOI] [PubMed] [Google Scholar]
- Banks, D.P., J. Plescia, D.C. Altieri, J. Chen, S.H. Rosenberg, H. Zhang, and S.C. Ng. 2000. Survivin does not inhibit caspase-3 activity. Blood. 96:4002–4003. [PubMed] [Google Scholar]
- Beltrami, E., J. Plescia, J.C. Wilkinson, C.S. Duckett, and D.C. Altieri. 2004. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J. Biol. Chem. 279:2077–2084. [DOI] [PubMed] [Google Scholar]
- Bolton, M.A., W. Lan, S.E. Powers, M.L. McCleland, J. Kuang, and P.T. Stukenberg. 2002. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 13:3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis, E., S.G. Hymowitz, and A.G. Cochran. 2007. The mitotic regulator Survivin binds as a monomer to its functional interactor Borealin. J. Biol. Chem. 282:35018–35023. [DOI] [PubMed] [Google Scholar]
- Brito, D.A., Z. Yang, and C.L. Rieder. 2008. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J. Cell Biol. 182:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde, J.-M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 67:179–188. [DOI] [PubMed] [Google Scholar]
- Buerstedde, J.-M., and S. Takeda. 2006. Reviews and Protocols in DT40 Research. Springer, New York.
- Burkard, M.E., C.L. Randall, S. Larochelle, C. Zhang, K.M. Shokat, R.P. Fisher, and P.V. Jallepalli. 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 104:4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., X. Yan, Y. Wu, H. Hu, Q. Li, T. Zhou, S. Jiang, and L. Yu. 2006. Survivin mutant (Surv-DD70, 71AA) disrupts the interaction of Survivin with Aurora B and causes multinucleation in HeLa cells. Biochem. Biophys. Res. Commun. 346:400–407. [DOI] [PubMed] [Google Scholar]
- Carvalho, A., M. Carmena, C. Sambade, W.C. Earnshaw, and S.P. Wheatley. 2003. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116:2987–2998. [DOI] [PubMed] [Google Scholar]
- Chalamalasetty, R.B., S. Hummer, E.A. Nigg, and H.H. Sillje. 2006. Influence of human Ect2 depletion and overexpression on cleavage furrow formation and abscission. J. Cell Sci. 119:3008–3019. [DOI] [PubMed] [Google Scholar]
- Chantalat, L., D.A. Skoufias, J.P. Kleman, B. Jung, O. Dideberg, and R.L. Margolis. 2000. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol. Cell. 6:183–189. [PubMed] [Google Scholar]
- Colnaghi, R., C.M. Connell, R.M. Barrett, and S.P. Wheatley. 2006. Separating the anti-apoptotic and mitotic roles of survivin. J. Biol. Chem. 281:33450–33456. [DOI] [PubMed] [Google Scholar]
- Conway, E.M., S. Pollefeyt, J. Cornelissen, I. DeBaere, M. Steiner-Mosonyi, K. Ong, M. Baens, D. Collen, and A.C. Schuh. 2000. Three differentially expressed survivin cDNA variants encode proteins with distinct antiapoptotic functions. Blood. 95:1435–1442. [PubMed] [Google Scholar]
- Cooke, C.A., M.M. Heck, and W.C. Earnshaw. 1987. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105:2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook, N.E., R.J. Clem, and L.K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval, B., A. Ferrand, N. Conte, C. Larroque, D. Hernandez-Verdun, C. Prigent, and D. Birnbaum. 2004. Aurora B -TACC1 protein complex in cytokinesis. Oncogene. 23:4516–4522. [DOI] [PubMed] [Google Scholar]
- Derry, W.B., L. Wilson, and M.A. Jordan. 1995. Substoichiometric binding of taxol suppresses microtubule dynamics. Biochemistry. 34:2203–2211. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., and C.A. Cooke. 1991. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of three distinct substages of metaphase and early events in cleavage furrow formation. J. Cell Sci. 98:443–461. [DOI] [PubMed] [Google Scholar]
- Engelsma, D., J.A. Rodriguez, A. Fish, G. Giaccone, and M. Fornerod. 2007. Homodimerization antagonizes nuclear export of survivin. Traffic. 8:1495–1502. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., C. James, G.I. Evan, and M.O. Hengartner. 1999. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr. Biol. 9:292–301. [DOI] [PubMed] [Google Scholar]
- Fukuda, S., R.G. Foster, S.B. Porter, and L.M. Pelus. 2002. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 100:2463–2471. [DOI] [PubMed] [Google Scholar]
