Abstract
A lack of oxygen can force many organisms to enter into recoverable hypometabolic states. To better understand how organisms cope with oxygen deprivation, our laboratory previously had shown that when challenged with anoxia, both the nematode Caenorhabditis elegans and embryos of the zebrafish Danio rerio enter into suspended animation, in which all life processes that can be observed by light microscopy reversibly halt pending the restoration of oxygen (P. A. Padilla and M. B. Roth, Proc. Natl. Acad. Sci. USA 98:7331-7335, 2001, and P. A. Padilla, T. G. Nystul, R. A. Zager, A. C. Johnson, and M. B. Roth, Mol. Biol. Cell 13:1473-1483, 2002). Here, we show that both sporulating and vegetative cells of the budding yeast Saccharomyces cerevisiae also enter into a similar state of suspended animation when made anoxic on a nonfermentable carbon source. Transcriptional profiling using cDNA microarrays and follow-on quantitative real-time PCR analysis revealed a relative derepression of aerobic metabolism genes in carbon monoxide (CO)-induced anoxia when compared to nitrogen (N2) gas-induced anoxia, which is consistent with the known oxygen-mimetic effects of CO. We also found that mutants deleted for components of the mitochondrial retrograde signaling pathway can tolerate prolonged exposure to CO but not to N2. We conclude that the cellular response to anoxia is dependent on whether the anoxic gas is an oxygen mimetic and that the mitochondrial retrograde signaling pathway is functionally important for mediating this response.
Molecular oxygen (O2) can be utilized by all organisms, except obligate anaerobes, in metabolic pathways that enable the extraction of chemical energy stored in nutrient compounds. When cells reliant on aerobic respiration suffer from poor oxygen availability, the cells respond by increasing anaerobic energy production, upregulating stress genes, and in the case of mammals, stimulating angiogenesis to increase oxygen delivery (60). In many species, it is thought that the delivery of oxygen to tissues is homeostatically adjusted in order to provide adequate oxygenation for energy generation (52), consistently with the idea that organisms must compensate for decreases in environmental oxygen levels in order to thrive or to merely survive. In addition to being a topic of interest in its own right, understanding how various cell types cope with a lack of oxygen can have important implications for human health, as the progression of several pathological conditions, including heart attack, stroke, and cancer, is associated with poor oxygenation to the affected tissues (35).
Many approaches in different systems have been undertaken to better understand the mechanisms that enable cells to survive a lack of oxygen. Among eukaryotic model organisms, the growth of yeasts under depressed oxygen levels has been of great interest historically, in large part due to the role of yeasts in the baking and brewing industries (6). Although budding yeast is a facultative anaerobe, continuous culturing under anaerobic conditions requires the addition of sterols (1) and unsaturated fatty acids (2) in the medium (since molecular oxygen is required to synthesize these compounds), as well as the activation of biochemical pathways to bypass those that require molecular oxygen (54). This highlights the importance of oxygen even for organisms classified as facultatively anaerobic.
The response of many metazoan species to decreased oxygen also has been extensively studied. These include many popular model organisms, such as nematodes (53), fruit flies (20), zebrafish (26), and mice (51). In addition, much work has been done on less well-studied systems, including brine shrimp (21), turtles (55), carp (15), sharks (37), and dogs (47). These organisms all appear to manifest physiological and behavioral changes that are consistent with a decrease in metabolism when exposed to lower-than-normal oxygen concentrations. From this veritable menagerie, it is clear that many species have evolved mechanisms to cope with a lack of oxygen at various levels of severity.
Our laboratory has been interested in the response of model systems to very severe oxygen deprivation and has demonstrated that two well-studied model organisms, the nematode Caenorhabditis elegans (40) and embryos of the zebrafish Danio rerio (39), enter into a reversible state of suspended animation when exposed to anoxia (operationally defined as an atmosphere containing less than 10 Pa of O2). Similarly to results reported for Drosophila embryos (16), all life processes observable by light microscopy are halted pending the restoration of oxygen. Moreover, it was found that the san-1 gene, which encodes a component of the mitotic spindle checkpoint, is required for anoxia-induced suspended animation in C. elegans embryos, as the depletion of the SAN-1 gene product by RNA-mediated interference resulted in chromosome missegregation and death (38).
To further elucidate the molecular mechanisms that underpin the process of anoxia-induced suspended animation, we turned to the budding yeast Saccharomyces cerevisiae, a model system that we show also enters into reversible suspended animation when exposed to anoxia on a nonfermentable carbon source. We carried out transcript microarray analysis on cells that were made anoxic on a nonfermentable substrate in order to identify pathways that may be important for survival under such conditions. We used two different anoxic gases, carbon monoxide (CO) and nitrogen (N2). As CO can mimic the presence of O2 by displacing the latter in the binding sites of many heme-containing proteins (reviewed in reference 41) while N2 cannot, we hypothesized that there would be marked differences in gene expression between the transcriptomes of cells exposed to each of the two anoxic gases. Consistently with the known oxygen-mimetic properties of CO, we found that exposure to this gas caused a coordinated derepression of aerobic metabolism genes when compared to a similar exposure to N2. Moreover, we found that mutants deleted for components of the mitochondrial retrograde signaling pathway recovered normally from prolonged exposure to CO but recovered poorly after similar exposure to N2. Our findings lead us to conclude that the response of yeast to anoxia is dependent on whether the applied anoxic gas is an oxygen mimetic and that the mitochondrial retrograde signaling pathway is functionally important for mediating the proper response.
MATERIALS AND METHODS
Anoxia-induced suspension of sporulation.
Diploid SK1 cells (MATa/MATα ho:LYS2/ho:LYS2 ura3/ura3 lys2/lys2 leu2:hisG/leu2:hisG arg4-nsp/arg4-bgl his4X:LEU2-URA3;his4B:LEU2) were grown for 24 h in 1 ml YPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C. This culture then was diluted 100-fold into YPA (1% yeast extract, 2% peptone, 3% potassium acetate) presporulation medium and grown for 48 h in a 1-liter baffled Erlenmeyer flask with shaking. Cells then were collected by centrifugation, washed once in sterile water, and resuspended in 100 ml sporulation medium (3% potassium acetate with appropriate supplements) with 0.2% antifoam to prevent excessive bubble formation. The culture then was split into two equal 50-ml portions, each of which was put into a sterile modified Scienceware gas wash bottle (Fisher Scientific, Pittsburgh, PA). These bottles were equipped with both fittings to enable the bubbling of gas directly into each culture and fine polyethylene (PE-10) tubing (Braintree Scientific, Braintree, MA) threaded through the stoppers that enabled sampling from each culture without disrupting the atmospheres within each bottle. The bottles were kept at 30°C throughout the experiment. The control culture was continuously bubbled with O2 (all gases used were from Airgas Nor Pac, Seattle, WA) at 100 cm3 per min. The test culture was bubbled with O2 up to the beginning of hour 6, with N2 (N2 was scrubbed with an Aeronex CE500KF14R inline inert gas purifier to remove trace O2 contamination) from hour 6 to the beginning of hour 18, and with O2 from hour 18 onward. Samples were collected every 3 h. Cells were collected by gentle centrifugation and fixed using 70% ethanol at −20°C. Cells then were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize chromatin, and the percent asci formation as well as the completion of meiosis I and II were quantified.
RNA extraction.
To obtain cells for RNA extraction, BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or rtg deletion cells (constructed de novo in the BY4741 background) were grown for 24 h at 30°C in 5 ml YPD with continuous tumbling on a rotator drum. Cells were collected by light centrifugation and washed once with sterile phosphate-buffered saline (PBS). Cells then were resuspended in ∼6 ml PBS. A volume of 0.5 ml of this suspension was pipetted onto each of 12 autoclaved nylon membranes (GE Water & Process Technologies, Trevose, PA) resting on the surface of YPA solid medium in 9.5-cm round petri dishes. Six of these plates then were sealed inside modified Pyrex crystallization dishes (Corning Inc., Lowell, MA) and flushed with hydrated N2 at 100 cm3/min. The other six plates were left in room air. All plates were incubated at 30°C. An analogous procedure was used to obtain CO-treated samples.
Cells were collected for RNA extraction at 15, 30, 45, 60, and 120 min and 24 h after the initiation of anoxic gas exposure. For each collection, membranes were removed from the anoxic plate and a room air control plate. Thus, the anoxic and normoxic cell samples collected at each time point were treated identically, except for the gaseous environment each sample was exposed to. Each membrane then was rolled into a tube shape and inserted into a Falcon 14-ml round-bottom tube that was preloaded with 10 ml ice-cold water. The tubes then were centrifuged at ∼0°C for 5 min to collect the cells. The water and membranes were discarded. Pellets were flash frozen in liquid nitrogen and stored at −80°C.
RNA extractions were initiated by resuspending each pellet in 400 μl TES (10 mM Tris-HCl, pH 7.5, 10 mM EDTA, 0.5% sodium dodecyl sulfate) and 400 μl acid phenol (prewarmed to 65°C) with vigorous vortexing. Cells were incubated at 65°C for 1 h, with vortexing every 10 min. Suspensions were transferred to microcentrifuge tubes and put on ice for 5 min and then centrifuged at top speed for 10 min at 4°C on an Eppendorf Minispin centrifuge. Aqueous phases were transferred to fresh tubes and extracted with 400 μl room temperature acid phenol twice more, followed by a single extraction with 400 μl chloroform. RNA then was precipitated from the aqueous phase using 40 μl 3 M sodium acetate, pH 5.3, and 1 ml ice-cold ethanol and centrifuged for 20 min. Pellets were washed with 0.5 ml ice-cold 80% ethanol and centrifuged for 5 min. RNA was redissolved in 20 μl water.
