Abstract
Acquisition of mitochondria by the ancestor of all living eukaryotes represented a crucial milestone in the evolution of the eukaryotic cell. Nevertheless, a number of anaerobic unicellular eukaryotes have secondarily discarded certain mitochondrial features, leading to modified organelles such as hydrogenosomes and mitosomes via degenerative evolution. These mitochondrion-derived organelles have lost many of the typical characteristics of aerobic mitochondria, including certain metabolic pathways, morphological traits, and, in most cases, the organellar genome. So far, the evolutionary pathway leading from aerobic mitochondria to anaerobic degenerate organelles has remained unclear due to the lack of examples representing intermediate stages. The human parasitic stramenopile Blastocystis is a rare example of an anaerobic eukaryote with organelles that have retained some mitochondrial characteristics, including a genome, whereas they lack others, such as cytochromes. Here we report the sequence and comparative analysis of the organellar genome from two different Blastocystis isolates as well as a comparison to other genomes from stramenopile mitochondria. Analysis of the characteristics displayed by the unique Blastocystis organelle genome gives us an insight into the initial evolutionary steps that may have led from mitochondria to hydrogenosomes and mitosomes.
Keywords: Blastocystis, hydrogenosome, mitochondrion, mitosome, reductive evolution, stramenopile
Introduction
The acquisition of the mitochondrion was a crucial step, perhaps even the decisive event, in the evolution of the eukaryotic cell from an archaeal ancestor (Martin and Müller 1998; Embley and Martin 2006). It is generally accepted that this organelle is derived from a symbiotic alpha-proteobacterium that over time became so integrated into the host cell that almost all its several hundred constituent proteins are now encoded in the nucleus. Only a small and variable subset of genes has been retained on a separate organellar genome that resides in the mitochondrial matrix (Gray et al. 2004). The genes present on this genome are almost exclusively involved in either electron transport or translation.
A significant number of eukaryotic metabolic pathways include steps that occur within the mitochondrion, including the tricarboxylic acid cycle, fatty acid and amino acid metabolism, and iron–sulfur center assembly. Despite their important role in metabolism, a number of anaerobic eukaryotes appear to lack mitochondria. Although previously thought to represent basal lineages that had diverged prior to the integration of the alpha-proteobacterial symbiont (Cavalier-Smith 1987), it is now known that all these putatively primitive eukaryotes contain organelles of mitochondrial ancestry, albeit in dramatically modified form in most cases (van der Giezen et al. 2005).
This secondary modification of the organelle has occurred independently many times and has followed at least two different paths. One path has resulted in conversion of the mitochondrion into a hydrogenosome. These organelles occur in a number of unrelated eukaryotic lineages. Hydrogenosomes were first described and are best known from trichomonad flagellates, but they have also been found in certain anaerobic fungi and ciliates, indicating convergent evolution (van der Giezen et al. 2005). In the latter two groups, the organelle conversion in the anaerobes must be relatively recent as closely related aerobic species with classical mitochondria can be identified. The terminal electron acceptor in hydrogenosomes is a proton, the end product is hydrogen, and the enzyme responsible is hydrogenase, with limited energy being produced by substrate-level phosphorylation (van der Giezen et al. 2005). Because the only specific defining characteristic of hydrogenosomes is the production of hydrogen, it remains to be seen how similar the organelles in these three groups are in other respects.
The second path of mitochondrial modification in anaerobic eukaryotes results in conversion into mitosomes. Like hydrogenosomes, mitosomes are found in unrelated lineages (including diplomonads, amoebae, and microsporidia), indicating multiple independent origins (Mai et al. 1999; Tovar et al. 1999, 2003; Williams et al. 2002). These organelles are defined largely by exclusion as they have no unique shared characteristic like hydrogen production. Mitosomes are therefore defined as organelles of mitochondrial ancestry that are not known to produce energy or hydrogen, and in some cases (e.g., in Entamoeba), their function remains completely unknown (van der Giezen et al. 2005).
Eukaryotes with true mitochondria that can function under anaerobic conditions also exist and include some fungi, some worms, and Euglena. These organisms are only facultatively anaerobic as they use oxygen when it is present and, therefore, presumably possess a normal complement of mitochondrial genes. In such organisms, nitrate or fumarate, for example, may be used as the terminal acceptor under anaerobic conditions (Tielens et al. 2002).
One characteristic shared by most hydrogenosomes and all mitosomes is the absence of the cristae that give classical mitochondria their instantly recognizable morphology under the electron microscope. Additionally, with one published exception, an organelle genome is absent from all these secondarily modified mitochondria. Although it is still incompletely sequenced, the cristate hydrogenosome of the ciliate Nyctotherus contains a genome that is known to encode several components of complex I of the mitochondrial electron transport chain as well as ribosomal proteins (Boxma et al. 2005). No trace of an organellar genome has been detected in any mitosome (León-Avila and Tovar 2004).