- Gassmann, R., A. Carvalho, A.J. Henzing, S. Ruchaud, D.F. Hudson, R. Honda, E.A. Nigg, D.L. Gerloff, and W.C. Earnshaw. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, D.A., and C. Henriques. 2006. Centrifugal elutriation as a means of cell cycle phase separation and synchronisation. Subcell. Biochem. 40:359–361. [DOI] [PubMed] [Google Scholar]
- Giodini, A., M.J. Kallio, N.R. Wall, G.J. Gorbsky, S. Tognin, P.C. Marchisio, M. Symons, and D.C. Altieri. 2002. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 62:2462–2467. [PubMed] [Google Scholar]
- Goto, H., T. Kiyono, Y. Tomono, A. Kawajiri, T. Urano, K. Furukawa, E.A. Nigg, and M. Inagaki. 2006. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8:180–187. [DOI] [PubMed] [Google Scholar]
- Grossman, D., P.J. Kim, J.S. Schechner, and D.C. Altieri. 2001. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc. Natl. Acad. Sci. USA. 98:635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse, A., M. Mishima, and M. Glotzer. 2005. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15:778–786. [DOI] [PubMed] [Google Scholar]
- Honda, R., R. Korner, and E.A. Nigg. 2003. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 14:3325–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash, A.A., U.R. Klein, D. Lindner, J. Ebert, E.A. Nigg, and E. Conti. 2007. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 131:271–285. [DOI] [PubMed] [Google Scholar]
- Jiang, X., C. Wilford, S. Duensing, K. Munger, G. Jones, and D. Jones. 2001. Participation of Survivin in mitotic and apoptotic activities of normal and tumor-derived cells. J. Cell. Biochem. 83:342–354. [DOI] [PubMed] [Google Scholar]
- Jones, G., D. Jones, L. Zhou, H. Steller, and Y. Chu. 2000. Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J. Biol. Chem. 275:22157–22165. [DOI] [PubMed] [Google Scholar]
- Kawamura, K., N. Sato, J. Fukuda, H. Kodama, J. Kumagai, H. Tanikawa, Y. Shimizu, and T. Tanaka. 2003. Survivin acts as an antiapoptotic factor during the development of mouse preimplantation embryos. Dev. Biol. 256:331–341. [DOI] [PubMed] [Google Scholar]
- Kelling, J., K. Sullivan, L. Wilson, and M.A. Jordan. 2003. Suppression of centromere dynamics by Taxol in living osteosarcoma cells. Cancer Res. 63:2794–2801. [PubMed] [Google Scholar]
- Kim, J.Y., J.Y. Chung, S.G. Lee, Y.J. Kim, J.E. Park, K.S. Yoo, Y.H. Yoo, Y.C. Park, B.G. Kim, and J.M. Kim. 2006. Nuclear interaction of Smac/DIABLO with Survivin at G2/M arrest prompts docetaxel-induced apoptosis in DU145 prostate cancer cells. Biochem. Biophys. Res. Commun. 350:949–954. [DOI] [PubMed] [Google Scholar]
- Klein, U.R., E.A. Nigg, and U. Gruneberg. 2006. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell. 17:2547–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer, S.K., C. Bier, N. Habtemichael, and R.H. Stauber. 2006. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 7:1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer, S.K., C. Bier, P. Schlag, J. Fritzmann, W. Dietmaier, F. Rodel, L. Klein-Hitpass, A.F. Kovacs, C. Doring, M.L. Hansmann, et al. 2007. The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle. 6:1502–1509. [PubMed] [Google Scholar]
- Lens, S.M., R.M.F. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R.H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22:2934–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens, S.M., G. Vader, and R.H. Medema. 2006. The case for Survivin as mitotic regulator. Curr. Opin. Cell Biol. 18:616–622. [DOI] [PubMed] [Google Scholar]
- Li, F. 2003. Survivin study: what is the next wave? J. Cell. Physiol. 197:8–29. [DOI] [PubMed] [Google Scholar]
- Li, F., G. Ambrosini, E.Y. Chu, J. Plescia, S. Tognin, P.C. Marchisio, and D.C. Altieri. 1998. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 396:580–584. [DOI] [PubMed] [Google Scholar]
- Li, F., E.J. Ackermann, C.F. Bennett, A.L. Rothermel, J. Plescia, S. Tognin, A. Villa, P.C. Marchisio, and D.C. Altieri. 1999. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1:461–466. [DOI] [PubMed] [Google Scholar]
- Ling, X., and F. Li. 2004. Silencing of antiapoptotic survivin gene by multiple approaches of RNA interference technology. Biotechniques. 36:450–460. [DOI] [PubMed] [Google Scholar]