Quantification of percentage of budded cells.
BY4741 cells were plated onto nylon membranes on solid YPA similarly to the procedure for RNA extraction. Cells were incubated at 30°C under continuous perfusion with either CO or N2 for 2 days. Cells were washed off the membranes as described for RNA extraction and then were fixed in 4% formaldehyde in 0.1 M potassium phosphate, pH 7.5, for 15 min at room temperature with continuous tumbling. Cells were washed twice in 0.1 M potassium phosphate, pH 7.5, supplemented with 1.2 M sorbitol and resuspended in the same buffer. More than 600 cells were counted from each sample for each of three biological replicates.
Microarray processing and analysis.
Microarray processing steps described here, up to and including the scanning of hybridized slides, were carried out by the DNA Array laboratory at the Fred Hutchinson Center. Four micrograms of total RNA from each sample was used as the substrate for the Ambion Amino Allyl MessageAmp protocol (Ambion Inc., Austin, TX). Dye-coupled products from the in vitro transcription step were hybridized to yeast open reading frame (ORF) microarray slides (bearing 6,229 yeast ORFs) that were printed in house. After incubation and washes, slides were scanned on a GenePix 4000B scanner (MDS Analytical Technologies, Toronto, Canada), and images were returned to the authors for analysis.
Array images were processed using Genepix Pro 6 (MDS Analytical Technologies). A Lowess normalization procedure was applied using GeneTraffic (Iobion Informatics, La Jolla, CA). T-profiler (10) was used to identify upstream consensus motif and gene ontology (GO) enrichment patterns within the array data. This online tool (http://www.t-profiler.org) utilizes the Student t test to derive an E value that reflects the degree of difference in the mean log2-transformed expression ratio of a predefined group of genes and the mean for the rest of the genome. Student t tests are calculated for each gene group, in each data set, and at each time point in a time course. An E value of <0.05 is considered indicative of a statistically significant difference in gene expression. As this approach compares the mean expression ratios of groups of genes, all genes within each group contribute to the evaluation of statistical significance, not just those genes that are judged to be differentially expressed on an individual basis. MEME (http://meme.sdsc.edu/meme/meme.html) (3, 5) and MAST (http://meme.sdsc.edu/meme/mast.html) (4) were used for de novo consensus motif identification and genomewide upstream sequence enrichment searches, respectively.
Quantitative real-time PCR (qRT-PCR).
To initiate cDNA synthesis for each sample, 5 μg total RNA was combined with 225 pmol random primers in water to a total volume of 18.5 μl and incubated at 70°C for 10 min. Samples then were immediately chilled on ice for 10 min. A cocktail of the following reagents in the appropriate multiple of these proportions was prepared: 6 μl 5× first-strand buffer, 3 μl 0.1 M dithiothreitol, 0.6 μl 25 mM each deoxynucleoside triphosphate, and 1.9 μl SuperScript II. A volume of 11.5 μl of this cocktail then was added to each RNA-primer mix and incubated for 2 h at 42°C. Reaction mixtures then were incubated at 95°C for 5 min to inactivate the reverse transcriptase. Two units of RNase H was added to each reaction, which then were incubated at 37°C for 20 min to degrade the template RNA. Finally, samples were incubated at 95°C for 5 min to inactivate the RNase H.
For qRT-PCR, each reaction mixture consisted of the following: 19.92 μl water, 3.0 μl 10× PCR buffer, 0.9 μl 50 mM MgCl2, 1.5 μl 2.5 mM each deoxynucleoside triphosphate, 0.03 μl Sybr green, 1.5 μl cDNA reaction mix, 0.15 μl Taq, and 3 μl 30 μM each gene-specific primer. A reaction cocktail consisting of the common components sufficient for the required number of reactions was set up and then dispensed into each well of a 96-well PCR dish. All reagents for this procedure were from Invitrogen (Carlsbad, CA). qRT-PCRs were carried out on a Bio-Rad iQ5 thermocycler, with a 5-min step at 94°C followed by 40 repeats of the following steps: 94°C for 30 s, 55°C for 30 s, 72°C for 60 s, 78°C for 10 s, and plate read. PCR products were analyzed on a 3% agarose gel to verify the size of each product and the absence of side products. The automated detection of the qRT-PCR threshold cycle by iQ5 software was applied with reactions utilizing the same primer pairs grouped together for each of the 16 primer pairs. The manual adjustment of threshold cycle detection was necessary in a few cases in which the software failed to correctly distinguish signal from background.
Identification of mutants sensitive to prolonged anoxia.
We searched the Saccharomyces Genome Database (SGD) for all genes that are annotated under the GO term signal transduction, as well as all genes annotated under subordinate terms. In total, we found 174 nonessential genes with corresponding deletion strains in the MATa deletion set. Each of these strains was inoculated into 200 μl liquid YPD and grown for 2 days at 30°C. Strains then were spotted at 1,000-fold dilution in PBS onto solid YPA medium and incubated for 4 days under continuous perfusion with hydrated CO or N2 at 100 cm3/min in modified crystallization dishes. Control plates were maintained in room air. Candidate strains for retesting were identified by comparing plates after the formerly anoxic cells were allowed to recover in air.
Candidate strains from the initial phenotypic test were pregrown in the same manner. Tenfold serial dilutions were spotted onto solid YPA medium and subjected to the same phenotypic testing procedure as described above using three biological replicates. rtg1, rtg2, and rtg3 deletion strains were constructed de novo and verified by PCR using standard methods (http://www.fhcrc.org/science/labs/gottschling/yeast/). rtg1Δ and rtg3Δ also were verified by testing for previously described glutamate and aspartate auxotrophies (24). The anoxia phenotypes of each of the rtg deletion strains then were confirmed by serial dilution spot tests using four biological replicates. Pregrowth by overnight culture in 5 ml YPD on a rotator drum can be substituted for 2-day growth in the 96-well dish format with similar phenotypic results.
Microarray accession number.
Microarray data were deposited at the NCBI GEO, under data set accession number GSE12004.
RESULTS
Yeast cells enter suspended animation when made anoxic on a nonfermentable carbon source.
In order to utilize budding yeast as a model for studying anoxia-induced suspended animation, we first had to demonstrate that it is possible to reversibly halt life processes observable via light microscopy by withdrawing oxygen. Since budding yeast can grow anaerobically on a fermentative carbon source, we used a nonfermentable carbon source in order to test whether anoxia-induced suspended animation is a conserved response to the lack of oxygen. Meiosis and sporulation comprise a well-studied developmental process in yeast that can be induced experimentally by transferring diploid cells to a defined medium that lacks nitrogen sources and contains only a nonfermentable carbon source (reviewed in reference 36). Meiosis and sporulation result in the formation of four haploid gametes (i.e., spores) enclosed in the remnant of the diploid cell, with this entire assembly being referred to as an ascus. Since meiosis and sporulation comprise a dynamic process whose progression can be easily monitored cytologically, we determined whether it was possible to reversibly halt this process by imposing anoxia.
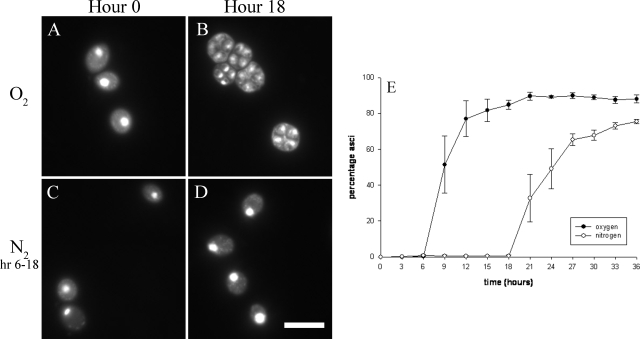
We utilized the efficiently sporulating SK1 strain for this series of experiments. Based on previously published results (44) and our own observations, SK1 does not form asci until more than 6 h after transfer to sporulation medium. We therefore attempted to stop sporulation by applying anoxia at the beginning of hour 6 after the transfer to sporulation medium, as the sporulation process should be well under way by that point, but hour 6 is still early enough that asci are not yet formed. We found that it is indeed possible to reversibly halt sporulation by perfusing the culture with N2. When cells were made anoxic at the beginning of hour 6 (after being allowed to initiate sporulation in the presence of oxygen up to that time) and maintained in anoxia up to hour 18, the majority of anoxic cells remained mononucleate and were unable to complete the sporulation process without O2 (Fig. 1C and D). Only upon the restoration of oxygen (at hour 18 of the experiment) do the cells continue with the sporulation process, which reaches a maximum of 75.5% asci formation by the end of the experiment (Fig. 1E). This value is 85.7% of the value observed in control cultures, which reach a maximum of 88.1% asci formation. In contrast to the cultures that were reversibly suspended by N2, control cultures that are allowed to sporulate normally had essentially completed sporulation by hour 18 (Fig. 1A, B, and E).
FIG. 1.
Anoxia-induced suspension of meiosis and sporulation in SK1 cells. SK1 cells allowed to initiate sporulation (A) in the continuous presence of oxygen have completed sporulation by hour 18, forming over 80% asci (B). In contrast, cells allowed to initiate sporulation in the presence of oxygen (C) but made anoxic from hours 6 to 18 did not complete the sporulation process (D). The scale bar represents 10 μm. (E) A plot of the percent asci as a function of time shows that cells made anoxic from hour 6 to hour 18 rapidly resumed sporulation when oxygen is restored at hour 18. Error bars represent standard errors of the means for four independent trials. Three hundred cells from each culture were counted at each time point.