Eukaryotes with morphologically classical mitochondria that are obligate anaerobes are rare. Blastocystis is a genetically diverse anaerobic parasite and a member of the diverse eukaryotic group known as the stramenopiles or heterokonts, which includes aerobic organisms like diatoms, oomycetes, and brown algae (Silberman et al. 1996). Blastocystis is a common member of the mammalian gut flora although whether it causes symptoms in humans is still a point of active debate (Tan et al. 2002). The mitochondrion of Blastocystis resembles what might be postulated as an intermediate stage during the conversion of classical mitochondria into hydrogenosomes or mitosomes. Electron microscopy showed that cristae are present (Stenzel and Boreham 1996), as are a transmembrane potential (Zierdt et al. 1988; Nasirudeen and Tan 2004) and DNA (Matsumoto et al. 1987; Nasirudeen and Tan 2004), but cytochrome-mediated electron transport and certain typical metabolic pathways (Zierdt 1986) appear to be absent.
In a recent publication, we reported a preliminary characterization of the Blastocystis mitochondrion-like organelle (MLO) through the analysis of expressed sequence tags (ESTs) sequences, confocal microscopy, and the partial sequence of the organellar DNA (Stechmann et al. 2008). Although hydrogenase protein was detected, hydrogen production has not been demonstrated, and so the definition of the organelle as a mitochondrion, mitosome, or hydrogenosome remains uncertain. As expected from the early work of Zierdt (1986), no evidence for complexes III and IV of the electron transport chain was found, in either the EST sequences or the MLO genome data. However, only a small segment of the MLO genome was sequenced (less than 6 kb), which encoded two subunits of reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase, seven transfer RNAs (tRNAs), and the ribosomal RNAs. As a result, our understanding of the genome was very incomplete. For example, the presence of genes encoding cytochrome or adenosine triphosphatase (ATPase) subunits on the MLO genome could not be ruled out even though they were not detected among the ESTs, as they may not have been expressed in the cells used for library construction or could have been present as pseudogenes, indicating a recent loss of function.
To complete our picture of the Blastocystis MLO and its genome, we here report the entire annotated sequence of the organelle genome from isolates representing two subtypes of Blastocystis and compare their structure and content with the mitochondrial genomes from aerobic stramenopiles sequenced to date. These are the first complete organellar genomes from obligately anaerobic protists, and the results illustrate one possible intermediate stage along the evolutionary pathway from mitochondrion to hydrogenosome or mitosome.
Materials and Methods
Blastocystis Origin and Culture
DNA from Blastocystis sp. NandII (ATCC50177) was a gift from Alexandra Stechmann and Tetsuo Hashimoto (Dalhousie University, Canada). Blastocystis sp. DMP/02-328 was grown at 36 °C with a mixed bacterial flora in LYSGM with 5% adult bovine serum. Subtyping of Blastocystis sp. DMP/02-328 indicated that this isolate belongs to subtype 4, whereas Blastocystis sp. NandII is subtype 1 (Stechmann et al. 2008). Both are of human origin.
DNA Extraction
Harvested Blastocystis sp. DMP/02-328 cells were concentrated and partially separated from bacteria using Histopaque-1077 (Sigma-Aldrich Ltd, Gillingham, Dorset, UK). The Blastocystis-enriched band at the medium–Histopaque interphase was recovered and washed with phosphate-buffered saline. Total DNA extraction was carried out on the resulting pellet using the Gentra Puregene Cell Kit (QIAGEN Ltd, Crawley, UK).
DNA Amplification by Polymerase Chain Reaction
The small segments of MLO DNA previously sequenced (Stechmann et al. 2008) were used as the starting points to design primers for inverse polymerase chain reaction (PCR) at both ends of the segment for both isolates.
Based on the restriction map of the available sequence, a set of restriction enzymes generating overhanging ends (BstBI, ClaI, EcoRI, HindIII, MfeI, NdeI, XbaI) or blunt ends (RsaI) was used for Southern blot hybridization (data not shown). Those enzymes generating short- to medium-sized fragments were used to digest the Blastocystis DNA. The resulting restriction fragments were self-ligated with T4 DNA ligase (Promega UK Ltd, Southampton, UK) and used as templates for the subsequent inverse PCRs.
In addition, degenerate primers for direct PCR were designed for other genes likely to be present in the genome (e.g., those encoding NADH dehydrogenase [nad] subunits), based on an alignment of gene sequences from other stramenopiles (see below).
