Abstract
The histone H3 amino terminus, but not that of H4, is required to prevent the constitutively bound activator Cha4 from remodeling chromatin and activating transcription at the CHA1 gene in Saccharomyces cerevisiae. Here we show that neither the modifiable lysine residues nor any specific region of the H3 tail is required for repression of CHA1. We then screened for histone H3 mutations that cause derepression of the uninduced CHA1 promoter and identified six mutants, three of which are also temperature-sensitive mutants and four of which exhibit a sin− phenotype. Histone mutant levels were similar to that of wild-type H3, and the mutations did not cause gross alterations in nucleosome structure. One specific and strongly derepressing mutation, H3 A111G, was examined in depth and found to cause a constitutively active chromatin configuration at the uninduced CHA1 promoter as well as at the ADH2 promoter. Transcriptional derepression and altered chromatin structure of the CHA1 promoter depend on the activator Cha4. These results indicate that modest perturbations in distinct regions of the nucleosome can substantially affect the repressive function of chromatin, allowing activation in the absence of a normal inducing signal (at CHA1) or of Swi/Snf (resulting in a sin− phenotype).
In eukaryotic organisms, genomic DNA is packaged by nucleosomes into chromatin. Contacts between histone proteins and DNA in the nucleosome limit the accessibility of DNA elements to binding factors. This packaging of DNA into chromatin has led to the evolution of a layer of regulatory complexity not available to prokaryotes (38).
Specific residues in the histone proteins participate in transcriptional regulation by a variety of mechanisms. Covalent modifications at a large number of sites are important not only for regulation of transcription but also for regulation of repair, replication, and recombination (6). These modification sites are principally located in the flexible histone amino termini, but some are also located in the structured interior of the nucleosome, at surface-accessible residues (2, 4). Modifications of these residues affect transcription by altering the affinities of transcription factors and cofactors for the chromatin template or by directly affecting chromatin structure (2). Chromatin structure can also be altered to regulate transcription by ATP-utilizing remodeling complexes, such as the Swi/Snf complex (21). Specific amino acid residues within the histone fold domains of histones H3 and H4, which comprise the innermost core of the nucleosome, are required to form nucleosomes that are repressive to transcription in the absence of remodeling, as mutations in these residues partially bypass the requirement for Swi/Snf for the activation of genes such as SUC2, GAL1-10, and HO in Saccharomyces cerevisiae (11, 14). Structural studies demonstrate that these Swi/Snf-independent (sin−) mutations alter histone-histone or histone-DNA interactions in the nucleosome (26). Similarly, targeted mutagenesis of residues in histone H4 implicated in interactions between the (H3-H4)2 tetramer and the H2A-H2B dimer resulted in a variety of transcriptional defects in yeast (35).
Our lab previously studied the role of the amino terminus of the histone H3 protein in transcription in Saccharomyces cerevisiae (budding yeast) at a genomewide scale and found that it is mainly repressive to transcription (33). Repression via the H3 amino terminus depends heavily on the modifiable lysine residues and, for many genes, on the histone deacetylase Rpd3, which targets the histone H3 and H4 amino termini (33, 34). Interestingly, CHA1, which encodes a serine/threonine dehydratase and is induced in the presence of high levels of serine or threonine, is derepressed in H3Δ1-28 yeast in the absence of serine or threonine, but this derepression does not depend on Rpd3 (33). Derepression of CHA1 in H3Δ1-28 yeast requires the activator Cha4, which binds the CHA1 promoter even in the absence of serine or threonine (22, 33). This suggests that the histone H3 amino terminus prevents activation by Cha4 by a mechanism that is potentially independent of the modifiable lysine residues.
To gain new insight into CHA1 repression mediated by histone H3, we tested whether the modifiable lysine residues in the tail were required for this effect and found that they were not. We then conducted a screen for H3 mutants that cause similar derepressing effects on the uninduced CHA1 promoter to those seen in the H3Δ1-28 mutant, and we report here the identification of six mutations that cause significant derepression of the CHA1 promoter. Three of the mutants are temperature sensitive and four are sin− mutants, showing that their effects are not unique to the CHA1 promoter. Furthermore, the mutations are dispersed throughout the length of histone H3, indicating that several residues that can be altered without affecting viability and which were not previously identified as affecting transcriptional regulation participate in the formation of repressive chromatin. Since derepression is observed at the CHA1 promoter and depends on the constitutively bound activator Cha4, these results also demonstrate that nucleosomes modulate transcription at steps occurring subsequent to activator binding.
MATERIALS AND METHODS
Yeast strains and growth.
Yeast strains used for this study are listed in Table 1. The parent strain used in the screen was derived from MX1-4C, which is of the S288C background (24). MX1-4C was transformed with a lys2::KanMX PCR fragment amplified from the yeast deletion collection mutant (8) to generate RMY310. To introduce a CHA1-LYS2 reporter gene into this strain, we constructed the integrating plasmid pRS403CHA1LYS2. First, the LYS2 coding sequence was excised from the plasmid ILys-8, a generous gift of Steve Hanes, by use of EcoRI and SalI and then ligated with pRS403 (3), also cut with EcoRI and SalI. The CHA1 promoter was then amplified as a 400-bp fragment, using primers that introduce EcoRI sites at both ends, and ligated into this plasmid, creating a fusion construct in which two additional amino acids were added to the N terminus of the LYS2 message, under the control of the CHA1 promoter. Yeast strain RMY320 was then constructed by transforming RMY310 with pRS403CHA1LYS2 partially digested with NheI. RMY330, used in later rounds of screening, was generated by transforming RMY320 with pRS305CHA1MEL1 digested with AflII. To construct pRS305CHA1MEL1, the CHA1-MEL1 fusion from pBM150CHA1MEL1 (32) was first inserted as a BamHI-EcoRI fragment into pRS414 (3) and was then cloned into pRS305 (36) as a SacI-XhoI fragment. The CHA1 promoter was activated by growing yeast in the presence of 1 mg/ml serine for at least 3 h or by including 1 mg/ml serine in plate assays. Growth was done at 30°C unless otherwise indicated.
TABLE 1.
Yeast strains used for this study
| Strain | Genotype | Reference |
|---|---|---|
| CY1-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pMS337[CEN ARS LEU2 HHT1-HHF1] | 44 |
| CY2-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pMS358[CEN ARS LEU2 hht1-2(Δ1-28)-HHF1] | 44 |
| CY3-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pCY318[CEN ARS LEU2 hht1(Δ1-20)-HHF1] | 44 |
| CY4-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pCY328[CEN ARS LEU2 hht1(Δ1-15)-HHF1] | 44 |
| CY5-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pCY338[CEN ARS LEU2 hht1(Δ1-10)-HHF1] | 44 |
| CY6-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pCY348[CEN ARS LEU2 hht1(Δ1-5)-HHF1] | 44 |
| CY7-4C | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pCY358[CEN ARS LEU2 hht1-3(K4,9,14,18,23,27Q)-HHF1] | 44 |
| MX1-4C | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pMS329 (CEN ARS URA3 HHT1-HHF1) | 24 |
| RMY301 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pDM18/19 (CEN ARS TRP1 HHT1-HHF1) | This study |
| RMY310a | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 lys2Δ::KanMX Δ(hht1-hhf1)(hht2-hhf2) pMS329 (CEN ARS URA3 HHT1-HHF1) | This study |
| RMY320a | MATα ura3-52 leu2-3,112 lys2Δ::KanMX trp1-289 his3Δ1::PCHA1LYS2::HIS3 Δ(hht1-hhf1)(hht2-hhf2) pMS329 (CEN ARS URA3 HHT1-HHF1) | This study |
| RMY330a | MATα ura3-52 leu2-3,112::PCHA1MEL1::LEU2 lys2Δ::KanMX trp1-289 his3Δ1::PCHA1LYS2::HIS3 Δ(hht1-hhf1) (hht2-hhf2) pMS329 (CEN ARS URA3 HHT1-HHF1) | This study |
| RMY341a | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pDM18/19 (CEN ARS TRP1 HHT1-HHF1) [URA3-ARS1] | This study |
| RMY350a | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) snf5Δ::KanMX pMS329 (CEN ARS URA3 HHT1-HHF1) | This study |
| RMY360a | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) cha4Δ::KanMX pMS329 (CEN ARS URA3 HHT1-HHF1) | This study |
For strains carrying mutant H3 alleles, the last digit of the strain name (from 0 to 7) designates the allele and plasmid that carries it, as follows: 0, pMS329; 1, HHT2 on pDM18/19; 2, hht2-3NT on pDM18/19; 3, hht2-2NT on pDM18/19; 4, hht2-RG on pDM18/19; 5, hht2-FE on pDM18/19; 6, hht2-AG on pDM18/19; 7, hht2-AT on pDM18/19; 8, hht2-RG-AT on pDM18/19.
