Abstract
Sex is orchestrated by the mating-type locus (MAT) in fungi and by sex chromosomes in plants and animals. In fungi, two patterns of sexuality occur: bipolar with a single, typically biallelic sex determinant that promotes inbreeding, and tetrapolar with two unlinked, often multiallelic sex determinants that restrict inbreeding. Multiallelism in either bipolar or tetrapolar mating systems promotes outcrossing. Cryptococcus neoformans is a pathogenic bipolar yeast with two unusually large MAT alleles (a/α) spanning >100 kb, ∼100-fold larger than many other fungal MAT loci. Based on comparative genomic analysis, this unusual MAT locus is hypothesized to have evolved from an ancestral tetrapolar system. In this model, the unlinked homeodomain (HD) transcription factor and pheromone/receptor tetrapolar loci acquired additional sex-related genes and then fused via chromosomal translocation, forming an intermediate transitional mating system (which we term tripolar), which then underwent recombination and gene conversion to fashion the extant bipolar MAT alleles. To experimentally validate this model, C. neoformans was engineered to have a tetrapolar mating system by relocating the MAT SXI1α and SXI2a HD genes to an unlinked genomic locale. Genetic and molecular analyses revealed that this modified organism could complete a tetrapolar sexual cycle. Analysis of progeny generated from bipolar, tripolar, and tetrapolar crosses provides direct experimental evidence that the tripolar state confers decreased fertility and therefore may represent an unstable evolutionary intermediate. These findings illustrate how transitions between outcrossing and inbreeding preference occur by involving sex determinant linkage and collapse from multiallelic to biallelic sex determination, providing insights into both fungal sex evolution and early steps in sex chromosome evolution.
Despite the fact that sex is almost universal among organisms, the process and mechanisms of sex determination are highly diverse. In vertebrates, sex determination can be either genetic or environmental. For example, sex in mammals and birds is governed by the XX/XY or ZZ/ZW sex chromosome system, respectively, while it is temperature determined in certain species of fish and turtles (4, 31). Surprisingly, recent studies revealed that in some species, the distinction between the two modes is less distinct. For instance, the lizard Pogona vitticeps and the common frog Rana temporaria were shown to possess both temperature and chromosomal sex determination systems (23, 29). A different dosage-dependent mechanism is adopted in invertebrates such as flies and worms, in which sex is determined by the X chromosome-to-autosome ratio (27).
In simple eukaryotes such as fungi, sex is also genetically controlled. However, unlike the seemingly more-complex sex-determining process controlled by sex chromosomes in mammals, sex in fungi is much more simplified, being governed by a delimited, sex-specific region of the genome called the mating-type locus (MAT). Studies of the structure and function of MAT loci in different fungal lineages have provided insight regarding how sex evolves (8, 16). The MAT loci encode sex-determining transcription factors that exhibit conserved motifs: homeodomain, α-box domain, or high-mobility-group domain. Despite great variation in the modes of sexual reproduction between different fungal species, mating types are determined by two predominant MAT paradigms: bipolar and tetrapolar. In bipolar species, MAT is a single locus with two idiomorphs (in ascomycetes) or two or multiple alleles (in basidiomycetes). In tetrapolar species, MAT occurs as two unlinked loci that are often multiallelic. Thus, bipolar fungi typically have two alternative mating types and the tetrapolar fungi, depending on the number of alleles that exist, may have up to hundreds and even thousands of mating types (18). Examples of bipolar multiallelic sex determination, such as in Coprinellus disseminatus, are also known (17).
The MAT locus of the basidiomycetous yeast Cryptococcus neoformans is of special interest because mating type is correlated with virulence (14, 19). In addition, in contrast to many fungal MAT loci that are limited in size, ranging from ∼700 to several thousand base pairs, the C. neoformans MAT locus is >120 kb and encodes more than 20 genes (22). How this MAT gene cluster evolved from a simpler ancestral form is an intriguing question. Studies of MAT in C. neoformans and the sibling species C. gattii have revealed that MAT is highly rearranged between mating types and even closely related species. Furthermore, MAT-specific genes exhibit different phylogenetic histories, suggesting that they were acquired into the locus at different time points during evolution and then subjected to more recent gene conversion, resulting in gene strata of different evolutionary ages. These lines of evidence have led to the hypothesis that this unusual MAT locus evolved from a simpler, ancestral tetrapolar mating system with two physically unlinked MAT loci (7).