Two types of PCR amplification were performed: a standard reaction using BioTaq (Bioline Ltd, London, UK) or one using TaKaRa Premix Taq (Lonza Biologics PLC, Wokingham, UK) for those reactions giving a negative result in the standard reaction as well as to obtain long PCR products (>2 kb). Because different combinations of primers were employed, some of which were degenerate, a touchdown amplification strategy was used. Normal touchdown PCR conditions consisted of 94 °C for 2 min, followed by 10 cycles of 94 °C for 15 s, 60 °C (decreasing by 0.5 °C per cycle) for 30 s, and 72 °C for 2 min, and followed by 20 cycles of 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 2 min. Long PCR conditions consisted of 94 °C for 2 min, followed by 10 cycles of 94 °C for 15 s, 60 °C (decreasing by 0.5 °C per cycle) for 30 s, and 68 °C for 4–5 min, and followed by 20 cycles of 94 °C for 15 s, 55 °C for 30 s, and 68 °C for 4–5 min (increasing by 5 s per cycle).
After PCR product sequencing, the process was repeated with new specific primers to “walk” along the MLO DNA in two directions: outward through inverse PCR and inward through direct PCR to cover the gaps between separate regions. To obtain the latter, pairs of specific primers were combined in all the possible ways and direct long PCR carried out in order to generate a PCR product linking two contigs.
Sequencing, Assembly, and Genome Analysis
PCR products were gel purified (Qiaquick Gel Extraction kit; QIAGEN Ltd), and direct sequencing of PCR products was performed on an ABI3730 with ABI Prism BigDye Terminator v3.1 reagents (Applied Biosystems, Warrington, UK). Sequences were assembled and analyzed with the Staden Package software programs (Staden et al. 2000). Blast searches were conducted to identify genes, pseudogenes, and structural RNAs. tRNA gene prediction was performed under the different search modes and models implemented in tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/; Lowe and Eddy 1997). Genes were annotated manually, based on Blast and alignment similarity (Altschul et al. 1997) to genes in other sequenced stramenopile mitochondrial genomes, and G + C content was calculated using GeneRunner v3.05 (http://www.generunner.com) (supplementary table S1, Supplementary Material online). The presence of group I and II introns was tested using the intron prediction tool RNAweasel (http://megasun.bch.umontreal.ca/RNAweasel; Lang et al. 2007). The GenBank accession numbers of the Blastocystis sp. DMP/02-328 and NandII MLO genomes analyzed in the present work are EF494739 and EF494740, respectively.
Sequence Alignments
For mitochondrial gene sequence alignments, all the stramenopile mitochondrial genomes sequenced to date were used: Thalassiosira pseudonana (DQ186202), Cafeteria roenbergensis (AF193903), Chrysodidymus synuroideus (AF222718), Ochromonas danica (AF287134), Phytophthora infestans (U17009), Phytophthora sojae (DQ832717), Phytophthora ramorum (DQ832718), Saprolegnia ferax (AY534144), Desmarestia viridis (AY500367), Dictyota dichotoma (AY500368), Fucus vesiculosus (AY494079), Laminaria digitata (AJ344328), and Pylaiella littoralis (AJ277126).
Nucleotide sequences were either directly aligned, in the case of structural RNAs, or first translated into amino acid sequences, in the case of protein-encoding genes, using the ClustalW tool implemented in the MEGA4 package (Kumar et al. 2004). The latter alignment was in turn used as a template to align the corresponding nucleotides to reduce ambiguities.
Amino acid sequence alignments were used to design degenerate primers based on conserved regions within the proteins. This allowed us to obtain additional sequences outside the existing contigs and, thus, additional sites for inverse PCR “walking.”
Phylogenetic Analyses
From the orthologous open-reading frames (ORFs) shared by Blastocystis MLO DNA and the other stramenopile genomes (see above), those producing unambiguous alignments (i.e., nad genes) were selected. For phylogenetic analyses, a range of other mitochondrial genomes were selected, based on the similarity of their nad gene content, as well as five alpha-proteobacterial genomes. Nucleotide sequences were concatenated, translated into amino acids, and aligned with ClustalW and MUSCLE v3.6 (Edgar 2004). The resulting alignments were edited manually to remove regions of ambiguous homology. Maximum likelihood (ML) and Bayesian inference analyses were carried out using Phyml_v2.4.5 (Guindon and Gascuel 2003) and MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001), respectively, using the best of the amino acid substitution models derived from ProtTest 1.4 (Abascal et al. 2005): a CpREV model using a gamma distribution, with the proportion of invariable sites estimated from the data and eight categories of substitution. In all, 400 bootstrap resamplings were performed to estimate statistical support for branches in the optimal ML tree. Bayesian analysis used four Markov chain Monte Carlo (MCMC) strands, 1,000,000 generations, with trees sampled every 100 generations. A consensus tree was produced after excluding an initial burn-in of 25% of the samples, as recommended.
Selection Analyses
To evaluate the type of selection operating on each gene (i.e., positive, purifying, or absence of either), we carried out an analysis using a Z-test of selection, implemented in MEGA3. The model used was modified Nei–Gojobori, one-tail with α = 0.025, and Jukes–Cantor as a nucleotide substitution model.