Mutant screen.
To generate histone H3 mutations, error-prone PCR was used in conjunction with plasmid gap repair (see Fig. 2A) (5, 10). Plasmid pDM18 (a gift from Andrea Duina and Fred Winston) (5), which harbors the HHF2-HHT2 locus, was used as template in a first-round PCR performed in buffer containing 0.5 mM MnSO4 (5 mM MgCl2, 0.5 mM MnCl2, 10 mM Tris, pH 8.3, 50 mM KCl, 0.05% NP-40, 0.05% Tween 20). The first-round PCR used the following two sets of primers: OD24 (5′-GGATCCCCCGGGGGTAATATGTAGACAGTGATT-3′) and OD4 (5′-GGGCGTCCTACGGATGGGAGTTGG-3′), whose product encompasses the entire HHT2 gene plus 264 bp 5′ and 303 bp 3′ of the gene (OD4-OD24 fragment); and OD24 and OD14 (5′-GTAACTTTCTGATCAACAGTTCAGTAG-3′), whose product encompasses 264 bp 5′ of the start ATG and 200 bp 3′ of the start ATG (OD14-OD24 fragment). Product from the first-round PCR was used as a template in the second-round PCR, using the same primers but without MnSO4 in the reaction mix. OD4-OD24 and OD14-OD24 fragments from second-round PCRs were then mixed with an AflII-RsrI fragment or an AgeI-RsrI fragment derived from the pDM18 plasmid, both of which retain the TRP1 and CEN sequences but have either the entire HHT2 gene or the 5′ end of it up to 120 bp removed. The mixture was used to transform yeast strain RMY320 (or RMY330, in later experiments). Transformants were selected on CSM−Trp medium, and the URA3-marked plasmid harboring wild-type HHT2 was then eliminated using counterselection with 5-fluoroorotic acid.
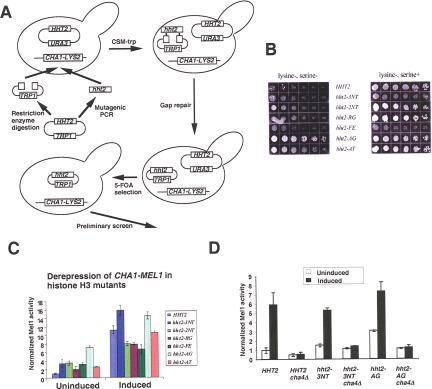
FIG. 2.
Identification of histone H3 mutants that derepress the uninduced CHA1 promoter. (A) Schematic of screen. (B) Growth of the parent strain (RMY321) and mutants (RMY322 to -327) on CSM−Lys±Ser. Cells were serially diluted threefold and were grown at 30°C for 72 h. (C) H3 mutant strains and the corresponding wild-type strain (RMY331 to -337) were grown in CSM−Ura−Trp, with or without 1 mg/ml serine, and the activity of the CHA1-MEL1 reporter gene was measured. Mel1 activities were normalized to that of the uninduced wild-type strain, and the SD is indicated for each (n = 3). (D) Cha4 is required for derepression of the CHA1 promoter in hht2-3NT and hht2-AG mutant yeast. Mel1 activities were normalized to that of the uninduced wild-type strain, and the SD is indicated for each (n = 3).
The resulting transformants, which potentially harbored viable mutations in histone H3, were replica plated on CSM−Lys−Ser and CSM−Lys+Ser media and grown at 30°C for 48 h. Colonies showing similar rates of growth with and without serine were then subjected to liquid Mel1 assay to monitor the activity of α-galactosidase, the product of the MEL1 gene, following transformation with pBM150CHA1MEL1 (31, 32) or in later screens using the integrated CHA1-MEL1 reporter gene, without and with serine induction. Plasmids were isolated from transformants passing this test and retransformed into RMY320 to verify that the mutation conferring the derepressed phenotype was plasmid borne. Finally, the HHT2 gene was sequenced from these confirmed mutants.
Enzyme assay and analysis of chromatin structure.
Measurement of α-galactosidase activity, which measures the activity of the MEL1 gene product, was performed as described previously (31). At least three independent yeast clones were assayed for each reported value. Chromatin was prepared, digested with micrococcal nuclease (MNase) or (for the PHO5 promoter) ClaI, and analyzed as described previously (13, 37). All chromatin mapping experiments were done at least twice. The CHA1 promoter was probed from the BamHI site 602 bp downstream of the starting ATG, and the ADH2 promoter was probed relative to a SacI site that is 656 bp 5′ of the starting ATG (33).
Tiling array experiments were performed and analyzed as previously described (16), except that the arrays used were Affymetrix 1.0R yeast tiling arrays. This necessitated the use of a different bpmap file, Sc03b_MR_v04.bpmap, from that used previously for analysis with Affymetrix tiling array software (TAS). This bpmap excludes probes not passing specific quality control criteria (e.g., likely to display cross-hybridization with other probes for the same DNA), and the resulting nucleosome occupancy profiles display gaps as a result. Two hybridizations were performed with nucleosomal DNA from wild-type yeast and three were performed with nucleosomal DNA from hht2-AG yeast; intensities were averaged and compared to averaged intensities from three control genomic DNA hybridizations reported previously (16), using TAS. Regions showing P values of <0.001 were identified using the interval analysis option in TAS, with the width set to 80 bp in steps of 40 bp. Results were viewed in the Integrated Genome Browser (http://www.affymetrix.com/support/developer/tools/download_igb.affx).
Topology assay.
The URA3-ARS1 yeast plasmid was made as follows. The URA3 gene was amplified from pRS426 (3) by PCR, and BamHI and HindIII sites were included in the primers to place sites near the ends of the amplified fragment. The BamHI-HindIII fragment from the PCR was then ligated with a 440-bp BamHI-HindIII fragment from pUC19TALS that included the ARS1 replication origin (30), yielding the 1.65-kb URA3-ARS1 plasmid among the ligation products, which were used directly to transform yeast. Transformants were selected on CSM−Ura medium, and topoisomer analysis was done as described previously (25, 37).
Western blotting.