Parallel studies of the MAT loci of smut fungi have revealed a similar scenario. The majority of species in the smut fungal lineage are tetrapolar, with Ustilago maydis as a paradigmatic example. The closely related species U. hordei has a bipolar mating system. Analysis of the U. hordei MAT locus revealed that the homeodomain and the pheromone/receptor MAT loci have been fused into an ∼500-kb, nonrecombining region of the genome, possibly via chromosomal translocation (1, 3, 21). A similar fused MAT arrangement in a more distantly related dandruff-associated fungus, Malassezia globosa, arose independently (2, 35).
Here, we recapitulated the ancestral tetrapolar, and the hypothesized tripolar, intermediate mating system of C. neoformans by using genetically engineered strains. The sex-determining, MAT homeodomain genes SXI1α or SXI2a that reside in the MAT locus were deleted, and the wild-type genes were reintroduced at a genetic locus unlinked to MAT on a different chromosome. By manipulating the structure of MAT to mimic a tetrapolar system, we provide direct experimental evidence to support models of the evolution of MAT, with implications for transitions commonly observed in sexual reproduction from tetrapolar to bipolar sexuality, involving outcrossing to inbreeding modes of reproduction. Our findings also mirror early events hypothesized to occur in sex chromosome evolution in which sex determinants arise on autosomes and then are linked to form nascent sex-determining gene clusters (5, 25).
MATERIALS AND METHODS
Strains, plasmids, and media.
Strains and plasmids used in this study are listed in Table 1. Mating reactions of the desired strains were established by coculturing the opposite mating-type cells on Murashige and Skoog (MS) medium minus sucrose (Sigma-Aldrich) or 5% V8 juice agar medium (pH 5).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| C. neoformans var. grubiia | ||
| H99 | MATα | 28 |
| KN99a | MATa | 24 |
| F99 | MATα ura5 | 24 |
| JF99a | MATaura5 | This study |
| JF135 | MATα sxi1α::NEO ura5 | This study |
| JF271 | MATasxi2a::NAT ura5 | This study |
| JF306 | MATα sxi1α::NEO ura5 SXI1α::URA5 | This study |
| JF289 | MATasxi2a::NAT ura5 SXI2a::URA5 | This study |
| YPH153 | MATaSXI1α::URA5 (MATaSXI1α) | This study |
| YPH510 | MATα SXI2a::URA5 (MATα SXI2a) | This study |
| YPH227 | MATasxi2a::NAT ura5 SXI1α::URA5 | This study |
| YPH220 | MATα sxi1α::NEO ura5 SXI2a::URA5 | This study |
| YPH716 | MATα sxi1α::NEO ura5 SXI2a::URA5 (H99 mitochondria) | This study |
| C. neoformans var. neoformans | ||
| JEC20 | MATa | 19 |
| JEC21 | MATα | 19 |
| Plasmid | ||
| pJAF7 | URA5 AmpR | This study |
| pJAF34 | SXI1α URA5 AmpR | This study |
| pJAF35 | SXI2aURA5 AmpR | This study |
Serotype A, congenic to H99 and KN99a.
Strain construction.