Results
Physical and Gene Map, Gene Content, and Structure of Blastocystis sp. DMP/02-328 and NandII MLO DNA
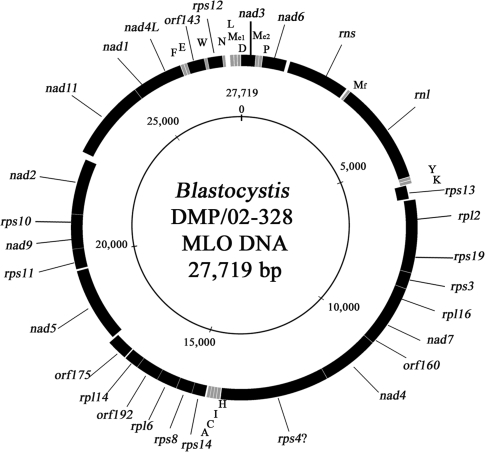
The MLO genome consists of a circular mapping molecule of 27,719 and 28,382 bp in Blastocystis sp. DMP/02-328 and NandII, respectively. The synteny between the two MLO genomes is complete, with no evidence of rearrangements having occurred since the divergence of subtypes 1 and 4, represented by Blastocystis sp. NandII and DMP/02-328, respectively. The genome consists of 45 genes, of which 27 are ORFs (23 of them with identifiable orthologs) and the rest structural RNA genes (the small and large subunit ribosomal RNAs, plus a set of 16 tRNAs, including two distinct tRNA-Mete genes). The structure of the Blastocystis sp. DMP/02-328 MLO genome is displayed in figure 1. It is a highly compact genome, with coding regions representing 96% of its total length and overlapping genes being a common feature.
FIG. 1.—
Gene and physical map of the Blastocystis MLO genome. Black blocks represent genes and ORFs that are transcribed clockwise (outside of circle) or counterclockwise (inside of circle). Gray blocks represent tRNA genes. tRNA genes are identified by their linked amino acid (single-letter code). Mf and Me1/Me2 are initiator and elongator methionyl tRNAs, respectively. The inner circle shows the size scale. The map was created using GenomeViz 1.1 [Ghai et al. 2004]). Because of overlapping coding regions (see text and supplementary table S2, Supplementary Material online), gaps between adjacent genes are not always present.
The extraordinary genetic diversity in Blastocystis previously reported (Stensvold et al. 2007) has also been detected in the MLO genome. The sequence divergence, which for some genes exceeds 50% between the two subtypes, is higher than that found between the mitochondrial genomes of many congeneric species or even distinct genera. The overall nucleotide identity between the Blastocystis sp. DMP/02-328 and NandII MLO genomes is 75.4%, or 71.6% and 64.3% for nucleotides and amino acids, respectively, when only protein-coding genes are considered. The gene-by-gene percent identity is shown in supplementary table S1 (Supplementary Material online).
As previously noted (Stechmann et al. 2008), the average G + C content of the MLO genomes of 21.9% and 19.9% in Blastocystis sp. DMP/02-328 and NandII, respectively (table 1), is remarkably lower than that of nuclear encoded genes, a commonly observed characteristic of mitochondrial (Lang et al. 2005) and other reduced genomes (such as those of obligate bacterial symbionts when compared with free-living bacteria [Moran 2002]). However, this G + C content is not evenly distributed along the genome, being higher in genes encoding structural RNAs than in those encoding proteins (supplementary table S1, Supplementary Material online). The G + C content of the 38 orthologous genes shared by Blastocystis and one representative per genus of all the stramenopiles included in this study is shown in table 1. Blastocystis displays the lowest overall G + C content.
Table 1.