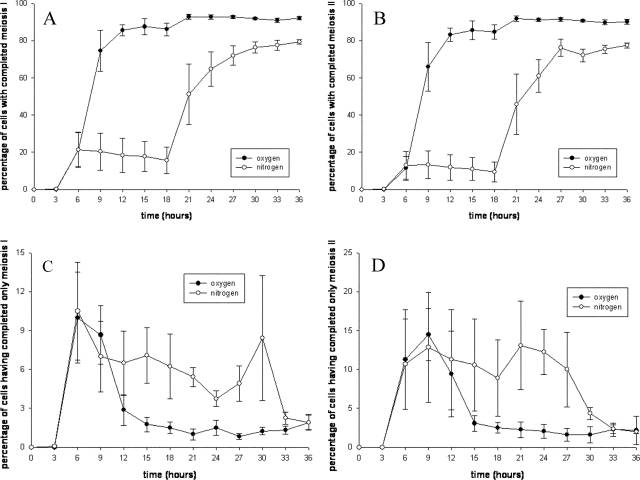
We also found that cells can be reversibly halted after having completed either of the two meiotic divisions. Figure 2A and B show that the cells are not accumulating at one particular stage when experiencing anoxia and suggest that stoppage at multiple stages within meiosis and sporulation is recoverable. Indeed, we found that the percentage of cells that had completed meiosis I but had not yet proceeded further when the sample was taken decreases over time, such that by the end of the experiment the percentage of such cells is the same for both cultures (Fig. 2C). The same is true of cells that had completed meiosis II but had not yet formed asci (Fig. 2D). If the cells that had been arrested immediately after meiosis I or II had not resumed sporulation, then the relative percentages of such cells would be expected to increase over the course of the experiment or at least stay the same. Thus, the observed decrease (over the time course) in the percentage of cells having just completed meiosis I or II suggests that many of the cells that had been arrested immediately after meiosis I or II in anoxia resumed sporulation upon reoxygenation, albeit with less synchronicity than that of the controls. In addition, spore dissections showed that spore viability was high: 82% for spores derived from cells that were made anoxic and 93% for control spores (data not shown). Taken together, these results demonstrate that anoxia-induced suspended animation is a conserved response to severe oxygen deprivation in sporulating yeast.
FIG. 2.
(A) Total percentage of cells that have completed meiosis I does not increase in anoxia (i.e., from hour 6 to hour 18 in the nitrogen samples). Note that the percentages plotted in this panel include cells that have completed at least meiosis I; hence, cells that have completed meiosis II or formed asci are included as well. (B) Total percentage of cells that have completed meiosis II also does not increase in anoxia. The percentages plotted in this panel include cells that have completed at least meiosis II; cells that have formed asci therefore are included. (C) In the nitrogen samples, the percentage of cells that have only completed meiosis I but have not yet completed meiosis II decreases over the course of the experiment, suggesting that cells arrested in anoxia after having completed only meiosis I can resume sporulation upon reoxygenation. (D) A similar decrease in the percentage of cells that have only completed meiosis II over the course of the experiment suggests that cells arrested in anoxia after having completed only meiosis II can resume sporulation upon reoxygenation. Data are from the four trials shown in Fig. 1E; error bars represent standard errors of the means.
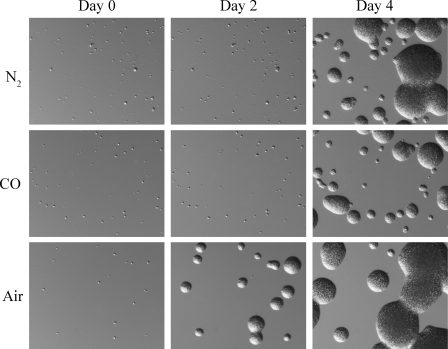
While it is clear from cytological evidence that sporulating yeast can undergo anoxia-induced suspended animation, we also wished to determine if the same were true of vegetative cells. If so, then any potential screening to identify genes that are required for suspended animation would be much easier to carry out using vegetative cells of any of the extant deletion sets (18), which were constructed in an S288c genetic background. Accordingly, we tested whether the haploid BY4741 strain (the S288c derivative parental strain of the MATa deletion set) also can undergo anoxia-induced suspended animation. In order to observe the growth of colonies originating from single cells, we spotted BY4741 cells at low density onto solid medium containing only a nonfermentable carbon source, acetate. When cells were deposited onto acetate medium and made anoxic for 2 days, colonies did not form as long as the cells were kept anoxic (Fig. 3). In fact, most cells did not divide over the 2 days of anoxia, and the few cells that divided did so only once (compare N2 and CO days 0 and 2). In contrast, cells left in room air clearly had undergone multiple rounds of cell division after 2 days (compare air day 0 to day 2). When the anoxic cells were restored to room air, they resumed cell divisions, forming colonies similar in size and appearance to those of the control cells after 2 days in air (compare N2 and CO day 4 to air day 2). These results show that it is possible to reversibly stop the growth of budding yeast by exposure to anoxia on a nonfermentable carbon source. Thus, our laboratory has shown that anoxia-induced suspended animation is a response to the extreme lack of oxygen that is conserved across three well-studied model species, budding yeast (both sporulating and vegetative cells), nematodes, and zebrafish.
FIG. 3.
BY4741 cells were spotted at low density onto solid YPA medium. One group of cells was continuously perfused with an atmosphere of pure N2 for 2 days and then was returned to room air to restart growth (top row). A second group of cells was similarly perfused with CO for 2 days and then returned to room air (middle row). A control group of cells was kept in room air for 4 days (bottom row). Cells that were made anoxic for 2 days halt their cell divisions but readily restart growth after return to room air.
Vegetative cells in suspended animation can reversibly arrest while in a budded state.
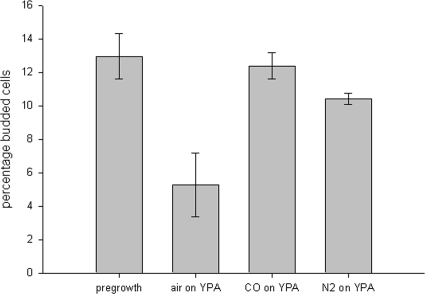
Yeast cells, when exposed to various conditions not conducive to continued growth, transiently arrest the cell cycle in G1. These conditions include starvation for carbon, nitrogen, sulfur, phosphorus, and potassium (22); exposure to mating pheromone (11); elevated temperature (25); oxidative stress (59); and osmotic stress (7). To determine whether cells in anoxia-induced suspended animation exhibit a similar arrest in G1, we quantified the percentage of budded cells from populations made anoxic with CO or N2 for 2 days on acetate medium (Fig. 4). We found that 12.4% of cells treated with CO were in a budded state, while 10.4% of cells treated with N2 were budded. These percentages are similar to the 13.0% budded cells observed in the overnight pregrowth cultures, with P = 0.728 compared to results for CO treatment and P = 0.138 compared to results for N2 treatment (by Student t test).
FIG. 4.
Cells in suspended animation can reversibly arrest while in a budded state. A total of 13.0% of BY4741 cells from overnight pregrowth cultures were budded. Similarly, among cells reversibly arrested in CO or nitrogen on acetate medium for 2 days, 12.4 and 10.4%, respectively, were arrested in a budded state. Note that only 5.3% of cells growing in the presence of air on acetate were budded, reflecting the slower cell cycle on nonfermentable medium. Data are from three biological replicates; error bars represent standard errors of the means.
Note that cells growing on acetate in the presence of room air exhibited a lower percentage of budded cells (5.3%). Compared to the pregrowth, CO-, and N2-treated samples, the respective P values are 0.030, 0.026, and 0.056. Cells growing on nonfermentable medium require almost fourfold more time to complete a cell cycle than cells growing on fermentable medium (63). The lower percentage of budded cells likely is due to the increased time required for cells in nonfermentable medium to grow to a size that is sufficient to pass START, resulting in more cells being in an unbudded state at any particular time. The similarity in percent budded cells between pregrowth and anoxic samples, combined with the dissimilarity between aerobic and anoxic samples on acetate, suggest that budded cells made anoxic on acetate reversibly arrest as such. In contrast to cells exposed to other stresses, these budded cells are unable to complete the cell cycle in progress and, thus, do not arrest in G1.
Vegetative cells retain high viability after continuous, prolonged anoxia.
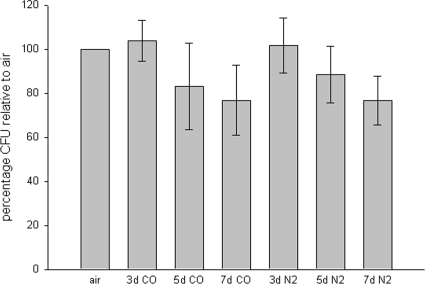
To assess the viability of cells in prolonged anoxia-induced suspended animation, we plated BY4741 cells at low density onto solid YPA medium and kept cells in continuous anoxia for up to 7 days (Fig. 5). Cells retained high viability (76.8% relative to room air controls) after even 1 week of continuous exposure to either CO or N2, as judged by the ability to form colonies after anoxia. Thus, vegetative yeast have a robust ability to withstand prolonged arrest in a nonproliferating state while anoxic on nonfermentable medium.
FIG. 5.
Cells retain high viability after up to 1 week of continuous anoxia. BY4741 cells were plated at low density onto solid YPA medium and continuously perfused with either CO or nitrogen for the indicated number of days and then allowed to recover in room air before colonies were counted. Data are from three biological replicates; error bars represent standard errors of the means.