- Mahotka, C., M. Wenzel, E. Springer, H.E. Gabbert, and C.D. Gerharz. 1999. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 59:6097–6102. [PubMed] [Google Scholar]
- McNeish, I.A., R. Lopes, S.J. Bell, T.R. McKay, M. Fernandez, M. Lockley, S.P. Wheatley, and N.R. Lemoine. 2005. Survivin interacts with Smac/DIABLO in ovarian carcinoma cells but is redundant in Smac-mediated apoptosis. Exp. Cell Res. 302:69–82. [DOI] [PubMed] [Google Scholar]
- Mirza, A., M. McGuirk, T.N. Hockenberry, Q. Wu, H. Ashar, S. Black, S.F. Wen, L. Wang, P. Kirschmeier, W.R. Bishop, et al. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 21:2613–2622. [DOI] [PubMed] [Google Scholar]
- Muchmore, S.W., J. Chen, C. Jakob, D. Zakula, E.D. Matayoshi, W. Wu, H. Zhang, F. Li, S.C. Ng, and D.C. Altieri. 2000. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell. 6:173–182. [PubMed] [Google Scholar]
- O'Connor, D.S., D. Grossman, J. Plescia, F. Li, H. Zhang, A. Villa, S. Tognin, P.C. Marchisio, and D.C. Altieri. 2000. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc. Natl. Acad. Sci. USA. 97:13103–13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, H., C. Bakal, A. Shahinian, A. Elia, A. Wakeham, W.K. Suh, G.S. Duncan, M. Ciofani, R. Rottapel, J.C. Zuniga-Pflucker, and T.W. Mak. 2004. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J. Exp. Med. 199:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennati, M., G. Colella, M. Folini, L. Citti, M.G. Daidone, and N. Zaffaroni. 2002. Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatin-induced apoptosis. J. Clin. Invest. 109:285–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennati, M., M. Binda, G. Colella, M. Zoppe, M. Folini, S. Vignati, A. Valentini, L. Citti, M. De Cesare, G. Pratesi, et al. 2004. Ribozyme-mediated inhibition of survivin expression increases spontaneous and drug-induced apoptosis and decreases the tumorigenic potential of human prostate cancer cells. Oncogene. 23:386–394. [DOI] [PubMed] [Google Scholar]
- Petersen, J., and I.M. Hagan. 2003. S. pombe Aurora kinase/Survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13:590–597. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., and M.K. Balasubramanian. 1999. S. pombe Pbh1p: an inhibitor of apoptosis domain containing protein is essential for chromosome segregation. FEBS Lett. 460:187–190. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., and M.K. Balasubramanian. 2002. Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics. 160:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J.A., S.W. Span, C.G. Ferreira, F.A. Kruyt, and G. Giaccone. 2002. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein Survivin. Exp. Cell Res. 275:44–53. [DOI] [PubMed] [Google Scholar]
- Rodriguez, J.A., S.M. Lens, S.W. Span, G. Vader, R.H. Medema, F.A. Kruyt, and G. Giaccone. 2006. Subcellular localization and nucleocytoplasmic transport of the chromosomal passenger proteins before nuclear envelope breakdown. Oncogene. 25:4867–4879. [DOI] [PubMed] [Google Scholar]
- Rosa, J., P. Canovas, A. Islam, D.C. Altieri, and S.J. Doxsey. 2006. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell. 17:1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud, S., K. Samejima, D.F. Hudson, S.H. Kaufmann, and W.C. Earnshaw. 2006. Genetic analysis of apoptotic execution. In Reviews and Protocols in DT40 Research. Vol. 40. J.M. Buerstedde and S. Takeda, editors. Springer, New York. 75–90. [DOI] [PubMed]
- Ruchaud, S., M. Carmena, and W.C. Earnshaw. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812. [DOI] [PubMed] [Google Scholar]
- Samejima, K., S. Tone, and W.C. Earnshaw. 2001. CAD/DFF40 nuclease is dispensable for high molecular weight DNA cleavage and stage I chromatin condensation in apoptosis. J. Biol. Chem. 276:45427–45432. [DOI] [PubMed] [Google Scholar]
- Scrittori, L., D.A. Skoufias, F. Hans, V. Gerson, P. Sassone-Corsi, S. Dimitrov, and R.L. Margolis. 2005. A small C-terminal sequence of Aurora B is responsible for localization and function. Mol. Biol. Cell. 16:292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S., B.J. Sung, Y.S. Cho, H.J. Kim, N.C. Ha, J.I. Hwang, C.W. Chung, Y.K. Jung, and B.H. Oh. 2001. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 40:1117–1123. [DOI] [PubMed] [Google Scholar]