Comparison of Some Genomic Features among 13 Mitochondrial Genomes from Stramenopiles
| Thalassiosira pseudonana | Cafeteria roenbergensis | Chrysodidymus synuroideus | Ochromonas danica | Phytophtora infestans | Saprolegnia ferax | Desmarestia viridis | Dictyota dichotoma | Fucus vesiculosus | Laminaria digitata | Pylaiella, littoralis | Blastocystis sp. DMP/02-328 | Blastocystis sp. NandII | |
| Total length (bp) | 43,827 | 43,159 | 34,119 | 41,035 | 37,957 | 46,930 | 39,049 | 31,617 | 36,392 | 38,007 | 58,507 | 27,719 | 28,382 |
| G + C content (%) | |||||||||||||
| Total | 30.1 | 27.3 | 24.1 | 26.2 | 22.3 | 23.1 | 36.6 | 36.5 | 34.5 | 35.1 | 38.0 | 21.9 | 19.9 |
| Shared genesa | 30.0 | 27.7 | 25.6 | 27.1 | 24.4 | 24.7 | 38.7 | 37.4 | 36.7 | 37.5 | 38.0 | 22.9 | 21.2 |
| Intergenic regions | 30.1 | 18.0 | 12.5 | 21.9 | 10.7 | 6.8 | 26.5 | 24.4 | 23.4 | 23.7 | 37.4 | 10.8 | 10.9 |
| Protein-coding regions | 29.0 | 25.9 | 22.9 | 24.7 | 21.0 | 20.9 | 35.6 | 35.4 | 33.3 | 34.0 | 37.1 | 20.7 | 17.9 |
| Structural RNA genes | 35.3 | 37.9 | 33.9 | 36.8 | 35.6 | 35.2 | 45.1 | 42.9 | 43.5 | 45.2 | 42.9 | 28.7 | 30.0 |
| Gene number | 61 | 58 | 62 | 75 | 67 | 77 | 68 | 66 | 67 | 67 | 79 | 45 | 45 |
| Protein coding | 35 | 34 | 37 | 44 | 40 | 43 | 39 | 38 | 38 | 39 | 52 | 27 | 27 |
| Structural RNAs | 27 | 24 | 25 | 31 | 27 | 24 | 28 | 27 | 28 | 27 | 26 | 18 | 18 |
| Average length ORFs (nt) | 804 | 1049 | 718 | 692 | 706 | 743 | 784 | 645 | 743 | 756 | 855 | 793 | 813 |
| Average length Intergenic region (nt) | 160 | 29 | 38 | 62 | 58 | 57 | 46 | 20 | 39 | 50 | 127 | 28 | 33 |
Consists of all Blastocystis genes except orf143, orf160, orf175, orf192, which have no clear homologs, and one of the two tRNA-Mete genes. Additionally, rps11 and rps10 are excluded due to their absence in certain other stramenopiles.
The standard genetic code is used in the translation of Blastocystis MLO-encoded proteins (supplementary table S3, Supplementary Material online). Like most other stramenopiles, except Pylaiella littoralis (Fontaine et al. 1995), there is no evidence of group I and II introns in either Blastocystis MLO genome.
Some features displayed by the MLO genome from Blastocystis are unique when compared with the true mitochondrial genomes of the other stramenopiles sequenced so far. As expected from a shorter genome, the gene repertoire of Blastocystis is reduced (see table 1). This is due to the complete absence of genes encoding cytochrome oxidase subunits I, II, and III (cox1–cox3), cytochrome b (cob), and all the F0F1-ATPase subunits (atp6, atp8, atp9, and in some species atp1) encoded by the mitochondrial DNA (mtDNA) in other stramenopiles.
In spite of the reduction in gene content, the Blastocystis MLO genome retains four unidentified ORFs (named orf143, orf160, orf175, and orf192 after the predicted number of amino acids in Blastocystis sp. DMP/02-328), suggesting that they serve an essential function, even though they have no clear homologs in the databases. None of them have conserved functional domains, and they show little or no similarity to known proteins in Blast searches, and only two are represented among the ESTs. orf160 shows a putative ATG start codon in Blastocystis sp. DMP/02-328 but has two early in-frame stop codons in Blastocystis sp. NandII. In addition, it has the lowest level of amino acid conservation between homologous genes in the two Blastocystis sp. MLO genomes; this is the most notable divergence in genome architecture between the two. It seems probable that these genes encode divergent ribosomal proteins, of which several are present in the majority of stramenopile mtDNAs but not identified in the MLO genomes (table 2).
Table 2.
Gene Content in Stramenopile mtDNAsa
| Genesb | Thalassiosira pseudonana | Cafeteria roenbergensis | Chrysodidymus synuroideus | Ochromonas danica | Phytophtora infestans | Saprolegnia ferax | Desmarestia viridis | Dictyota dichotoma | Fucus vesiculosus | Laminaria digitata | Pylaiella, littoralis | Blastocystis sp. |
| rns, rnl | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| trnA-W | 25 | 22 | 23 | 24 (29) | 25 | 25 (30) | 25 | 24 | 25 | 24 | 23 | 15 (16) |
| atp6, atp8 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| cob | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| cox1–cox3 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| nad1–nad7, nad9 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| nad11 | ▪ | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| atp1 | □ | ▪ | □ | □ | ▪ | ▪ | □ | □ | □ | □ | □ | □ |
| atp9 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rps2 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rps3 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps4 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ? |
| rps7 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rps8 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps10 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps11 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps12, rps14, rps19 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rpl2 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rpl5 | ▪ | □ | □ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rpl6, rp14, rp16 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| ymf16 | □ | □ | ▪ | □ | ▪ | □ | □ | □ | □ | □ | □ | □ |
| ORFs | 1 | 4 | 5 | 10 (12) | 4 | 4 | 4 | 3 | 3 | 4 | 16 | 4 |
The table is based on the gene content reported by Chesnick et al. (2000), with the addition of more recently sequenced stramenopile mtDNAs and the MLO.
Filled boxes–genes present; open boxes–genes absent.