Broad similarity in the transcriptional responses to both anoxic gases.
To better understand the molecular underpinnings enabling anoxia-induced suspended animation, we wished to identify sets of genes that are differentially expressed as cells undergo reversible deanimation when made anoxic on a nonfermentable carbon source. These data should provide insight into patterns of gene expression that define the transcriptional response of cells undergoing suspended animation, allowing us to draw comparisons to previously published microarray studies of yeast undergoing anaerobiosis (29, 30, 31, 57) and other stress conditions (12, 17). Accordingly, we carried out microarray analysis on cells collected over six time points (15, 30, 45, 60, and 120 min and 24 h) during anoxia using each of the two anoxic gases.
We elected to use T-profiler (10) to analyze the microarray data. This online tool readily identifies groups of genes, with related GO annotations, that are differentially expressed when comparing two conditions without the need to apply cutoffs that exclude a large proportion of the expression data from further consideration (see Materials and Methods). We found broad similarity in the transcriptional responses to anoxia caused by each of the two gases. Specifically, two groups of genes were upregulated at most time points in both anoxic gases: genes encoding cell wall components and genes grouped under the heading cellular component unknown (Table 1). It has been shown that yeasts upregulate many cell wall genes when undergoing anaerobiosis (29, 30, 31, 57), apparently to remodel the cell wall's composition. Our results are consistent with these previous findings.
TABLE 1.
Gene groups that were identified by T-profiler as being significantly upregulated during the CO and N2 time coursesa
| Time point | CO
|
N2
|
||||
|---|---|---|---|---|---|---|
| E < 10−6 | 10−6 ≤ E < 10−4 | 10−4 ≤ E < 0.05 | E < 10−6 | 10−6 ≤ E < 10−4 | 10−4 ≤ E < 0.05 | |
| 15 min | Ribosome component, protein synthesis, biosynthesis, mitochondrion component | Macromolecule synthesis, transporter activity, cell wall component, oxidoreductase activity, oxidative phosphorylation | Ribonucleoprotein complex, ion transport, translational elongation, proton-transporting ATP synthase, purine nucleoside triphosphate metabolism | Cellular component unknown | Cell wall component, ribosome component | Transporter activity, mitochondrial electron transport chain, structural molecule activity, carrier activity, multichaperone complex component |
| 30 min | Oxidative phosphorylation, energy pathways, mitochondrion component, mitochondrial electron transport chain | Oxidoreductase activity, electron transport | Proton-transporting ATP synthase, purine nucleoside triphosphate metabolism, aerobic respiration, cell wall component, coenzyme metabolism, carbohydrate metabolism, ribonucleotide biosynthesis | Cellular component unknown, cell wall component, plasma membrane component, transporter activity | Membrane component | Extracellular protein, siderochrome transport |
| 45 min | Cellular component unknown | Cellular component unknown | Cell wall component | |||
| 60 min | Cellular component unknown | Cell wall component | Oxidoreductase activity, energy pathways, carbohydrate metabolism, membrane component, proton transporter activity | Cellular component unknown | Peroxide oxidoreductase activity | |
| 120 min | Cellular component unknown | Cell wall component | Response to external stimulus | Cellular component unknown | Response to external stimulus, response to abiotic stimulus | |
| 24 h | Transporter activitiy, mitochondrion component | Membrane component, cellular component unknown | Cell wall component, aerobic respiration, mitochondrial electron transport chain, glycosyl hydrolase activity, energy pathways, oxidative phosphorylation, glucosidase activity, aspartate amino acid catabolism | Cellular component unknown, cell wall component, transporter activity, mitochondrion component | Glycosyl hydrolase activity, glucosidase activity, membrane component | Carbohydrate transport, plasma membrane component, amine transport |
Gene groups are binned by E values, with groups having lower E values listed first within each bin at each time point.
In the hopes of shedding light on the role(s) of the group of uncharacterized ORFs that is upregulated under both anoxic gases, consensus motif identification searches were carried out on this group of genes. Using MEME (3, 5), an online consensus motif discovery tool, we identified three consensus motifs that are enriched in the upstream regions of 55 of the most highly upregulated genes in this group, genes with a mean of at least twofold induction across the six time points. We then used MAST (4), a related online tool, to search for the enrichment of these three consensus motifs in the upstream regions of all yeast genes. This search found 122 genes with E < 10−6. Using GO tools at the SGD, we found that 63 of these 122 genes coded for products with roles in nucleobase, nucleoside, nucleotide, and nucleic acid metabolic processes. Forty-eight of these 63 genes are found in Ty-transposable elements. Due to the high degree of sequence similarity among transposable element genes (32), the possibility of significant cross-hybridization in microarray studies makes it difficult to derive an accurate assessment of overrepresentation relative to the genome. This difficulty notwithstanding, it is nonetheless possible that the activation of retrotransposition is a part of the transcriptional response to oxygen deprivation, as transposable elements are mobilized when cells are exposed to various stresses (32). We also note that a similar oxygen limitation-dependent upregulation of transposable elements has been reported in fission yeast (50).
The responses to both anoxic gases also were very similar among the groups of genes that become relatively less abundant in anoxia. The large majority of these genes are involved in transcription or translation, along with related metabolic processes. These groups include ribosome and nucleolar components as well as ribosome biogenesis, RNA metabolism, protein metabolism, amino acid metabolism, and RNA ligase activities (Table 2). As the cells are unable to grow when made anoxic, it is not surprising to find a profound and prolonged decrease in the abundance of such transcripts. Based solely on our own data, it is unclear to what degree these genes are relatively induced in the reference (air) cells compared to being repressed in the anoxic cells. However, based on previously published observations (46), it is known that for cells transferred from glucose to a nonfermentable carbon source (glycerol), there is apparently little relative change in the transcript abundance of ribosome biogenesis genes for at least 60 min after the transfer. Additionally, as shown in reference 46, such transcripts actually become less abundant after prolonged growth in the nonfermentable medium. Given the findings in reference 46 and the fact that the downregulated gene groups all are of interrelated function, it is probable that the relative change in gene expression seen across these gene groups is due more to coordinate repression in anoxia than to coordinate induction in air.
TABLE 2.
Gene groups that were identified by T-profiler as being significantly downregulated during the CO and N2 time coursesa
| Time point | CO
|
N2
|
||||
|---|---|---|---|---|---|---|
| E < 10−6 | 10−6 ≤ E < 10−4 | 10−4 ≤ E < 0.05 | E < 10−6 | 10−6 ≤ E < 10−4 | 10−4 ≤ E < 0.05 | |
| 15 min | Nucleolus, nucleus, ribosome biogenesis, rRNA metabolism, RNA metabolism, cytoplasm biogenesis | Nucleic acid binding, cell growth and maintenance, cell proliferation | Nucleolus, ribosome biogenesis, rRNA metabolism, nucleus, RNA metabolism, nucleic acid metabolism, cytoplasm biogenesis | Cell growth and maintenance, nucleic acid binding | RNA modification, sulfur amino acid metabolism, ATP-dependent helicase activity, protein folding | |
| 30 min | Ribosome biogenesis, nucleolus, rRNA metabolism, RNA metabolism, nucleus | Nucleic acid binding, RNA helicase activity, cell growth and maintenance | Ribosome biogenesis and component, nucleolus, rRNA metabolism, RNA metabolism, protein metabolism, nucleic acid metabolism, cytoplasm biogenesis, nucleus, macromolecule metabolism, structural molecule, nucleic acid binding | RNA modification, translation, tRNA ligase activity | RNA ligase activity, RNA helicase activity, amino acid metabolism, translation regulator activity, protein folding, tRNA modification, amine metabolism, translation initiation factor activity | |
| 45 min | Ribosome biogenesis, nucleolus, ribosome component, rRNA metabolism, RNA metabolism, biosynthesis | Cytoplasm biogenesis, amino acid metabolism, amine metabolism | RNA helicase activity, macromolecule metabolism, nucleic acid binding, RNA ligase activity, tRNA ligase activity, organic acid metabolism, translation, catalytic activity | Ribosome component and biogenesis, nucleolus, rRNA metabolism, RNA metabolism, structural molecule activity, protein metabolism, macromolecule metabolism, nucleic acid binding, translation | tRNA ligase activity, RNA ligase activity, RNA helicase activity, nucleus | Amino acid metabolism, rRNA modification, tRNA modification, amine metabolism, cell growth and maintenance, nucleocytoplasmic transport, organic acid metabolism, regulation of translational fidelity |
| 60 min | Ribosome component, ribosome biogenesis, nucleolus, rRNA metabolism, RNA metabolism, biosynthesis, structural molecule, nucleic acid metabolism, protein metabolism, nucleus, nucleic acid binding | RNA polymerase activity, RNA helicase activity, amino acid metabolism, macromolecule metabolism, amine metabolism | Amino acid metabolism, tRNA modification, RNA ligase activity, translation, tRNA ligase activity, aromatic compound metabolism, cell growth and maintenance, organic acid metabolism | Ribosome component and biogenesis, nucleolus, RNA metabolism, rRNA metabolism, cytoplasm biogenesis, protein metabolism, nucleic acid metabolism, macromolecule metabolism | Cell growth and maintenance, tRNA ligase activity, RNA ligase activity, RNA modification, nucleic acid binding, amino acid metabolism, helicase activity, translation | Amine biosynthesis, ATPase activity, tRNA modification, organic acid metabolism |
| 120 min | Ribosome component, protein biosynthesis, biosynthesis, structural molecule activity, macromolecule biosynthesis, nucleolus, rRNA metabolism, translation, cytoplasm biogenesis, RNA metabolism | Nucleic acid binding, tRNA ligase activity, RNA ligase activity, RNA modification, regulation of translational fidelity, mitochondrion component | Nucleic acid metabolism, amino acid metabolism, translation factor activity, RNA helicase activity, amine metabolism, tRNA modification, organic acid metabolism, translational initiation | Ribosome component, structural molecule activity, protein metabolism, ribosome biogenesis, macromolecule metabolism, rRNA metabolism, translation, nucleolus, cytoplasm biogenesis, RNA metabolism | tRNA ligase activity, RNA ligase activity, RNA modification | Nucleic acid binding, regulation of translational fidelity, amino acid metabolism, ergosterol biosynthesis, tRNA modification, organic acid metabolism, nucleic acid metabolism, amino acid metabolism, helicase activity, translation regulator activity, amine metabolism, sterol biosynthesis |
| 24 h | Ribosome biogenesis, nucleolus, rRNA metabolism, ribosome component, cytoplasm biogenesis, nucleus, nucleic acid metabolism, protein biosynthesis | Nucleic acid binding, macromolecule biosynthesis, RNA helicase activity | ATPase activity, protein metabolism, amino acid metabolism, acid anhydride hydrolase activity, amine metabolism | Ribosome biogenesis, nucleolus, rRNA metabolism, RNA metabolism, nucleus, nucleic acid biosynthesis, amino acid metabolism, ATPase activity, amine metabolism, translation | Acid anhydride hydrolase activity, nucleic acid binding | RNA helicase activity, protein metabolism, organic acid metabolism, branched chain amino acid metabolism |
Gene groups are binned by E values, with groups having lower E values listed first within each bin at each time point.