Two optical density units (at 600 nm) of cells (A600 = ∼1.0) were precipitated and washed with 50 mM Tris (pH 7.5), 10 mM NaN3 on ice. After being resuspended in 70 μl of ESB (2% sodium dodecyl sulfate [SDS], 80 mM Tris [pH 6.8], 10% glycerol, 1.5% dithiothreitol, 0.1 mg/ml bromphenol blue), cells were transferred to microcentrifuge tubes for a 3-min incubation at 100°C. Glass beads were added to reach the meniscus, and the samples were vortexed at top speed for 2 min. An additional 30 μl of ESB was added, and the samples were heated to 100°C for 1 min. Following standard SDS-polyacrylamide gel electrophoresis, proteins were electroblotted on a Millipore polyvinylidene difluoride membrane. The membrane was blocked with Blotto buffer (5% nonfat milk in phosphate-buffered saline [PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 2 mM KH2PO4]) and incubated for 3 h at 4°C. For Western blotting against histone H3, the membrane was incubated at 4°C overnight with antibody directed against the carboxy terminus of histone H3 (Upstate 07-690; 1/15,000). After being washed by PBST (PBS plus 0.05% Tween 20), the blots were incubated for 3 h at room temperature with horseradish peroxidase-linked anti-rabbit antibody. After washing of the blots, an ECL Enhanced kit (GE Healthcare) was used to illuminate the reactive band.
Microarray accession number.
Microarray data have been deposited at GEO (accession number GSE12163).
RESULTS
The repressive function of the H3 amino terminus toward CHA1 transcription is dispersed.
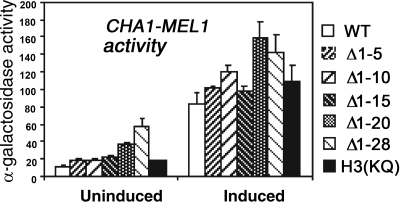
Repression of CHA1 under noninducing conditions requires the histone H3 amino terminus (33). To determine whether this repression depends on a specific region or on specific residues of the H3 amino terminus, we tested the activity of a CHA1-MEL1 reporter gene in yeast strains with successively longer truncations of the H3 amino terminus, as well as in a mutant strain in which the lysine residues in the H3 tail were mutated to glutamine [H3(KQ) mutant] (44). The results indicate that no single region of the H3 tail is responsible for this repressive function, as significantly increased expression was observed in the absence of induction upon removal of amino acids 1 to 5, 16 to 20, and 21 to 28 (Fig. 1). Modest increases in expression of the induced CHA1-MEL1 reporter were also seen for most of the same tail deletions, in a similar graded fashion. Furthermore, although many genes that are derepressed in H3Δ1-28 yeast are also derepressed in the H3(KQ) mutant (34), we were surprised to find that the CHA1-MEL1 reporter gene was expressed at wild-type levels under both uninduced and induced conditions in this mutant and also in an S10A mutant (Fig. 1 and data not shown). We concluded that the repressive function of the H3 amino terminus on CHA1 expression under uninduced conditions is dispersed over most of the amino terminus and that regulation of the CHA1 promoter does not depend on the modifiable lysine residues.
FIG. 1.
The repressive effect of the H3 amino terminus is dispersed and independent of the lysine residues. Yeast strains harboring wild-type histone H3 (CY1-4C) or H3 mutants having successive truncations of the amino terminus, as indicated (CY2-, CY3-, CY4-, CY5-, and CY6-4C), or having lysines mutated to glutamines (CY7-4C) were transformed with the reporter gene plasmid pBM150CHA1MEL1. Three independent transformants of each strain were grown in complete synthetic medium lacking uracil and leucine (CMS−Ura−Leu), with or without 1 mg/ml serine, and Mel1 activity was measured. Standard deviations (SD) are indicated with error bars.
Identification of histone H3 mutations that derepress the uninduced CHA1 promoter.
To gain new insight into the regulation of CHA1 repression by histone H3, we conducted a screen to identify histone H3 mutations that mimic the N-terminal deletion that causes CHA1 activation under noninducing conditions. The strategy used was similar to that employed by Duina and Winston (Fig. 2A) (5). We began with a strain, RMY320, having both chromosomal copies of the genes encoding histones H3 and H4 deleted and carrying the HHF2-HHT2 locus on a CEN plasmid with a URA3 marker. In addition, this yeast strain harbors a chromosomal LYS2 gene driven by the CHA1 promoter, allowing robust growth of the parent strain on CSM−Lys medium in the presence, but not absence, of serine (Fig. 2B). Error-prone PCR was used to amplify the HHT2 gene, which was introduced into RMY320 together with the gapped pDM18 plasmid. Resulting TRP+ transformants were replica plated on plates containing 5-fluoroorotic acid to eliminate the URA3-marked plasmid harboring the wild-type HHT2 gene and subsequently tested for growth on CSM−Lys±Ser plates (Fig. 2B). Transformants showing similar growth on CSM−Lys in the presence and absence of serine were chosen for further analysis, including analysis of their effect on expression of a CHA1-MEL1 reporter gene, as described in Materials and Methods.
Using this protocol, screening of approximately 8,000 transformants resulted in identification of six histone H3 mutants that caused significant derepression of the uninduced CHA1 promoter (Table 2). Expression of a CHA1-MEL1 reporter gene (in which the MEL1-encoded α-galactosidase is under the control of the CHA1 promoter) in strains bearing these mutations was derepressed two- to sevenfold over wild-type repressed levels in the absence of serine (Fig. 2C). In all of the mutants, increased expression was still observed upon serine induction, though for some the induced level was significantly decreased compared to that of the wild-type strain. Interestingly, the effects of these mutants on the uninduced and induced levels of CHA1-MEL1 expression were not strictly correlated (e.g., compare effects of the 3NT and 2NT mutations or of the FE and AT mutations on uninduced and induced levels of CHA1-MEL1 expression) (Fig. 2C), indicating that the effects of these mutations on uninduced and induced levels of expression occur by distinct mechanisms.
TABLE 2.
Histone H3 mutants that derepress the CHA1 promoter
| Mutant | Mutation(s) in H3 sequence | Location of mutation | ts mutant | sin− mutant |
|---|---|---|---|---|
| hht2-3NTa | K4E, V35A, K37R | N terminus | − | − |
| hht2-2NTb | T11I, G33C | N terminus | + | + |
| hht2-RG | R69G | Histone fold domain, α-helix 1 | − | + |
| hht2-FEc | F104S, E105G | Histone fold domain, α-helix 2 | + | + |
| hht2-AG | A111G | Histone fold domain, α-helix 2 | + | + |
| hht2-AT | A111T | Histone fold domain, α-helix 2 | − | − |
Separated K4E and V35A K37R (the latter was not separated into individual mutations) mutants each show modest derepressing effects, which are approximately additive in the hht-3NT mutant.
Separated T11I and G33C mutants show only modest derepressing effects compared to the parental mutant. Furthermore, neither mutation causes temperature sensitivity in the host strain or is a sin− mutation (data not shown).
Separated F104S and E105G mutants show only modest derepressing effects compared to the parental mutant. Furthermore, neither mutation causes temperature sensitivity in the host strain. The F104S mutant shows no sin− phenotype, and the E105G mutant is a weak sin− mutant (data not shown).