The sxi1α and sxi2a mutants were generated with the dominant selectable markers NEO or NAT, respectively, using an overlap PCR approach as previously described (9). The 5′ flanking region of SXI1α was amplified with primers JOHE9212/JOHE9213, and the 3′ flanking region was amplified with primers JOHE9214/JOHE9215. Similarly, the 5′ and 3′ flanking regions of SXI2a were amplified with primer pairs JOHE10020/JOHE10021 and JOHE10022/JOHE10023, respectively. The SXI1α::NEO and the SXI2a::NAT deletion cassettes were introduced into serotype A strains MATα ura5 (F99) and MATa ura5 (JF99a) by biolistic transformation. PCR and Southern analyses were used to verify the proper deletion of the target genes. The sxi1α and sxi2a mutants were designated strains JF135 and JF271. To reconstitute the wild-type SXI1α gene at the ura5 locus, a 6,041-bp XbaI/SphI fragment containing the wild-type SXI1α gene was first cloned into vector pUC19 and further subcloned into the XbaI/EcoICRI sites of plasmid pJAF7, which has the wild-type URA5 gene. Strain JF135 was biolistically transformed with the resulting plasmid pJAF34 in circular form to generate strain JF306 in which the wild-type SXI1α gene resides at the ura5 locus. Transformants were screened by PCR and Southern analysis to confirm the integration of the SXI1α-URA5 allele at ura5. The wild-type SXI2a gene was reintroduced into the ura5 locus using a similar approach. A 7,126-bp SXI2a KpnI/SacII fragment was cloned into the vector pBluescript SK(−) and further subcloned into the SacII/SmaI sites of plasmid pJAF7. Strain JF271 was transformed with this resulting plasmid, pJF35, to generate strain JF289, in which the wild-type SXI2a gene has been integrated into the ura5 locus. Southern analysis showed that multiple copies of SXI2a were tandemly integrated. Further multiple attempts to generate strains with a single-copy integration have been unsuccessful thus far, for unknown reasons. To generate the strain sxi1α SXI2a (YPH716) with H99-type mitochondria, the sxi1α::NEO mutant JF135 was transformed with the circular plasmid pJAF35, which carries the wild-type SXI2a gene. Transformants were then screened by PCR and Southern analyses to confirm SXI2a integration. Strains YPH153 and YPH510 were progeny derived from the “α” × a or “a” × α tripolar cross, respectively, and strains YPH227 and YPH220 were progeny derived from the “α” × “a” tetrapolar cross. Sequences of primers used in this study are listed in Table S1 in the supplemental material.
Micromanipulation of meiotic basidiospores.
Basidiospores were isolated with a micromanipulator as described previously (11). DNA was extracted from the germinated yeast cells, and PCR was performed to determine the genotype of each isolate.
Mitochondrial DNA inheritance.
Mitochondrial DNA inheritance among progeny was determined by PCR with primer pair Da3/Da20 (34).
RESULTS
Ectopically expressed SXI1α and SXI2a homeodomain genes are functional.
To physically unlink the homeodomain protein genes from the MAT locus, the SXI1α and SXI2a genes encoded by MAT were deleted with a NEO or a NAT dominant selectable marker in serotype A, MATα ura5 or MATa ura5 strain backgrounds. The resulting sxi1α and sxi2a mutants failed to mate. This mating defect is due to the lack of a functional Sxi1α/Sxi2a heterodimer that initiates the downstream sexual developmental cascade (12, 13). Next, the wild-type SXI1α and SXI2a genes were reintroduced into the sxi1α and sxi2a mutants, respectively, at the ura5 locus with the wild-type URA5 gene as a selectable marker, resulting in strains JF306 and JF289 in which the SXI1α and SXI2a transgenes (chromosome 7) are physically unlinked from the MAT locus (chromosome 4).