Another distinctive feature of the Blastocystis MLO genome is the existence of an ORF of more than 920 amino acids for which no methionine start codon has been identified and which diverges significantly between the two isolates. Its N-terminal third shows weak similarity to rps4 in other stramenopiles. Part of this sequence appears in the same transcript as nad4 in one of the EST identified by Stechmann et al. (2008) (EF512301). rps4 in most other sequenced stramenopiles ranges from 153 to 269 amino acids length. Although larger than this, the putative Blastocystis rps4 is still far shorter than that of C. roenbergensis (1641 amino acids). The existence of alternative start codons has been reported for rps4 in some plant mitochondrial genomes (Bergthorsson et al. 2004), but we do not have empirical evidence (i.e., transcripts, ESTs) to confirm the rps4 start codon in Blastocystis.
Comparison of Organellar Genome Features between Blastocystis and Other Stramenopiles
The repertoire of tRNA genes is, by far, the smallest of any known stramenopile. The tRNA genes in the Blastocystis MLO genome are mostly clustered in a few locations rather than being evenly distributed along the genome. There are also two different elongator tRNA-Met genes instead of the single copy found in all other stramenopiles. As in most protists, many fungi, plants, and even a few animals, the number of organelle genome–encoded tRNA genes is not sufficient to support mitochondrial translation. This implies that the missing mitochondrial tRNAs are imported from the cytosol (Schneider and Maréchal-Drouard 2000). The number of tRNAs that are imported depends on the species, the most extreme cases being those of Trypanosoma brucei and Leishmania spp. (Simpson et al. 1989; Hancock and Hajduk 1990) and Plasmodium falciparum (Feagin 2000), which must import the complete set of tRNAs from the cytosol. Although far from being this extreme, the Blastocystis MLO genome still represents the most dramatic case of tRNA gene loss observed within the stramenopiles, with only 60–65% of the normal stramenopile tRNA gene complement being present.
Because the mitochondrial gene order is extremely varied among stramenopile mtDNAs, it is not surprising that their organization in Blastocystis does not resemble any of the others, despite the synteny within Blastocystis. However, a few traces of conserved blocks of ribosomal protein genes, previously reported in distantly related bacteria and mtDNA (Hauth et al. 2005), still remain in Blastocystis. These include the cluster rpl2-rps19-rps3-rpl16, as well as the block formed by rps14-rps8-rpl6. The latter is preceded in Blastocystis by orf175, which has weak similarity to rpl5, the gene normally (although not universally) found upstream of rps14-rps8-rpl6 in stramenopiles.
Phylogenetic Position of Blastocystis
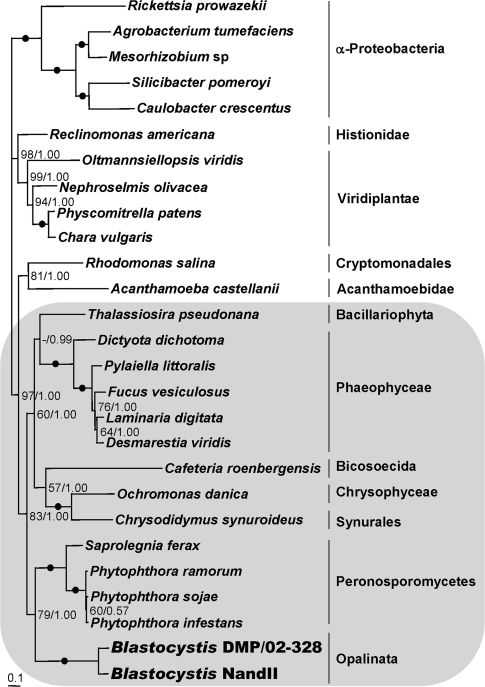
The ML tree based on the amino acid sequences of nine nad genes is shown in figure 2 (nad11 was excluded as it is not present in all organelle genomes analyzed). The topology of the Bayesian tree was identical. Trees using either method strongly support the clustering of Blastocystis sp. DMP/02-328 and NandII as a monophyletic clade, with weaker support for the Peronosporomycetes, represented by S. ferax and Phytophthora spp., as a sister group. This result suggests that oomycete mitochondrial genomes are the closest relatives of the Blastocystis MLO, in agreement with the analysis of Stechmann et al. (2008), which was based on a smaller number of MLO genes. Our trees support the monophyly of the Phaeophyceae, the Peronosporomycetes, and the clade consisting of the Chrysophyceae and Synurales. The branch lengths also agree with the relative rate test of substitution (Moran 1996; data not shown) in reflecting intermediate values of acceleration in substitution rates in Blastocystis sp., compared with the more accelerated C. roenbergensis and the slower remaining stramenopiles.