Patterns of upstream consensus motif enrichment suggest similarity to other stress-induced transcriptional responses.
In addition to identifying sets of genes that are differentially expressed in anoxia, we also used T-profiler to identify consensus motifs that are associated with genes that show differential expression. Similarly to the procedure for identifying gene group enrichment, genes are assigned to groups based upon common consensus motifs, and the t statistic and associated E value are calculated. The results from these searches are summarized in Table 3. Among genes that are upregulated in anoxia, there is a consistent enrichment for those with MSN2/MSN4 consensus motifs (AGGGG, CCCCT, or HRCCCYTWDT) (27) throughout all time points. This is to be expected, as Msn2 and Msn4 are transcription factors that regulate the expression of many genes that are upregulated in response to myriad stresses (34, 49). Conversely, among genes that are downregulated, there is consistent enrichment for the PAC (CGATGAG) and rRPE motifs (AAAATTT). The PAC motif (motif M3b in reference 56) is associated with RNA binding and processing genes, while the rRPE motif (motif mRRPE in reference 42) is found upstream of genes with roles in ribosomal biogenesis. In addition, genes with the RAP1 motif (CCRTACA), which have roles in ribosome biogenesis (9), and another motif that is annotated as rRNA related by T-profiler (GCGATGAGMTGARAW), also are significantly downregulated at all time points after 30 min. These patterns of consensus motif enrichment are consistent with previously published observations of the transcriptional response to various stresses (17, 12) as well as the patterns of gene group enrichment described in this paper.
TABLE 3.
Consensus motifs whose associated genes were found to be upregulated or downregulated by T-profiler analysisa
| Time point | Upregulated motifs enriched in:
|
Downregulated motifs enriched in:
|
||
|---|---|---|---|---|
| CO | N2 | CO | N2 | |
| 15 min | CWTCC (GCR1, 0.0039), CCCCT (MSN2-4, 0.0060), CGCNNNNNNNNNNNNN NBCGB (unknown, 0.030) | AGGGG (MSN2-4, 6.8 × 10−5), TCTCC (ADR1, 0.016), TCCGYGGA (PDR3, 0.033), TCCGYGGR (unknown, 0.049) | CGATGAG (PAC, 0.0012) | AAAATTT (rRPE, 3.0 × 10−9), CGATGAG (PAC, 3.7 × 10−7), GCGATGAGMTGARAW (rRNA?, 0.0046) |
| 30 min | CCCCT (MSN2-4, 2.0 × 10−6), AGGGG (MSN2-4, 9.2 × 10−6) | AGGGG (MSN2-4, 2.8 × 10−9), CCCCT (MSN2-4, 1.4 × 10−6), TATAWAW (TBP, 2.7 × 10−4), TCCGYGGR (unknown, 3.1 × 10−4), TCCGYGGA (PDR3, 5.1 × 10−4), TGCACCC (RCS1, 0.0085) | CGATGAG (PAC, 8.2 × 10−6), AAAATTT (rRPE, 2.6 × 10−4) | AAAATTT (rRPE, 3.4 × 10−11), CGATGAG (PAC, 2.0 × 10−7), CCRTACA (RAP1, 2.0 × 10−5), GCGATGAGMTGARAW (rRNA?, 0.0019), YCGTNNNNMRYGAY (ABF1, 0.0055) |
| 45 min | AGGGG (MSN2-4, 1.6 × 10−7), CCCCT (MSN2-4, 0.001), TCCGYGGA (PDR3, 0.012), HRCCCYTWDT (MSN2-4, 0.025), TCCGYGGR (unknown, 0.031) | AGGGG (MSN2-4, < 10−15), CCCCT (MSN2-4, < 10−15), TATAWAW (TBP, 2.8 × 10−6), TCCGYGGA (PDR3, 5.5 × 10−6), TCCGYGGR (unknown, 6.1 × 10−5), HRCCCYTWDT (MSN2/4, 2.3 × 10−4), CCNNNWWRGG (MCM1, 0.018), TCTCC (ADR1, 0.029), TCCGCGG (unknown, 0.030) | AAAATTT (rRPE, 2.1 × 10−10), CGATGAG (PAC, 2.0 × 10−8), GCGATGAGMTGARA W (rRNA?, 0.0030), CCRTACA (RAP1, 0.0055) | AAAATTT (rRPE, 3.9 × 10−11), CGATGAG (PAC, 1.2 × 10−9), CCRTACA (RAP1, 2.5 × 10−9), GCGATGAGMTGARAW (rRNA?, 0.0060) |
| 60 min | AGGGG (MSN2-4, < 10−15), CCCCT (MSN2-4, < 10−15), TATAWAW (TBP, 0.0015), HRCCCYTWDT (MSN2/4, 0.0092), TCCGYGGA (PDR3, 0.041), CCNNNWWRGG (MCM1, 0.044) | AGGGG (MSN2-4, < 10−15), CCCCT (MSN2-4, < 10−15), TATAWAW (TBP, 1.8 × 10−5), TCCGYGGA (PDR3, 0.0022), HRCCCYTWDT (MSN2/4, 0.020) | AAAATTT (rRPE, < 10−15), CGATGAG (PAC, 1.1 × 10−13), CCRTACA (RAP1, 7.2 × 10−5), GCGATGAGMTGARA W (rRNA?, 7.8 × 10−4), TGACTCA (GCN4, 0.025) | AAAATTT (rRPE, 8.2 × 10−8), CGATGAG (PAC, 3.5 × 10−7), CCRTACA (RAP1, 0.0022) |
| 120 min | AGGGG (MSN2-4, 2.4 × 10−8), TCCGYGGA (PDR3, 1.2 × 10−6), CCCCT (MSN2-4, 1.4 × 10−5), TCCGYGGR (unknown, 1.2 × 10−4), TATAWAW (TBP, 0.0046), HRCCCYTWDT (MSN2/4, 0.018) | CCCCT (MSN2-4, 2.9 × 10−8), AGGGG (MSN2-4, 4.2 × 10−7), TCCGYGGA (PDR3, 7.0 × 10−6), TCCGYGGR (unknown, 1.3 × 10−4), TATAWAW (TBP, 0.0021), HRCCCYTWDT (MSN2/4, 0.027), TCCGCGG (unknown, 0.049) | CCRTACA (RAP1, < 10−15), AAAATTT (rRPE, 6.6 × 10−10), CGATGAG (PAC, 1.2 × 10−4) | CCRTACA (RAP1, < 10−15), AAAATTT (rRPE, 9.4 × 10−10), CGATGAG (PAC, 2.8 × 10−4) |
| 24 h | CCCCT (MSN2-4, 2.9 × 10−8), AGGGG (MSN2-4, 8.2 × 10−6), TCCGYGGA (PDR3, 3.8 × 10−4), TCCGYGGR (unknown, 0.0011), CGGNNNNNNNNNNNCCG (GAL4, 0.0036), CGGNNNNNNNNNNCCG (PUT3, 0.0060), TGCACCC (RCS1, 0.010), GCAYGTG (INO4, 0.019), ATGYGRAWW (NBF, 0.025), TCCGCGG (unknown, 0.039) | TCCGYGGA (PDR3, 3.1 × 10−4), AGGGG (MSN2-4, 7.8 × 10−4), CGGNNNNNNNNNNCCG (PUT3, 0.0010), TCCGYGGR (unknown, 0.0018), CGGNNNNNNNNNNNCCG (GAL4, 0.016), ATGYGRAWW (NBF, 0.035), TCCGCGG (unknown, 0.039), CCCCT (MSN2-4, 0.041), CGGGGTA (MIG1, 0.044) | AAAATTT (rRPE, 4.9 × 10−11), CGATGAG (PAC, 2.3 × 10−7), CCRTACA (RAP1, 0.0092) | AAAATTT (rRPE, 2.1 × 10−7), CCRTACA (RAP1, 4.2 × 10−5), TGACTCA (GCN4, 5.5 × 10−5), CGATGAG (PAC, 8.6 × 10−4) |
Motifs that were enriched by treatment with either gas at each time point are listed, along with the cognate transcription factor (if known) and the E value in parentheses. Besides A, C, G, and T, the other one-letter codes represent different nucleotides as follows: R for purines, Y for pyrimidines, N for any nucleotide, W for weak (A or T), S for strong (C or G), M for amino (A or C), K for keto (G or T), B for any nucleotide that is not A, H for any not G, D for any not C, and V for any not T.