Previous work from our lab showed that derepression of the uninduced CHA1 promoter in H3Δ1-28 yeast requires Cha4, the dedicated activator of the CHA1 gene (12, 33). We tested two H3 mutants, the hht2-3NT and hht2-AG mutants, for the ability to derepress the CHA1 promoter in a cha4Δ background. The hht2-3NT mutant was chosen because its mutated residues are located in or near the amino-terminal domain, and one of them, H3 K4, is known to be modified by trimethylation at the 5′ ends of open reading frames (ORFs) that are actively transcribed by RNA polymerase II; the hht2-AG mutant was chosen because it has the strongest derepressing effect measured by Mel1 assay. Expression assays using the CHA1-MEL1 reporter showed that CHA4 deletion eliminated the increased activity of the uninduced CHA1 promoter in these two mutants (Fig. 2D). Since Cha4 is constitutively bound to the CHA1 promoter (33), we concluded that, like the case for the H3Δ1-28 mutant, these mutations alter histone H3 in a way that prevents normal inhibition of bound Cha4 at the CHA1 promoter under noninducing conditions.
The derepression caused by the identified H3 mutations could be due to indirect effects if, for example, the mutations led to elevated intracellular levels of serine or threonine (28). We measured intracellular levels of amino acids and found that serine and threonine levels did not differ between mutant and wild-type yeast (data not shown). Thus, as for the depression of CHA1 observed in H3Δ1-28 yeast, the effect of these histone mutations is likely to occur via a direct effect on the CHA1 promoter (33).
Sequencing of the six confirmed mutants revealed that the mutations were dispersed throughout the amino acid sequence of histone H3 (Table 2). Four of the mutants have two or three distinct point mutations, and separation of these resulted in substantially diminished effects on CHA1 repression (data not shown). Interestingly, two distinct mutations at A111 showed substantially different effects on CHA1 repression as well as on ts and sin phenotypes (Fig. 2C; see Fig. 4).
FIG. 4.
sin− phenotypes of histone H3 mutants. Strains expressing wild-type histone H3 or histone H3 mutants and deleted for the SNF5 gene (RMY351 to -357) were serially diluted threefold and grown on glucose- or galactose-only medium at 30°C for 72 h. The control SNF5+ strain was RMY301.
Identification of ts and sin− mutants.
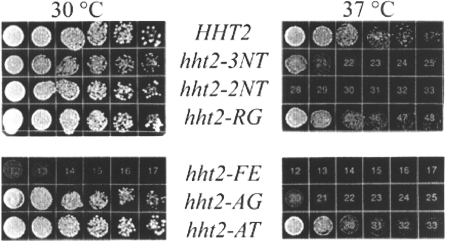
We noted that two mutants, the hht2-3NT and -FE mutants, caused somewhat slower growth of the host strains at 30°C (Fig. 2B and data not shown). We tested all mutants for temperature sensitivity on rich medium at 37°C and found that three mutants, the hht2-3NT, -2NT, and -AG mutants, showed pronounced temperature sensitivity at 37°C (Fig. 3).
FIG. 3.
Temperature sensitivity of histone mutants. Cells from wild-type and H3 mutant (RMY331 to -337) strains were serially diluted threefold and were grown on yeast extract-peptone-dextrose at 30°C for 48 h or at 37°C for 72 h.
A group of histone mutants called sin− mutants were previously identified by the ability to alleviate transcriptional defects of certain genes when the nucleosome remodeling complex Swi/Snf is inactive. sin− mutants were identified in all four core histone genes, including histone H3 E105K, R116A, R116H, T118A, T118H, and T118I mutants (14, 35). We tested the H3 mutants identified in our screen for the sin− phenotype. SNF5, which encodes a component of the Swi/Snf nucleosome remodeling complex essential for activity, was deleted from wild-type yeast and our H3 mutant strains, and the resulting strains were grown on medium having galactose as the carbon source (43). Control experiments showed that all six histone mutant strains were capable of robust growth on galactose-containing medium (data not shown). The snf5Δ strain expressing wild-type histone H3 grew poorly on galactose, as expected (Fig. 4) (11, 27). Two of the H3 mutants showed little or no growth on galactose in the absence of Snf5, but four H3 mutants, the hht2-2NT, -RG, -FE, and -AG mutants, exhibited significantly improved growth in the absence of Snf5 on galactose-containing medium (Fig. 4), thus qualifying as sin− mutants. swi/snf mutant yeast cells are also defective in activation of SUC2 transcription in low-glucose medium (11, 27), and we found partial suppression of this defect in hht2-AG snf5Δ yeast, further supporting classification of the hht2-AG mutant as a sin− mutant (data not shown).
H3 mutants that derepress CHA1 do not strongly alter minichromosome topology.
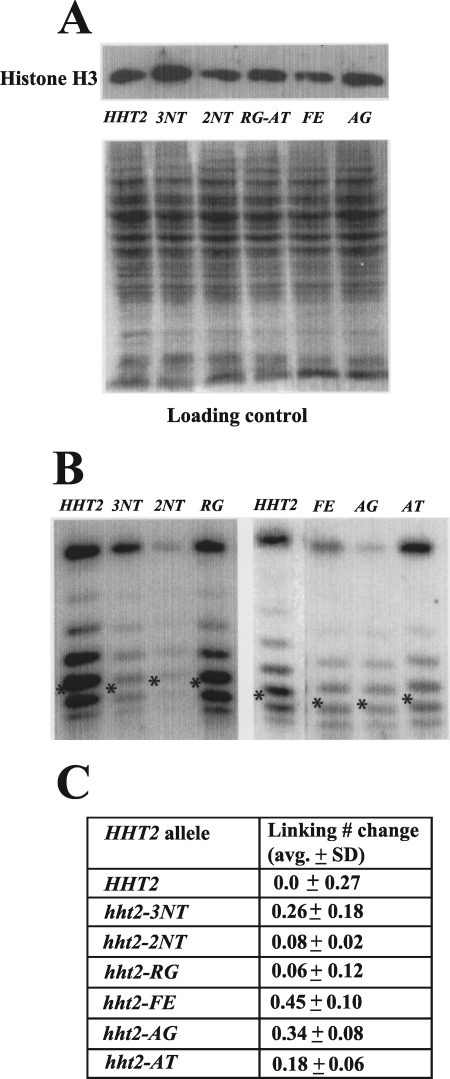
A reduced histone protein level has been shown to cause changes in yeast transcription; for example, loss of one copy of the H2A-H2B coding sequence can relieve the effect of the lack of Swi/Snf nucleosome remodeling activity in yeast (11). Thus, the effect of our H3 mutants on CHA1 promoter regulation could be caused by altered expression levels. To test this possibility, we performed Western blots using whole protein extracts prepared from strains having wild-type or mutant H3. Antibody that recognizes an epitope within the carboxy terminus of histone H3 was used because none of the residues in this region was mutated in our six characterized mutants.
Results from the Western blot showed that none of the six identified mutants had a significantly lower protein level than that of the wild type (Fig. 5A); indeed, all of them had a very similar protein level to that of the wild type, and the hht2-3NT mutant had a slightly higher protein level. We concluded that the derepression of the CHA1 promoter caused by these histone H3 mutants was not caused by changes in their expression.
FIG. 5.
Histone H3 mutations do not grossly affect protein level or nucleosome structure. (A) Histone mutants are expressed at levels similar to that of wild-type histone H3. Cell lysates from yeast strains harboring wild-type histone H3 (RMY331) or the indicated mutants (RMY332, RMY333, RMY335, RMY336, and RMY338) were electrophoresed in a 10% polyacrylamide gel, subjected to Western blotting, and visualized using an antibody against the C terminus of histone H3. The loading control was done by staining the SDS-PAGE gel after protein transfer. (B) Histone mutations cause little change in minichromosome topology. Genomic DNAs from strains harboring URA3-ARS1 and expressing wild-type histone H3 (RMY341) or the indicated H3 mutants (RMY342 to -347) were extracted using glass beads. DNA samples were run in a 1.5% agarose gel containing 40 μg/ml chloroquine; following Southern blotting, the minichromosomes were visualized using a probe corresponding to the URA3 coding sequence. Asterisks indicate the center of the Gaussian distribution of topoisomers. The band at the top of each lane is nicked circular URA3-ARS1 plasmid, and faster-migrating topoisomers represent more positively supercoiled species. (C) Linking number change (toward more positively supercoiled values) of the URA3-ARS1 minichromosome in yeast harboring the indicated histone H3 mutants relative to that in the wild type. Averages and SD were determined using at least three independent clones.