FIG. 2.—
ML tree of nad proteins. This tree was obtained using the edited alignment produced by MUSCLE v. 3.6, the CpREV+I+G+F amino acid substitution model, and is based on the concatenated amino acid sequences of the nine NADH dehydrogenase genes present in all of the stramenopiles included (i.e., nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9; see Materials and Methods) as well as in the 12 taxa used as outgroups. Numbers beside the internal nodes are the ML bootstrap values from 400 resamplings obtained with Phyml and the Bayesian MCMC posterior probability values. Black circles indicate 100% bootstrap support and 1.00 posterior probability values. Support values over 50% are shown adjacent to the corresponding nodes. Values below 50% are represented by a dash (–). The clade containing all the stramenopiles is shaded. Taxonomic classification follows the nomenclature of Adl et al. (2005).
Discussion
The completion of the MLO genome from two different Blastocystis subtypes has revealed some distinctive features that combine to make this organism's organelle unique. The MLO genome is missing several protein-encoding genes that are present in the mtDNAs of all other stramenopiles studied to date. However, these are not a random selection but primarily consist of all the genes encoding cytochrome and ATPase subunits. The absence of MLO-encoded subunits of the ubiquinol-cytochrome c reductase complex (complex III) and cytochrome c oxidase (complex IV) confirms the metabolism inferences made by Stechmann et al. (2008), which were based on the absence of any evidence for these complexes among their EST sequences. The absence of complexes III and IV in Blastocystis, together with the absence of F0F1-ATPase subunits, also agrees with previous biochemical studies (Zierdt 1986; Zierdt et al. 1988; Nasirudeen and Tan 2004) and represents a unique case to date among completely sequenced mitochondrial genomes. Our motivation for sequencing two MLO genomes from distinct subtypes was to see whether evidence for ongoing degeneration existed, but, at least in the MLO genome, no evidence for this was found. The absence of pseudogenes in the MLO genomes suggests that the gene loss is not recent.
Stechmann et al. (2008) proposed a metabolic map of the Blastocystis MLO based primarily on a library of EST sequences. The six nuclear encoded subunits of complex I they identified among the ESTs and the total of 10 now identified in the completed MLO genome reinforce the apparent importance of maintaining this complex in Blastocystis. It is likely to be responsible for generating the proton gradient in these organelles detected with MitoLight and Rhodamine 123 staining (Zierdt et al. 1988; Nasirudeen and Tan 2004) and MitoTracker (Hamblin et al. 2008; Stechmann et al. 2008). In Blastocystis, it is likely that complex I pumps protons derived from NADH as it passes electrons via an unidentified quinone to alternative oxidase (AOX).
Despite being incompletely sequenced, some traits of the Nyctotherus ovalis hydrogenosome genome (Boxma et al. 2005) are known to be shared with the MLO of Blastocystis. In both cases, a set of subunits from complexes I and II of the electron transport chain are present (Boxma et al. 2005; Hamblin et al. 2008; Stechmann et al. 2008), whereas no components of complexes III and IV or the F0F1-ATPase have been found. The fact that the latter are typically present in classical mitochondria but are absent from the MLO, hydrogenosomes, and mitosomes suggests that the loss of these three complexes could be linked to the first steps in degenerative evolution of mitochondria in anaerobic organisms, whatever the ultimate outcome.
We propose that a likely scenario would be one in which the organism passed through a facultatively anaerobic stage. In contrast to known examples where nitrate or fumarate may be used as an alternative, the absence of the normal terminal electron acceptor (O2) would result in all electrons being passed through existing pathways, either to AOX or hydrogenase via a quinone or ferredoxin, respectively, or potentially both (Stechmann et al. 2008). Subsequent mutations affecting any component of complexes III and IV would no longer be lethal, resulting in a removal of selection for maintaining those genes and producing a now obligately anaerobic organelle. The reduced transmembrane proton gradient generated by complex I alone may no longer be sufficient to support both solute transport and adenosine triphosphate synthesis via the F0F1-ATPase, leading to loss of the latter. All trace of the now unnecessary genes would disappear from the MLO genome (and presumably the nucleus also).
This appears to be the stage of degeneration at which Blastocystis is presently found, and in the absence of evidence for hydrogen production, the MLOs are probably most accurately described as obligately anaerobic mitochondria. Whether all other mitochondrion-derived organelle-bearing organisms went through an identical stage is impossible to know, as is whether the Blastocystis organelle will eventually degenerate further. Compared with the trichomonad hydrogenosome (Carlton et al. 2007) and known mitosomes, the Blastocystis MLO has a much more complete mitochondrion-like metabolism (Stechmann et al. 2008). This suggests that electron transport is one of the first, if not actually the first, pathways to be affected during the degenerative evolution of the mitochondrion into the derived organelles.
Note
While this article was in revision, a paper appeared describing the MLO genome of another subtype of Blastocystis sp. (Wawrzyniak et al. 2008).