Relative derepression of gene groups involved in aerobic energy generation when cells are exposed to CO but not when exposed to N2.
In contrast to the broad similarity described in the preceding sections, we noted differences in gene expression between exposure to CO and exposure to N2 for the groups of genes that were upregulated early in the anoxia time course (Table 1). At 15 and 30 min of exposure to CO, many gene groups associated with aerobic energy generation were upregulated. These groups include oxidative phosphorylation (29 genes), proton-transporting ATP synthesis (16 genes), mitochondrial electron transport chain (22 genes), and aerobic respiration (61 genes). This is in marked contrast to what was observed for N2, for which, except for a significant increase in mitochondrial electron transport chain gene expression at the 15-min time point, these gene groups were not found to have significantly changed expression throughout the N2 time course.
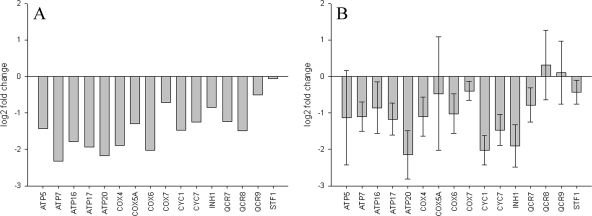
To verify the differences in gene expression between exposure to CO and N2, we carried out qRT-PCR on 16 transcripts with roles in aerobic energy generation. An analysis of microarray data had shown that these 16 transcripts were in relatively high abundance in CO-treated cells at 30 min in anoxia compared to that of the room air control. Further, the array data showed that most of these transcripts were at least twofold more abundant in CO-treated cells than in N2-treated cells (Fig. 6A). qRT-PCR analysis confirmed these findings for most of the 16 genes examined (Fig. 6B). We conclude that, consistently with the known oxygen-mimetic properties of CO, treating cells with this gas causes a coordinated relative derepression of genes involved in aerobic energy generation when compared to cells treated with N2.
FIG. 6.
(A) Log2-transformed change (n-fold), as determined by microarray analysis, comparing BY4741 N2-treated cells to CO-treated cells at 30 min in anoxia for 16 transcripts of genes involved in aerobic metabolism. These 16 transcripts were found to be relatively most abundant in CO-treated cells at 30 min in anoxia. Most of the transcripts are less abundant in N2 by at least twofold. (B) Log2-transformed change (n-fold), as determined by qRT-PCR analysis, comparing BY4741 N2-treated cells to CO-treated cells at 30 min in anoxia for the same 16 transcripts described in panel A. Similarly to the microarray results, most of the transcripts were found to be less abundant in N2. Data are from five replicate qRT-PCR runs; error bars represent standard errors of the means.
Mitochondrial retrograde signaling is required for recovery from prolonged exposure to N2 but not CO.
Since O2-dependent biochemistry would be equally impossible in the continuous presence of pure CO or N2, it is unlikely that intracellular signaling due to decreased flux through O2-requiring pathways would account for the relative differences in gene expression between CO and N2. Instead, we hypothesized that differences in intracellular signaling between CO and N2 more likely would be due to the ability of CO to mimic O2 by interacting with, for instance, heme-containing proteins (23). We therefore tested for differential sensitivity to prolonged anoxia by either CO or N2, using 174 strains from the MATa deletion set that each are deleted for a nonessential gene annotated as playing a role in signal transduction, according to GO on SGD. Essentially, we sought to identify signal transduction mutants that are sensitive to one anoxic gas but not the other.
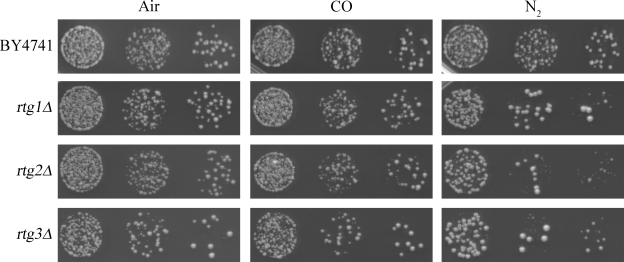
The three mutants that exhibited the most striking phenotype in regard to growth following exposure to anoxia were rtg1, rtg2, and rtg3. These deletion mutants exhibit poor recovery after exposure to N2 but normal recovery after exposure to CO (Fig. 7). RTG1, RTG2, and RTG3 function in the mitochondrial retrograde signaling pathway, which is thought to be important for metabolic reconfiguration to compensate for mitochondrial dysfunction (reviewed in reference 33). In contrast to the essentially normal recovery from CO, rtg1, rtg2, and rtg3 deletion mutants tend to permanently arrest as small microcolonies when attempting to recover from N2. For each rtg mutant, a small fraction of colonies manages to grow relatively large in the recovery from N2, possibly due to the accumulation of suppressing mutation(s).
FIG. 7.
Retrograde signaling is required for proper recovery from prolonged exposure to N2. Tenfold serial dilutions of each strain spotted onto solid YPA are shown after 7 days of growth in air (left column), 4 days of arrest in CO followed by 7 days of recovery in air (middle column), and 4 days of arrest in N2 followed by 7 days of recovery in air (right column). Each of these deletion strains recovers poorly after prolonged exposure to N2 but exhibit relatively normal recovery after similar exposure to CO. Rtg1, Rtg2, and Rtg3 are components of the so-called mitochondrial retrograde signaling pathway, which is thought to activate changes in nuclear gene expression to compensate for mitochondrial dysfunction.
Aerobic energy generation genes are relatively derepressed when retrograde signaling mutants are exposed to N2.
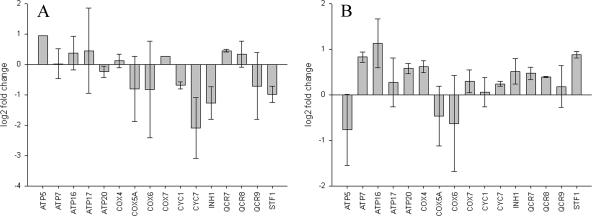
Given the N2-specific sensitivity of the retrograde signaling mutants and the fact that, in wild-type BY4741 cells, aerobic energy generation genes show differential regulation in CO compared to that in N2, we carried out qRT-PCR to determine if the pattern of gene expression in anoxia differed between the wild type and rtg mutants. We examined the strains deleted for RTG1 and RTG3, as each of these genes encodes a basic helix-loop-helix leucine zipper transcription factor. Together, Rtg1 and Rtg3 function as a heterodimer to effect nuclear gene expression as a result of signaling via the retrograde pathway (33). We found that in both rtg1 and rtg3 deletion mutants, the 16 aerobic energy generation genes previously described tend to be more derepressed at 30 min in N2 than in CO (compare Fig. 6 to Fig. 8). This contrast between the mutant and wild type is particularly striking when comparing rtg3Δ (Fig. 8B) to BY4741 (Fig. 6B). We conclude that the deletion of either the RTG1 or RTG3 transcription factor gene results in gene expression patterns upon exposure to N2 that are aberrant compared to those of the wild type.
FIG. 8.
(A) Log2-transformed change (n-fold), as determined by qRT-PCR, comparing rtg1Δ N2-treated cells to CO-treated cells at 30 min in anoxia for the 16 transcripts depicted in Fig. 6. Many of these aerobic metabolism genes are relatively derepressed in rtg1Δ when treated with N2. (B) A similar plot comparing rtg3Δ N2-treated cells to CO-treated cells at 30 min in anoxia for the same 16 transcripts. Most of these aerobic metabolism genes are more markedly derepressed in rtg3Δ than in rtg1Δ when treated with N2. Data for each panel are from two replicate qRT-PCR runs; error bars represent standard errors of the means.
DISCUSSION
Budding yeast is a model for anoxia-induced suspended animation.
Understanding the molecular mechanisms that enable cells to cope with a lack of oxygen is of high importance to human health while also being of great interest from a basic science perspective. To better elucidate such mechanisms, our laboratory previously characterized the phenomenon of anoxia-induced suspended animation in two model organisms, the nematode C. elegans and the zebrafish D. rerio. To determine if a similar response is conserved in a model species with even more facile gene-based research tools, we turned to the budding yeast S. cerevisiae. We demonstrated that both sporulating and vegetative yeast on a nonfermentable carbon source undergo anoxia-induced suspended animation in response to severe oxygen deprivation. The sporulation process can be suspended even after being well under way, with high recoverability, for up to 12 h. Vegetative cells can be maintained in a suspended state for at least 7 days, also with high recoverability (more than 75%). These results, combined with the demonstration of a similar phenomenon in Drosophila (16), show that anoxia-induced suspended animation is a response to severe oxygen deprivation that is conserved among four well-studied model organisms.