The effect of our identified histone mutations on chromatin structure was further assessed by analyzing the distribution of topoisomers of a stably replicating minichromosome in strains carrying wild-type or mutant H3. Each nucleosome reduces the linking number of a closed circular minichromosome by almost exactly one (that is, confers one negative supercoil), and it has been shown that histone mutants, as well as histone depletion, can alter supercoiling in minichromosomes (7, 17, 18, 25). We examined the impact of our mutations on the distribution of topoisomers of a small (1.65 kb), closed, circular minichromosome designated URA3-ARS1 and found little change in yeast harboring mutant H3 compared to the wild type (Fig. 5B). Quantitation showed that the Gaussian centers of the URA3-ARS1 topoisomer distributions in the H3 mutants shifted only modestly compared to the wild type (Fig. 5C). Thus, these histone H3 mutants do not strongly change chromatin structure in vivo, for example, by causing unwinding of DNA from the exit and entry points of the nucleosome (17).
The H3 A111G mutation causes chromatin remodeling of the uninduced chromosomal CHA1 promoter.
We next sought to determine the effect of the H3 hht2-AG mutant, which expresses the A111G mutation and shows the strongest derepressing effect of the six mutants identified here on the chromatin structure of the CHA1 promoter. The uninduced CHA1 promoter is characterized by an accessible upstream activating sequence, a nucleosomal TATA element, and strongly positioned nucleosomes across the coding sequence (23, 33). Upon induction, the TATA-containing nucleosome was disrupted, and positioning of downstream nucleosomes was altered as well (Fig. 6A, lanes 1 to 10) (23, 33). This remodeled state is constitutive in H3Δ1-28 yeast (33).
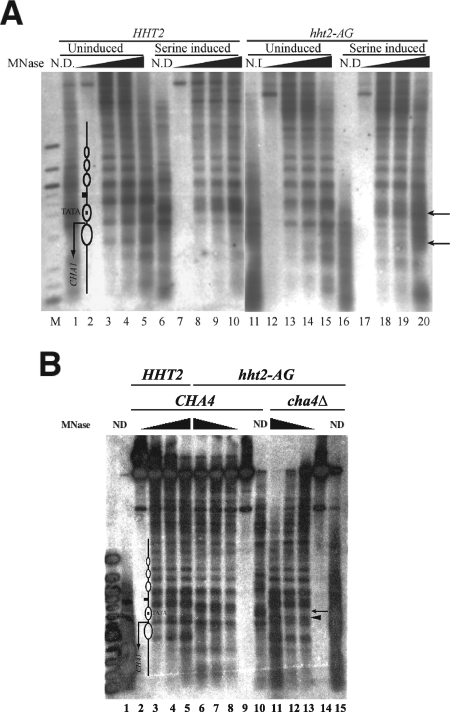
FIG. 6.
Perturbed chromatin structure of the CHA1 promoter in hht2-AG mutant yeast. (A) MNase cleavage sites were mapped for naked DNA and for chromatin from HHT2 (RMY301) and hht2-AG (RMY306) yeast grown in the presence or absence of serine, as indicated. Cleavage sites were mapped relative to the BamHI site 602 bp 3′ of the starting ATG. Chromatin was digested with MNase at 10 units/ml (lanes 3, 8, 13, and 18), 20 units/ml (lanes 4, 9, 12, and 19), and 50 units/ml (lanes 5, 10, 15, and 20); no-MNase controls are in lanes 2, 7, 12, and 17, and lanes 1, 6, 11, and 16 contain naked DNA digested with 2 units/ml MNase. Lane M contains 100-bp-ladder marker DNA (NEB). The upper arrow indicates a cleavage close to the TATA element that is protected in the inactive promoter, and the lower arrow indicates a cleavage between two positioned nucleosomes present in the inactive gene that is lost following nucleosome rearrangement in the active gene. The diagram in lane 2 shows the locations of nucleosomes in the inactive promoter and the TATA element; the small square between nucleosomes and upstream of the TATA element represents the CHA1 upstream activating sequence. (B) MNase cleavage sites were mapped from the BamHI site from chromatin prepared from HHT2 (RMY301) (lanes 2 to 5), hht2-AG (RMY306) (lanes 6 to 9), and cha4Δ::KanMX hht2-AG (RMY356) (lanes 11 to 14) yeast grown in the absence of serine. The leftmost lane contains a 100-bp marker (NEB). Samples were digested with MNase at 10 units/ml (lanes 3, 8, and 13), 20 units/ml (lanes 4, 7, and 12), and 50 units/ml (lanes 5, 6, and 11); lanes 2, 9, and 14 are no-MNase controls, and lanes 1, 10, and 15 contain naked DNA digested with 2 units/ml MNase. The structure of the uninduced promoter region is indicated on the left, with the ovals representing positioned nucleosomes. The arrowhead next to lane 12 indicates a cleavage site that is reproducibly cut more prominently in uninduced cha4Δ hht2-AG yeast than in uninduced HHT2 yeast, and the arrow above indicates a cleavage site in the region of the TATA element that is cleaved in hht2-AG yeast when Cha4 is present but not in cha4Δ hht2-AG yeast.
Mapping of MNase cleavage sites in chromatin from uninduced hht2-AG mutant yeast revealed increased accessibility of the TATA element region of the CHA1 promoter compared to the protection seen in uninduced wild-type yeast (Fig. 6A, upper arrow; compare lanes 13 to 15 to lanes 3 to 5). (Some variability in the extent of this remodeling was seen from experiment to experiment [Fig. 6B].) Remodeling was also evident in the region downstream of the transcription start site (Fig. 6A, lower arrow). Consistent with the increased activation observed upon induction in the mutant strain (Fig. 2C), increased chromatin remodeling was seen under inducing conditions (Fig. 6A, compare lanes 18 to 20 to lanes 13 to 15).
The remodeled state of the CHA1 promoter seen in wild-type yeast under inducing conditions and in H3Δ1-28 yeast under noninducing conditions depends on the activator Cha4 (23, 33), as does CHA1 promoter derepression in the hht-AG mutant (Fig. 2D). We therefore investigated whether the nucleosome remodeling caused by the hht2-AG mutant on the uninduced CHA1 promoter is also dependent on CHA4. The chromatin structure of the endogenous CHA1 promoter was analyzed as before in cha4::KanMX hht2-AG yeast, and the results showed that nucleosomes in the uninduced CHA1 promoter are positioned in a similar pattern to that seen for the uninduced wild-type strain. However, we reproducibly observed slightly enhanced digestion within the TATA-occupying nucleosome in cha4Δ hht2-AG yeast compared to that in the uninduced wild-type yeast (Fig. 6B, highlighted by a filled triangle in lane 13). Taken together, the results of Fig. 6 suggest that the hht2-AG mutation alters nucleosome structure in a way that prevents the chromatin-mediated inhibition of the constitutively bound activator Cha4 in the absence of an inducing signal (22, 33).