- Simpson, S. 1912. An investigation into the effects of seasonal changes on body temperature. Proc. Roy. Soc. Edin. 32:110–135. [Google Scholar]
- Song, Z., X. Yao, and M. Wu. 2003. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 278:23130–23140. [DOI] [PubMed] [Google Scholar]
- Song, Z., S. Liu, H. He, N. Hoti, Y. Wang, S. Feng, and M. Wu. 2004. A single amino acid change (Asp 53-> Ala53) converts Survivin from anti-apoptotic to pro-apoptotic. Mol. Biol. Cell. 15:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes, E.K., A. Uren, D. Vaux, and H.R. Horvitz. 2000. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 6:211–223. [DOI] [PubMed] [Google Scholar]
- Srinivasula, S.M., and J.D. Ashwell. 2008. IAPs: what's in a name? Mol. Cell. 30:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber, R.H., U. Rabenhorst, A. Rekik, K. Engels, C. Bier, and S.K. Knauer. 2006. Nucleocytoplasmic shuttling and the biological activity of mouse survivin are regulated by an active nuclear export signal. Traffic. 7:1461–1472. [DOI] [PubMed] [Google Scholar]
- Sun, C., D. Nettesheim, Z. Liu, and E.T. Olejniczak. 2005. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry. 44:11–17. [DOI] [PubMed] [Google Scholar]
- Tamm, I., Y. Wang, E. Sausville, D.A. Scudiero, N. Vigna, T. Oltersdorf, and J.C. Reed. 1998. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 58:5315–5320. [PubMed] [Google Scholar]
- Temme, A., M. Rieger, F. Reber, D. Lindemann, B. Weigle, P. Diestelkoetter-Bachert, G. Ehninger, M. Tatsuka, Y. Terada, and E.P. Rieber. 2003. Localization, dynamics, and function of survivin revealed by expression of functional survivinDsRed fusion proteins in the living cell. Mol. Biol. Cell. 14:78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A.G., T. Beilharz, M.J. O'Connell, S.J. Bugg, R. van Driel, D.L. Vaux, and T. Lithgow. 1999. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA. 96:10170–10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A.G., L. Wong, M. Pakusch, K.J. Fowler, F.J. Burrows, D.L. Vaux, and K.H. Choo. 2000. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 10:1319–1328. [DOI] [PubMed] [Google Scholar]
- Vader, G., J.J. Kauw, R.H. Medema, and S.M. Lens. 2006. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 7:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia, M.A., H. Huang, E. Dutil, D.A. Kaiser, T. Hunter, and J.P. Noel. 2000. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 7:602–608. [DOI] [PubMed] [Google Scholar]
- Wall, N.R., D.S. O'Connor, J. Plescia, Y. Pommier, and D.C. Altieri. 2003. Suppression of survivin phosphorylation on Thr34 by flavopiridol enhances tumor cell apoptosis. Cancer Res. 63:230–235. [PubMed] [Google Scholar]
- Walter, D., S. Wissing, F. Madeo, and B. Fahrenkrog. 2006. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 119:1843–1851. [DOI] [PubMed] [Google Scholar]
- Wang, Z., J. Sampath, S. Fukuda, and L.M. Pelus. 2005. Disruption of the inhibitor of apoptosis protein survivin sensitizes Bcr-abl-positive cells to STI571-induced apoptosis. Cancer Res. 65:8224–8232. [DOI] [PubMed] [Google Scholar]
- Wheatley, S.P., and I.A. McNeish. 2005. Survivin: a protein with dual roles in mitosis and apoptosis. Int. Rev. Cytol. 247:35–88. [DOI] [PubMed] [Google Scholar]
- Yan, H., J. Thomas, T. Liu, D. Raj, N. London, T. Tandeski, S.A. Leachman, R.M. Lee, and D. Grossman. 2006. Induction of melanoma cell apoptosis and inhibition of tumor growth using a cell-permeable Survivin antagonist. Oncogene. 25:6968–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., A. Welm, and J.M. Bishop. 2004. Cell division and cell survival in the absence of survivin. Proc. Natl. Acad. Sci. USA. 101:15100–15105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffaroni, N., A. Costa, M. Pennati, C. De Marco, E. Affini, M. Madeo, R. Erdas, A. Cabras, S. Kusamura, D. Baratti, et al. 2007. Survivin is highly expressed and promotes cell survival in malignant peritoneal mesothelioma. Cell. Oncol. 29:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., E. Bossy-Wetzel, and W. Jiang. 2005. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem. J. 389:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.