We found that vegetative yeast cells on nonfermentable medium rapidly halt their cell divisions when made anoxic. This result is reminiscent of transient cell cycle arrest under nutrient limitation (45) and prolonged arrest under starvation (14, 61, 62). However, cells that are made anoxic on nonfermentable medium do not uniformly arrest in G1. In contrast, cells exposed to various stimuli, including nutrient starvation (22), mating pheromone (11), elevated temperature (25), oxidative stress (59), and osmotic stress (7), tend to arrest in G1. We therefore conclude that the phenomenon of anoxia-induced suspended animation, which we have named in analogy to related phenomena in higher eukaryotes, is distinct from the growth arrest of wild-type yeast previously described.
In addition, note that depending on the genotype, not all types of starvation result in viable nonproliferative states. A striking example of this phenomenon was described by Botstein and colleagues, who worked with a strain that is auxotrophic for leucine and uracil. When starved for either phosphate or sulfur in liquid medium, the cells remained completely viable after 1 week, as judged by subsequent colony formation on solid YPD. In contrast, when the cells were starved for leucine for 1 week, there was a 10-fold decrease in viability. Similar starvation for uracil resulted in an even more severe viability decrease of more than 100-fold (8). Thus, reversible arrest in a viable nonproliferative state is not a default response to starvation. Instead, there are likely particular cellular mechanisms that enable cells to enter into and maintain a viable nonproliferative state, such as anoxia-induced suspended animation on nonfermentable medium.
Also, while a straightforward analogue for anoxia-induced suspended animation can be described when comparisons are made across model systems such as yeast, nematodes, fruit flies, and zebrafish, the responses to nutrient limitation or starvation are much more divergent across different species; for example, when comparing yeast to nematodes (48). This similarity in the response to oxygen deprivation among model species suggests the possibility that yeast that are made to assume an obligate aerobic lifestyle on nonfermentable medium serve as a useful model for studying conserved cellular responses to oxygen deprivation. Also, given the apparently conserved nature of this response to extremely low oxygen levels, it is curious to consider well-documented cases in the medical literature describing humans who have survived prolonged bouts of oxygen deprivation due to hypothermic circulatory arrest in various accidents with little or no adverse sequelae (58, 19).
Analysis of gene expression reveals a coordinated derepression of aerobic energy generation genes in CO but not in N2.
In order to identify genes that are involved in suspended animation, we carried out transcript microarray analysis on cells that were exposed to either CO or N2. Using the T-profiler tool, we found that while the gene group and consensus motif enrichment profiles were quite similar for the two anoxic gases, there were areas of marked difference between exposure to CO and exposure to N2. Specifically, multiple gene groups whose constituent genes have roles in aerobic energy generation were significantly upregulated at 15 and 30 min in CO but not in N2. We noted 16 genes that were upregulated by at least twofold in CO at 30 min based on the microarray data. Most of these 16 genes were at least twofold more abundant in CO than in N2, again based on the microarray data. We carried out qRT-PCR to verify these results and confirmed that most of these genes were indeed relatively derepressed in CO compared to their levels in N2. The derepression of these genes is consistent with the idea that CO acts as an oxygen mimetic, presumably by binding at the heme of hemoproteins (43), which effects signal transduction events that result in the observed changes in transcription. We propose that the presence of a high concentration of CO essentially fools the cells into sensing that there is abundant O2, and thus genes that encode proteins with roles in aerobic metabolism are coordinately derepressed compared to the expression levels in the presence of N2, which is not an oxygen mimetic.
Previously, Poyton and colleagues found that the anaerobic induction of two genes (CYC7 and OLE1) in N2 can be completely blocked by treatment with CO, while the induction of a third gene (COX5B) is partially blocked by CO in cells on galactose medium (28). They found that 11 other genes, previously shown to be oxygen regulated, showed no difference in expression after treatment with either anoxic gas. To our knowledge, no one has looked for differential gene regulation in CO and compared it to that of N2 on a genomewide scale. It is worth noting that the choice of medium perhaps has a strong influence on the likelihood of observing differences in gene expression. Since a lack of oxygen brought on by exposure to pure CO or N2 would not be expected to differentially affect anaerobic metabolism per se, it is possible that cells on a fermentable medium are less likely to manifest differences in gene expression than cells on a nonfermentable medium. This is because energy generation from a nonfermentable carbon source requires O2 as well as gene expression changes associated with the need to utilize O2. If so, then the application of an O2 mimetic, namely CO, while the cells are on a nonfermentable substrate would be more likely to elicit changes in gene expression that are normally O2 dependent than a similar CO exposure on a fermentable substrate. In addition, the use of nonfermentable medium essentially converts the yeast cells into obligate aerobes, thus making the yeast model more similar to the truly obligate aerobic cells of higher eukaryotes.
An analysis of the microarray data also showed that a general stress response (previously referred to as the environmental stress response by Gasch et al. [17] and the common environmental response by Causton et al. [12]) is likely activated by yeast that were made anoxic on a nonfermentable substrate. Specifically, we found that genes with MSN2/MSN4 motifs and genes encoding cell wall proteins were significantly upregulated at almost all time points, while genes with roles in transcription, translation, and many associated processes were significantly downregulated, again at nearly all time points. These genes that are similarly regulated between the two anoxic gases appear to form the core transcriptional response to anoxia, sharing much in common with other stress responses.
The mitochondrial retrograde signaling pathway is functionally important for recovery from prolonged exposure to N2 but not to CO.
Having found that aerobic energy generation genes tend to be relatively derepressed in CO, we set out to identify genes that are functionally important for enabling survival in one anoxic gas but not the other. We found that while mutants deleted for components of the mitochondrial retrograde signaling pathway were able to recover normally after prolonged exposure to CO, they recovered very poorly after a similar exposure to N2. We then found that in both the rtg1 and the rtg3 deletion mutants a number of aerobic energy generation genes tend to be derepressed in N2. This is in contrast to the relative repression of these same genes in wild-type cells. Thus, the disruption of mitochondrial retrograde signaling results in aberrant gene expression in an anoxic gas-dependent manner.
CO may cause divergent signals from two distinct oxygen-sensing pathways in budding yeast.
The molecular nature of oxygen sensing in budding yeast has been an area of active study for a long time. Multiple molecular mechanisms for oxygen sensing in yeast have been proposed. One such mechanism proposes that cells sense the intracellular level of some compound(s) that requires O2 for synthesis. Thus, the concentration of the compound(s) can be an effective proxy for oxygen concentration. Candidate compounds that can serve in this role are heme (64) and sterols (13). A distinct oxygen-sensing mechanism proposes that the binding of molecular oxygen to heme-containing proteins causes signal transduction events that result in changes in gene expression (43).
Based on our gene expression data, we propose a model in which both of these mechanisms are simultaneously functioning in budding yeast, such that each mechanism mediates a subset of the overall transcriptional response. First, recall that treatment with CO causes a derepression of aerobic metabolism genes relative to that seen in N2. As noted previously, this is consistent with hemoprotein occupancy-based signaling that mimics O2 binding. In addition, we note that other parts of the transcriptional response are very similar for both anoxic gases. Gene groups that are downregulated include ribosome biogenesis genes as well as genes involved in transcription, translation, and related biosynthetic pathways. As noted in Results, ribosome biogenesis genes are known to be downregulated only after prolonged growth on a nonfermentable medium (46). Thus, the downregulation seen in the anoxic samples relative to the reference air samples on acetate probably represents a greater degree of transcriptional repression than can be accounted for merely by the transitioning to growth on a nonfermentable carbon source. We also observed an upregulation of cell wall genes in both anoxic gases, similarly to previously published results (29, 30, 31, 57) for cells made anoxic on fermentable medium. Taking into consideration these common features of the transcriptional response to both anoxic gases, we propose that the signal that results in these similar patterns of gene expression originates from a mechanism that would be expected to respond similarly to both CO and N2, such as the depletion of some compound(s) that requires O2 for its synthesis.
In this model, treating cells with N2 results in signals from both mechanisms that are convergent, i.e., both signal a lack of O2, as N2 is not believed to bind in hemoproteins like O2 does, while the concentration of some compound(s) requiring O2 for synthesis should decrease. In contrast, treating cells with CO results in divergent signals, as the oxygen-mimetic properties of CO points to the (perceived) presence of O2, while the concentration of the compound(s) that require O2 in order to be synthesized still would decrease, pointing to the (actual) lack of O2. The difference in signaling between CO and N2 ultimately results in transcriptional responses that are similar across much of the transcriptome but are markedly different for aerobic metabolism genes. Since the differences between exposure to CO and N2 are most apparent in the aerobic metabolism genes, it is quite possible that such changes can be observed only when the cells are forced to generate energy aerobically by being put on a nonfermentable medium. As such, we propose that the experimental paradigm we arrived at to demonstrate the conservation of anoxia-induced suspended animation in budding yeast can be of considerable utility in continuing efforts to better understand the molecular mechanisms that mediate oxygen-regulated gene expression.
Acknowledgments
This work was supported by grant R01 GM48435 from the National Institutes of Health.