Effect of H3 A111G mutation on chromatin structure at other sites.
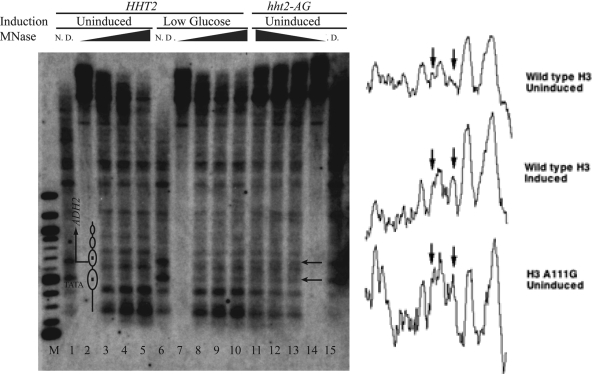
The constitutively remodeled state that we observed for the CHA1 promoter in hht2-AG yeast led us to examine the chromatin structures of two other inducible promoters that exhibit nucleosome remodeling during their activation, i.e., PHO5 and ADH2 (1, 40), to determine whether they were similarly perturbed by this mutation. The uninduced PHO5 promoter is packaged in well-positioned nucleosomes, one of which prevents cleavage of a ClaI site; this site becomes strongly accessible when PHO5 is active (1). We did not find any significant change in the accessibility of the ClaI site in the uninduced PHO5 promoter in hht2-AG yeast compared to that in the wild type (data not shown). In contrast, MNase digestion followed by indirect end-label probing of the ADH2 promoter revealed a constitutively remodeled state (Fig. 7). In particular, two nucleosomes that are well positioned in the uninduced ADH2 promoter, with one incorporating the TATA element and the other incorporating the starting ATG of the ADH2 ORF, are both perturbed by activation in wild-type yeast (when grown in low-glucose medium) (40) and in the uninduced hht2-AG mutant. In both cases, enhanced cleavage is observed in the region of the TATA-containing nucleosome, and cleavage sites that mark nucleosome boundaries in the uninduced promoter and coding sequence are less prominently cut (Fig. 7, compare lanes 8 to 13 to lanes 3 to 5; also see the densitometric scans in the right panel). Thus, the hht2-AG mutation causes nucleosome remodeling in the uninduced ADH2 promoter, altering its chromatin structure to a state similar to that seen in the induced wild-type strain.
FIG. 7.
Perturbed chromatin structure of the ADH2 promoter in hht2-AG yeast. MNase cleavage sites were mapped for naked DNA and for chromatin from HHT2 (RMY301) and hht2-AG (RMY306) yeast in the presence of 2% glucose (uninduced) and 0.05% glucose (low glucose), as indicated, for 2.75 h. Cleavage sites were mapped relative to the SacI site 656 bp 5′ of the starting ATG. Samples were digested with MNase at 10 units/ml (lanes 3, 8, and 13), 20 units/ml (lanes 4, 9, and 12), and 50 units/ml (lanes 5, 10, and 11); lanes 2, 7, and 14 are no-MNase controls, and lanes 1, 6, and 15 contain naked DNA digested with 2 units/ml MNase. Lane M contains 100-bp-ladder marker DNA. The arrows indicate a cleavage site close to the TATA element and a cleavage site close to the starting ATG site that are cleaved more strongly in induced wild-type or uninduced hht2-AG yeast than in the uninduced wild-type yeast. The diagram in lane 2 indicates the positions of nucleosomes in the uninduced promoter and the TATA element. To the right are densitometric scans of portions of lanes 3 (upper), 8 (middle), and 13 (lower), between 200 and 1,000 bp. The left and right arrows correspond to the upper and lower arrows next to lane 14, respectively. Note the peak indicated by the right arrows in the lower two scans and essentially absent from the upper scan and the stronger intensities of surrounding peaks, indicated by the left arrows, in the lower two scans than in the upper scan.
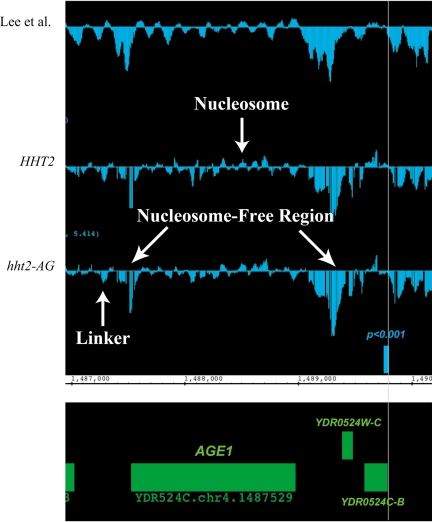
To determine the scope of changes in chromatin structure caused by the H3 A111G mutation on a global level, we compared the patterns of nucleosome occupancy in wild-type and hht2-AG mutant yeast genomewide by hybridizing mononucleosomal DNA from each strain to high-resolution tiling arrays (16). We obtained nucleosome occupancy profiles for three independent preparations of nucleosomal DNA from hht2-AG yeast and for two preparations of nucleosomal DNA from wild-type yeast (a third wild-type sample was of low quality and was not used). The nucleosome profiles generated by averaging the profiles for these replicate samples were in good agreement with that previously reported, although the signal-to-noise ratio was somewhat lower, and were in extremely good agreement with one another (Fig. 8). Thus, both samples showed nucleosome-depleted regions at promoters, consistent with earlier work, and evidence for well-positioned nucleosomes across much of the genome, particularly in regions occupied by ORFs. We obtained those regions showing the most significant differences between the A111G mutant and wild-type nucleosome occupancy profiles by interval analysis in TAS, employing an 80-bp sliding window moving over 40-bp increments, and found 115 regions with P values of <0.001. Close examination showed that these regions typically showed modestly increased or decreased nucleosome occupancy in the A111G mutant compared to that in the wild type, were fairly evenly distributed among ORF, promoter, and other (e.g., ARS) regions, and in general revealed relatively modest differences in chromatin structure (Fig. 8). Because these profiles were obtained for yeast growing in rich medium in order to compare the data with those reported previously, we did not expect to observe changes in the chromatin structure of CHA1, as the CHA1 promoter is activated by the serine present in rich medium. We concluded that although the A111G mutation (and other mutations reported here) may alter nucleosome structure in a way that predisposes some promoters toward a remodeled state, overall this mutation (and likely the others) causes only subtle changes in chromatin structure and does not result in gross alterations of nucleosome occupancy across the yeast genome.
FIG. 8.
Nucleosome occupancy measured by high-density tiling arrays over a region of yeast chromosome IV. The upper trace indicates nucleosome occupancy (measured as the log2 of the intensity of mononucleosomal DNA compared to that of a genomic DNA control), measured by Lee et al. (16), and the traces below show nucleosome occupancy in wild-type yeast (RMY301) and hht2-AG yeast (RMY306), as indicated, from the present study. Chromosomal coordinates are indicated, and the green rectangles represent known ORFs, as indicated. The small blue rectangle labeled “P < 0.001” is a region at which nucleosome occupancy differs between the wild type and the hht2-AG mutant, as can be seen from the individual traces (vertical line). Examples of a well-positioned nucleosome, nucleosome-free regions, and a linker region are indicated. The small gaps visible in the profiles from the current study are a processing artifact (see Materials and Methods).