Supplementary Material
Supplementary tables S1, S2, and S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grant 078566 from the Wellcome Trust awarded to CGC. We would like to thank Drs Andrew Roger and Alexandra Stechmann for providing us with the DNA from Blastocystis sp. NandII and also Dr Mark van der Giezen for his advice and for commenting on the manuscript.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Farmer MA, et al. (28 co-authors) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Altschul SE, Madden TL, Schäffer AA, Zhang J, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101:17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxma B, de Graaf RM, van der Staay GW, et al. (15 co-authors) An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, et al. (65 co-authors) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Eukaryotes with no mitochondria. Nature. 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- Chesnick JM, Goff M, Graham J, Ocampo C, Lang BF, Seif E, Burger G. The mitochondrial genome of the stramenopile alga Chrysodidymus synuroideus. Complete sequence, gene content and genome organization. Nucleic Acids Res. 2000;28:2512–2518. doi: 10.1093/nar/28.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Feagin JE. Mitochondrial genome diversity in parasites. Int J Parasitol. 2000;30:371–390. doi: 10.1016/s0020-7519(99)00190-3. [DOI] [PubMed] [Google Scholar]
- Fontaine JM, Rousvoal S, Leblanc C, Kloareg B, Loiseaux-de Goër S. The mitochondrial LSU rDNA of the brown alga Pylaiella littoralis reveals alpha-proteobacterial features and is split by four group IIB introns with an atypical phylogeny. J Mol Biol. 1995;251:378–389. doi: 10.1006/jmbi.1995.0441. [DOI] [PubMed] [Google Scholar]
- Ghai R, Hain T, Chakraborty T. GenomeViz: visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hamblin K, Standley DM, Rogers MB, Stechmann A, Roger AJ, Maytum R, van der Giezen M. Localization and nucleotide specificity of Blastocystis succinyl-CoA synthetase. Mol Microbiol. 2008;68:1395–1405. doi: 10.1111/j.1365-2958.2008.06228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K, Hajduk SL. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- Hauth AM, Maier UG, Lang BF, Burger G. The Rhodomonas salina mitochondrial genome: bacteria-like operons, compact gene arrangement and complex repeat region. Nucleic Acids Res. 2005;33:4433–4442. doi: 10.1093/nar/gki757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lang BF, Brinkmann H, Koski LB, Fujishima M, Gortz HD, Burger G. On the origin of mitochondria and Rickettsia-related eukaryotic endosymbionts. Jpn J Protozool. 2005;38:171–183. [Google Scholar]
- Lang BF, Laforest M-J, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- León-Avila G, Tovar J. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology. 2004;150:1245–1250. doi: 10.1099/mic.0.26923-0. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Yamada M, Yoshida Y. Light-microscopical appearance and ultrastructure of Blastocystis hominis, an intestinal parasite of man. Zentralbl Bakteriol Mikrobiol Hyg [A] 1987;264:379–385. doi: 10.1016/s0176-6724(87)80059-7. [DOI] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Nasirudeen AM, Tan KS. Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J Microbiol Methods. 2004;58:101–109. doi: 10.1016/j.mimet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Schneider A, Maréchal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- Silberman JD, Sogin ML, Leipe DD, Clark CG. Human parasite finds taxonomic home. Nature. 1996;380:398. doi: 10.1038/380398a0. [DOI] [PubMed] [Google Scholar]
- Simpson AM, Suyama Y, Dewes H, Campbell DA, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal K, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Hamblin K, Pérez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold CR, Suresh GK, Tan KS, Thompson RC, Traub RJ, Viscogliosi E, Yoshikawa H, Clark CG. Terminology for Blastocystis subtypes-a consensus. Trends Parasitol. 2007;23:93–96. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Stenzel DJ, Boreham PF. Blastocystis hominis revisited. Clin Microbiol Rev. 1996;9:563–584. doi: 10.1128/cmr.9.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KS, Singh M, Yap EH. Recent advances in Blastocystis hominis research: hot spots in terra incognita. Int J Parasitol. 2002;32:789–804. doi: 10.1016/s0020-7519(02)00005-x. [DOI] [PubMed] [Google Scholar]
- Tielens AGM, Rotte C, van Hellemond JJ, Martin W. Mitochondria as we don't know them. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Tovar J, León-Avila G, Sánchez L, Sutak R, Tachezy J, van der Giezen M, Hernández M, Müller M, Lucocq JM. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I, Roussel M, Diogon M, Couloux A, Texier C, Tan KSW, Vivarès CP, Delbac F, Wincker P, El Alaoui H. Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis. Int J Parasitol. 2008;38:1377–1382. doi: 10.1016/j.ijpara.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Williams BAP, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Zierdt CH. Cytochrome-free mitochondria of an anaerobic protozoan-Blastocystis hominis. J Protozool. 1986;33:67–69. doi: 10.1111/j.1550-7408.1986.tb05559.x. [DOI] [PubMed] [Google Scholar]
- Zierdt CH, Donnolley CT, Muller J, Constantopoulos G. Biochemical and ultrastructural study of Blastocystis hominis. J Clin Microbiol. 1988;26:965–970. doi: 10.1128/jcm.26.5.965-970.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.