We are grateful to the Breeden, Gottschling, and Tsukiyama laboratories at the Fred Hutchinson Center for gifts of strains and reagents, the use of equipment, and much technical advice. We also are thankful to Lazar Dimitrov, Harold Frazier, Kiersten Henderson, Dana Miller, Michael Petrascheck, Ashwin Unnikrishnan, and Joshua Veatch for the critical reading of the manuscript.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Andreasen, A. A., and T. J. Stier. 1953. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell Physiol. 4123-36. [DOI] [PubMed] [Google Scholar]
- 2.Andreasen, A. A., and T. J. Stier. 1954. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in a defined medium. J. Cell Physiol. 43271-281. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 228-36. [PubMed] [Google Scholar]
- 4.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 1448-54. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, T. L., N. Williams, C. Misleh, and W. W. Li. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34W369-W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, J. A. 2003. Beginnings of microbiology and biochemistry: the contribution of yeast research. Microbiology 149557-567. [DOI] [PubMed] [Google Scholar]
- 7.Bellí, G., E. Garí, M. Aldea, and E. Herrero. 2001. Osmotic stress causes a G1 cell cycle delay and downregulation of Cln3/Cdc28 activity in Saccharomyces cerevisiae. Mol. Microbiol. 391022-1035. [DOI] [PubMed] [Google Scholar]
- 8.Boer, V. M., S. Amini, and D. Botstein. 2008. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. USA 1056930-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boorsma, A., H. de Nobel, B. ter Riet, B. Bargmann, B. Stanley, K. J. Hellingwerf, and F. M. Klis. 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21413-427. [DOI] [PubMed] [Google Scholar]
- 10.Boorsma, A., B. C. Foat, D. Vis, F. Klis, and H. J. Bussemaker. 2005. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 33W592-W595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bücking-Throm, E., W. Duntze, L. H. Hartwell, and T. R. Manney. 1973. Reversible arrest of haploid yeast cells at the initiation of DNA synthesis by diffusible sex factor. Exp. Cell Res. 7699-110. [DOI] [PubMed] [Google Scholar]
- 12.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, B. S., and J. Rine. 2006. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drebot, M. A., C. A. Barnes, R. A. Singer, and G. C. Johnston. 1990. Genetic assessment of stationary phase for cells of the yeast Saccharomyces cerevisiae. J. Bacteriol. 1723584-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell, A. P., and J. A. Stecyk. 2007. The heart as a working model to explore themes and strategies for anoxic survival in ectothermic vertebrates. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 147300-312. [DOI] [PubMed] [Google Scholar]
- 16.Foe, V. E., and B. M. Alberts. 1985. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J. Cell Biol. 1001623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Véronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. André, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Güldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kötter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert, M., R. Busund, A. Skagseth, P. A. Nilsen, and J. P. Solbo. 2000. Resuscitation from accidental hypothermia of 13.7 degrees C with circulatory arrest. Lancet 355375-376. [DOI] [PubMed] [Google Scholar]
- 20.Gorr, T. A., M. Gassmann, and P. Wappner. 2006. Sensing and responding to hypoxia via HIF in model invertebrates. J. Insect Physiol. 52349-364. [DOI] [PubMed] [Google Scholar]
- 21.Hand, S. C. 1998. Quiescence in Artemia franciscana embryos: reversible arrest of metabolism and gene expression at low oxygen levels. J. Exp. Biol. 2011233-1242. [DOI] [PubMed] [Google Scholar]
- 22.Hartwell, L. H. 1974. Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 38164-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochachka, P. W., L. T. Buck, C. J. Doll, and S. C. Land. 1996. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc. Natl. Acad. Sci. USA 939493-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia, Y., B. Rothermel, J. Thornton, and R. A. Butow. 1997. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol. 171110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston, G. C., and R. A. Singer. 1980. Ribosomal precursor RNA metabolism and cell division in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 178357-360. [DOI] [PubMed] [Google Scholar]
- 26.Jonz, M. G., and C. A. Nurse. 2006. Ontogenesis of oxygen chemoreception in aquatic vertebrates. Respir. Physiol. Neurobiol. 154139-152. [DOI] [PubMed] [Google Scholar]
- 27.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423241-254. [DOI] [PubMed] [Google Scholar]
- 28.Kwast, K. E., P. V. Burke, B. T. Stahl, and R. O. Poyton. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. USA 965446-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwast, K. E., L. C. Lai, N. Menda, D. T. James, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, L. C., A. L. Kosorukoff, P. V. Burke, and K. E. Kwast. 2005. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol. Cell. Biol. 254075-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, L. C., A. L. Kosorukoff, P. V. Burke, and K. E. Kwast. 2006. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot. Cell 51468-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesage, P., and A. L. Todeschini. 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 11070-90. [DOI] [PubMed] [Google Scholar]
- 33.Liu, Z., and R. A. Butow. 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40159-185. [DOI] [PubMed] [Google Scholar]
- 34.Martínez-Pastor, M. T., G. Marchler, C. Schüller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 152227-2235. [PMC free article] [PubMed] [Google Scholar]
- 35.Michiels, C. 2004. Physiological and pathological responses to hypoxia. Am. J. Pathol. 1641875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. J. 1989. Sporulation in Saccharomyces cerevisiae, p. 489-550. In A. H. Rose and J. S. Harrison (ed.), The yeasts: metabolism and physiology of yeasts, vol. 3. Academic Press, Berkeley, CA. [Google Scholar]
- 37.Nilsson, G. E., and G. M. Renshaw. 2004. Hypoxic survival strategies in two fishes: extreme anoxia tolerance in the North European crucian carp and natural hypoxic preconditioning in a coral-reef shark. J. Exp. Biol. 2073131-3139. [DOI] [PubMed] [Google Scholar]
- 38.Nystul, T. G., J. P. Goldmark, P. A. Padilla, and M. B. Roth. 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science 3021038-1041. [DOI] [PubMed] [Google Scholar]
- 39.Padilla, P. A., and M. B. Roth. 2001. Oxygen deprivation causes suspended animation in the zebrafish embryo. Proc. Natl. Acad. Sci. USA 987331-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padilla, P. A., T. G. Nystul, R. A. Zager, A. C. Johnson, and M. B. Roth. 2002. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell 131473-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piantadosi, C. A. 2002. Biological chemistry of carbon monoxide. Antioxid. Redox Signal. 4259-270. [DOI] [PubMed] [Google Scholar]
- 42.Pilpel, Y., P. Sudarsanam, and G. M. Church. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29153-159. [DOI] [PubMed] [Google Scholar]
- 43.Poyton, R. O. 1999. Models for oxygen sensing in yeast: implications for oxygen-regulated gene expression in higher eucaryotes. Respir. Physiol. 115119-133. [DOI] [PubMed] [Google Scholar]
- 44.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26415-423. [DOI] [PubMed] [Google Scholar]
- 45.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle, p. 97-142. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces cerevisiae; life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Roberts, G. G., and A. P. Hudson. 2006. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Gen. Genomics 276170-186. [DOI] [PubMed] [Google Scholar]
- 47.Safar, P., S. A. Tisherman, W. Behringer, A. Capone, S. Prueckner, A. Radovsky, W. S. Stezoski, and R. J. Woods. 2000. Suspended animation for delayed resuscitation from prolonged cardiac arrest that is unresuscitable by standard cardiopulmonary-cerebral resuscitation. Crit. Care Med. 28N214-N218. [DOI] [PubMed] [Google Scholar]
- 48.Sawin, E. R., R. Ranganathan, and H. R. Horvitz. 2000. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26619-631. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 935777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sehgal, A., C. Y. Lee, and P. J. Espenshade. 2007. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 31389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Semenza, G. L. 2006. Regulation of physiological responses to continuous and intermittent hypoxia by hypoxia-inducible factor 1. Exp. Physiol. 91803-806. [DOI] [PubMed] [Google Scholar]
- 52.Semenza, G. L. 2007. Vasculogenesis, angiogenesis, and arteriogenesis: mechanisms of blood vessel formation and remodeling. J. Cell Biochem. 102840-847. [DOI] [PubMed] [Google Scholar]
- 53.Shen, C., and J. A. Powell-Coffman. 2003. Genetic analysis of hypoxia signaling and response in C. elegans. Ann. N. Y. Acad. Sci. 995191-199. [DOI] [PubMed] [Google Scholar]
- 54.Snoek, I. S., and H. Y. Steensma. 2007. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res. 241-10. [DOI] [PubMed] [Google Scholar]
- 55.Storey, K. B. 2007. Anoxia tolerance in turtles: metabolic regulation and gene expression. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 147263-276. [DOI] [PubMed] [Google Scholar]
- 56.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22281-285. [DOI] [PubMed] [Google Scholar]
- 57.ter Linde, J. J., H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 1817409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walpoth, B. H., B. N. Walpoth-Aslan, H. P. Mattle, B. P. Radanov, G. Schroth, L. Schaeffler, A. P. Fischer, L. von Segesser, and U. Althaus. 1997. Outcome of survivors of accidental deep hypothermia and circulatory arrest treated with extracorporeal blood warming. N. Engl. J. Med. 3371500-1505. [DOI] [PubMed] [Google Scholar]
- 59.Wanke, V., K. Accorsi, D. Porro, F. Esposito, T. Russo, and M. Vanoni. 1999. In budding yeast, reactive oxygen species induce both RAS-dependent and RAS-independent cell cycle-specific arrest. Mol. Microbiol. 32753-764. [DOI] [PubMed] [Google Scholar]
- 60.Wenger, R. H. 2002. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 151151-1162. [DOI] [PubMed] [Google Scholar]
- 61.Werner-Washburne, M., E. Braun, G. C. Johnston, and R. A. Singer. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57383-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Werner-Washburne, M., E. L. Braun, M. E. Crawford, and V. M. Peck. 1996. Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 191159-1166. [DOI] [PubMed] [Google Scholar]
- 63.Winderickx, J., I. Holsbeeks, O. Lagatie, F. Giots, J. Thevelein, and H. de Winde. 2003. From feast to famine; adaptation to nutrient availability in yeast. In S. Hohmann and W. H. Mager (ed.), Yeast stress responses. Springer-Verlag, New York, NY.
- 64.Zitomer, R. S., and C. V Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]