DISCUSSION
Previously, we reported that the histone H3 amino terminus, but not that of H4, is required to prevent the constitutively bound activator Cha4 from remodeling chromatin and activating transcription at the CHA1 gene in yeast (33). Here we report that in spite of the modifiable lysine residues in the H3 tail being required for its repressive function at the majority of affected genes (34), they are not required for repression of CHA1. To gain additional information about the repression of CHA1 by histone H3, we therefore conducted a screen for histone H3 mutations that cause derepression of the uninduced CHA1 promoter. We identified six H3 mutants that derepress both a CHA1-LYS2 reporter integrated into the LEU2 locus and a plasmid-borne CHA1-MEL1 reporter (Fig. 2B and C). We also examined expression of the endogenous CHA1 gene by Northern blotting and quantitative reverse transcription-PCR but found little derepression in either the H3 mutants identified here or the H3Δ1-28 mutant (data not shown). However, chromatin structure at the endogenous locus is altered both in H3Δ1-28 yeast (23) and in the A111G mutant (Fig. 6), indicating that these mutants do affect regulation of the endogenous CHA1 promoter. It is possible that transcriptional derepression in these mutants at the endogenous CHA1 locus may be prevented by the influence of the nearby silent-mating-type locus HML, which is within about 1 kb of the 3′ end of the CHA1 ORF. Consistent with this possibility, the endogenous CHA1 gene is derepressed in sir4Δ yeast (23), while a plasmid-borne CHA1-MEL1 reporter shows no such derepression (data not shown).
Three of the identified mutants are temperature sensitive and four exhibit a sin− phenotype. The mutant H3 proteins were expressed at levels similar to that of wild-type H3, indicating that the derepression caused by these mutations is not the result of altered chromatin structure caused by a decreased H3 protein level. The mutations did not cause gross alterations in nucleosome structure, as indicated by a lack of change in minichromosome topology for all six mutants and in the genomewide nucleosome occupancy profile for the strongly derepressing hht2-AG mutant. However, we did find that yeast harboring the hht2-AG mutation exhibited a constitutively “remodeled” chromatin structure at the uninduced CHA1 promoter that was equivalent to the active chromatin configuration in wild-type yeast. A similar finding was made for the ADH2 promoter, but the PHO5 promoter appeared not to be perturbed. Both transcriptional derepression and altered chromatin structure at CHA1 depended on the activator Cha4.
Our findings have implications for the mechanism of repression of the uninduced CHA1 gene as well as for the more generally repressive function of chromatin toward transcription. First, we have shown that mutation of all six lysine residues in the amino terminus of H3 to glutamine, mimicking the uncharged acetylated state, has no effect on CHA1 regulation. This was a surprising finding, given the strong requirement for the H3 amino terminus for CHA1 repression, but is consistent with the lack of requirement for Rpd3 for CHA1 repression or Gcn5 for its activation (23, 33). We also observed intermediate levels of derepression caused by shorter deletions of the H3 tail (Fig. 1); interestingly, a similar graded effect of H3 amino-terminal deletions was previously seen for transcriptional activation of several genes activated by Gcn4 (44). Taken together, our results suggest that the repressive function of the H3 amino terminus is dispersed and possibly independent of its charge. This dispersed repressive function may also explain our failure to find single point mutants in the amino terminus that mimic the H3Δ1-28 phenotype.
The nature of the six H3 mutants we identified that cause significant derepression of the CHA1 promoter suggests that subtle and dispersed defects in nucleosome structure can have significant effects on transcriptional regulation. Structural considerations provide some clues as to how these mutations may affect chromatin function. Two alleles, hht2-3NT and hht2-2NT, affect residues in the N-terminal region of histone H3, which is not resolved in the X-ray crystal structure of the nucleosome (20, 42). Some of the affected residues (V35 and K37 in hht2-3NT and G33 in hht2-2NT) fall in a region of H3 shown to be required for repression of basal transcription in yeast (17). However, the deletion of this region also substantially affects minichromosome topology (17), whereas the mutant alleles do not, so we do not believe that these mutations grossly affect basal repression. We speculate that the affected residues in these mutants alter the H3 tail in a way that directly affects CHA1 repression, perhaps by affecting its interaction with an unidentified repressor protein or perhaps with Cha4 itself.
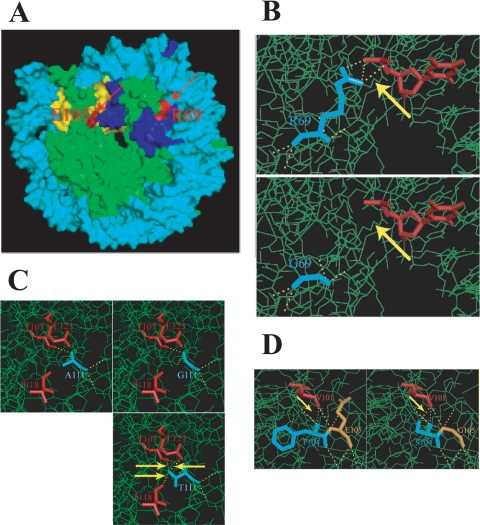
The hht2-RG allele encodes an R69G mutation. Residue R69 is located on the surface of the nucleosome and forms direct contact via its amine group with the phosphate backbone of nucleosomal DNA (Fig. 9A and B). The R69G mutation is predicted to cause a loss of the R69-DNA interaction but not of histone-histone interactions (Fig. 9B, yellow arrows). This loss of histone-DNA interaction may destabilize the nucleosome sufficiently to allow its remodeling at the CHA1 promoter under noninducing conditions and is likely also the reason for the hht2-RG mutant being a sin− mutant. Other sin− mutants have similarly been found to cause a loss of histone-DNA interaction when they are assembled into nucleosomes (26). Interestingly, though, whereas sin− mutations in the histones have been found in regions that might perturb histone-DNA contacts near the nucleosome dyad (superhelix loop [SHL] 0.5) or that would affect histone-histone interactions (19, 26, 29, 35), R69 makes its contact at SHL 1.5 (that is, 1.5 helical turns from the nucleosome dyad). This and the hht2-2NT mutant thus define new types of sin− mutants.
FIG. 9.
Structural features of histone H3 mutants. (A) Locations of R69 and E105 on the surface of the nucleosome core. R69 (red) forms direct contact with nucleosomal DNA (cyan), while E105 (red) does not. F104 and A111 are buried in the histone fold domain and thus are invisible. Yellow and blue, histone H3 proteins that have the labeled R69 and E105 residues, respectively; green, the other core histone proteins. (B) Closeup of the R69-DNA interaction and effect of R69G mutation. (C) Closeup of the region around A111 and replacement with glycine or threonine. (D) Closeup of the region around F104 and E105 and the F104S E105G (hht2-FE) mutation.
Residue A111 is buried in the nucleosome near the end of α-helix L2, and the A111G mutation found in the hht2-AG mutant is not expected to have a direct effect on histone-DNA interaction since this residue does not contact DNA (Fig. 9C). However, this residue is in close proximity to H3 T118; H3 T118 forms a hydrogen bond with H4 R45, whose side chain projects into the minor groove of DNA near the nucleosome dyad (near SHL 0.5), and specific mutations of H3 T118 and H4 R45 confer a sin− phenotype (14). We speculate that the decreased volume and/or perturbation of α-helix L2 caused by the A111G mutation affects DNA-histone contact near the nucleosome dyad, similarly to H3 T118 or H4 R45 sin− mutation, thus altering nucleosome stability (26). Consistent with this view, the more conservative A111T mutation (hht2-AT) has a reduced effect on CHA1 repression and does not display a sin− phenotype.
The hht2-FE allele encodes F104S and E105G mutations. F104 is buried in the histone fold domain, but E105 is on the nucleosomal surface (Fig. 9D). Neither F104 nor E105 has direct contact with DNA. The reduced side chain volumes caused by the F104S and E105G mutations may change the spacing of neighboring residues and elicit structural alterations in the nucleosome in a similar fashion to that of the A111G mutation. This notion is consistent with the finding that not only CHA1 derepression but also the ts and sin− phenotypes of the hht2-FE mutant are far weaker or absent in the corresponding single mutants.
As pointed out above, the hht2-2NT and hht2-RG mutants define new types of sin− mutants. Previous work has shown a range of effects of sin− mutants on nucleosome structure, varying from nearly negligible to strong effects on MNase susceptibility, minichromosome topology, and resolved structure (15, 26, 35, 41). Our results are most consistent with mild structural perturbations causing sin− phenotypes, and the occurrence of sin− mutations at distinct regions of the nucleosome indicates that various alterations in nucleosome structure permit at least partial bypass of normally obligatory chromatin remodeling. As discussed by Muthurajan et al. (26), a delicate balance must exist between the role of chromatin in DNA organization and the requirement for a dynamic structure that allows for transcription, replication, and other DNA transactions; this need must contribute to the extreme evolutionary conservation of the histones (39).
We focused on the hht2-AG (A111G) mutant, which shows strongest derepression of CHA1 and strong sin− and ts phenotypes, and found that this mutation causes a constitutively induced chromatin configuration at the CHA1 and ADH2 promoters. The alteration in chromatin structure at CHA1 depends on Cha4, leading us to hypothesize that in the uninduced hht2-AG mutant strain, the constitutively promoter-bound Cha4 protein has sufficient activity to remodel the unstable TATA-occupying nucleosome assembled with the hht2-AG mutant H3 and thus to derepress the CHA1 promoter. These findings also imply that chromatin is important for transcriptional repression downstream of activator binding, since Cha4 is constitutively bound.
Interestingly, in spite of the strongly remodeled chromatin structure observed in hht2-AG mutant yeast, only partial activation of the CHA1 promoter was observed (Fig. 2C); expression of ADH2, although its promoter chromatin structure resembles that of the active gene, was also not detectably altered in the hht2-AG mutant (data not shown). Similarly, we recently found that robust chromatin remodeling of the induced CHA1 promoter occurs in the absence of functional Mediator, but transcriptional activation and recruitment of Pol II and TATA-binding protein are greatly reduced (9). Taken together, the findings reported here provide additional evidence that chromatin remodeling and recruitment of the transcription machinery, although both dependent on transcriptional activators, occur via at least partially independent pathways (9, 32).
Acknowledgments
We thank Fred Winston and Andrea Duina for helpful discussions and materials, Corey Nislow for help and advice on the microarray experiments, and our colleagues at the Wadsworth Center for helpful discussions. We also acknowledge expert assistance from the Wadsworth Center Biochemistry and Microarray Core Facilities and particularly thank Mike Palumbo of the Computational Molecular Biology and Biostatistics Center.
This work was supported by NSF grant MCB0517825 to R.H.M.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Almer, A., H. Rudolph, A. Hinnen, and W. Horz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 52689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 3.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 111037-1043. [DOI] [PubMed] [Google Scholar]
- 5.Duina, A. A., and F. Winston. 2004. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell. Biol. 24561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 2712335-2349. [DOI] [PubMed] [Google Scholar]
- 7.Germond, J. E., B. Hirt, P. Oudet, M. Gross-Bellark, and P. Chambon. 1975. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc. Natl. Acad. Sci. USA 721843-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 9.He, Q., L. Battistella, and R. H. Morse. 2007. Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J. Biol. Chem. 2835276-5286. [DOI] [PubMed] [Google Scholar]
- 10.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 151999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 62288-2298. [DOI] [PubMed] [Google Scholar]
- 12.Holmberg, S., and P. Schjerling. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent, N. A., L. E. Bird, and J. Mellor. 1993. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 214653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruger, W., C. L. Peterson, A. Sil, C. Coburn, G. Arents, E. N. Moudrianakis, and I. Herskowitz. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 92770-2779. [DOI] [PubMed] [Google Scholar]
- 15.Kurumizaka, H., and A. P. Wolffe. 1997. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 176953-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 391235-1244. [DOI] [PubMed] [Google Scholar]
- 17.Lenfant, F., R. K. Mann, B. Thomsen, X. Ling, and M. Grunstein. 1996. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 153974-3985. [PMC free article] [PubMed] [Google Scholar]
- 18.Ling, X., T. A. Harkness, M. C. Schultz, G. Fisher-Adams, and M. Grunstein. 1996. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 10686-699. [DOI] [PubMed] [Google Scholar]
- 19.Luger, K. 2003. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 13127-135. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 21.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13136-142. [DOI] [PubMed] [Google Scholar]
- 22.Martens, J. A., P. Y. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 192695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreira, J. M., and S. Holmberg. 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 176028-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan, B. A., B. A. Mittman, and M. M. Smith. 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 114111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morse, R. H. 1999. Analysis of DNA topology in yeast chromatin. Methods Mol. Biol. 119379-393. [DOI] [PubMed] [Google Scholar]
- 26.Muthurajan, U. M., Y. Bao, L. J. Forsberg, R. S. Edayathumangalam, P. N. Dyer, C. L. White, and K. Luger. 2004. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23260-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neigeborn, L., and M. Carlson. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, J. O., M. A. Rodriguez, M. Praetorius-Ibba, T. Nilsson-Tillgren, I. L. Calderon, and S. Holmberg. 1997. Locus-specific suppression of ilv1 in Saccharomyces cerevisiae by deregulation of CHA1 transcription. Mol. Gen. Genet. 255561-569. [DOI] [PubMed] [Google Scholar]
- 29.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth, S. Y., A. Dean, and R. T. Simpson. 1990. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol. Cell. Biol. 102247-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan, M. P., R. Jones, and R. H. Morse. 1998. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 181774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan, M. P., G. A. Stafford, L. Yu, and R. H. Morse. 2000. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 205847-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 1004084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabet, N., S. Volo, C. Yu, J. P. Madigan, and R. H. Morse. 2004. Genome-wide analysis of the relationship between transcriptional regulation by Rpd3p and the histone H3 and H4 amino termini in budding yeast. Mol. Cell. Biol. 248823-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santisteban, M. S., G. Arents, E. N. Moudrianakis, and M. M. Smith. 1997. Histone octamer function in vivo: mutations in the dimer-tetramer interfaces disrupt both gene activation and repression. EMBO J. 162493-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stafford, G. A., and R. H. Morse. 1997. Chromatin remodeling by transcriptional activation domains in a yeast episome. J. Biol. Chem. 27211526-11534. [DOI] [PubMed] [Google Scholar]
- 38.Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 981-4. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan, S., D. W. Sink, K. L. Trout, I. Makalowska, P. M. Taylor, A. D. Baxevanis, and D. Landsman. 2002. The histone database. Nucleic Acids Res. 30341-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdone, L., G. Camilloni, E. Di Mauro, and M. Caserta. 1996. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol. 161978-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechser, M. A., M. P. Kladde, J. A. Alfieri, and C. L. Peterson. 1997. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 162086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 205207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, L., and F. Winston. 1997. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 254230-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, C., M. J. Palumbo, C. E. Lawrence, and R. H. Morse. 2006. Contribution of the histone H3 and H4 amino termini to Gcn4p- and Gcn5p-mediated transcription in yeast. J. Biol. Chem. 2819755-9764. [DOI] [PubMed] [Google Scholar]