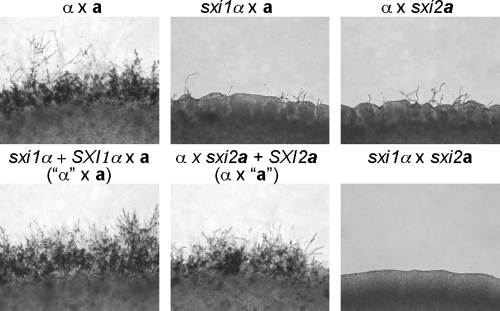
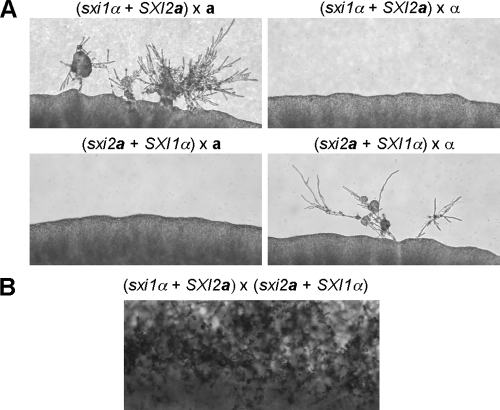
To analyze whether the reintroduced SXI1α and SXI2a genes at the ura5 locus complement the original mating defects, strains JF306 and JF289 were crossed to wild-type MATa (KN99a) and MATα (H99) strains, respectively. Mating assays showed that the reintroduced SXI1α and SXI2a genes restored the mating abilities of the sxi1α and sxi2a mutants; hyphae with basidia and long chains of basidiospores were observed (Fig. 1). Therefore, the SXI1α and SXI2a genes remain functional even though they are physically unlinked to the MAT locus. Strains JF306 and JF289 are referred to as the “α” and “a” strains, respectively, to indicate the fact that the homeodomain protein gene has been moved to reside at the ura5 locus.
FIG. 1.
The “a” and “α” strains are able to complete the sexual cycle when crossed to the wild-type α (H99) and a (KN99a) strains. Serotype A strains of the indicated genotypes were crossed on MS mating medium in the dark at 25°C. Mating colonies were photographed at ×100 magnification.
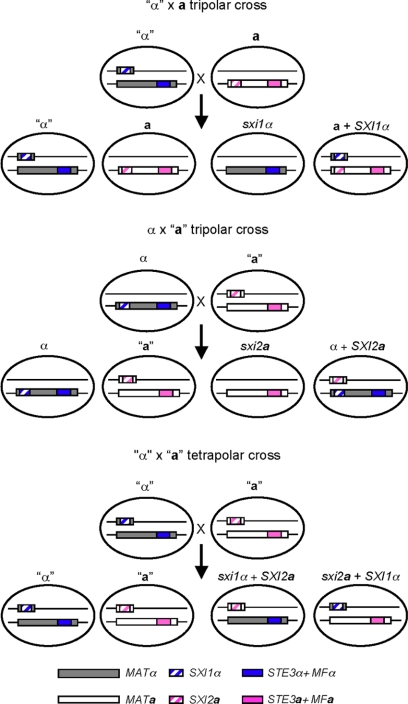
Crossing an “α” strain to a wild-type a strain—a tripolar cross.
To analyze the mating properties of the “α” strain in which the two components of MAT are genetically unlinked, strain JF306 was crossed to the MATa wild-type strain KN99a in what we term a tripolar cross (Fig. 2). We define the cross as “tripolar” because one of the parents has one contiguous MAT locus (one pole) while the other has two unlinked sex-determining regions (two poles). Basidiospores were randomly isolated by micromanipulation and germinated. The genotypes of the resulting yeast colonies were determined by PCR and growth assays on yeast-peptone-dextrose medium containing the drug G418. Four different genotypes were detected among the 48 progeny isolated, as expected based on Mendelian segregation: 12 were “α” strains, 10 were a strains, 9 were sxi1α strains, and the remaining 17 were a strains carrying the SXI1α gene at the ura5 locus (Table 2). The “α” and the a strains are parental types and exhibited the same mating properties as their parents when crossed to the wild-type a and α reference strains. The “α” cells are only compatible with a cells and not α cells, while the a cells only mate with α cells.
FIG. 2.
Diagrams depict chromosome segregation patterns in the tripolar and tetrapolar crosses. The SXI1α and SXI2a homeodomain genes were relocated at the URA5 genomic locus on another chromosome in the “α” and “a” strains. “STE3α+MFα” and “STE3a+MFa” represent the pheromone and pheromone receptor loci.
TABLE 2.
Segregation analysis of progeny derived from an “α” (JF306) × a (KN99a) tripolar cross
| Genotype | Phenotype | No. |
|---|---|---|
| “α” | Fertile | 12 |
| a | Fertile | 10 |
| sxi1α | Sterile | 9 |
| aSXI1α | Sterile | 17 |
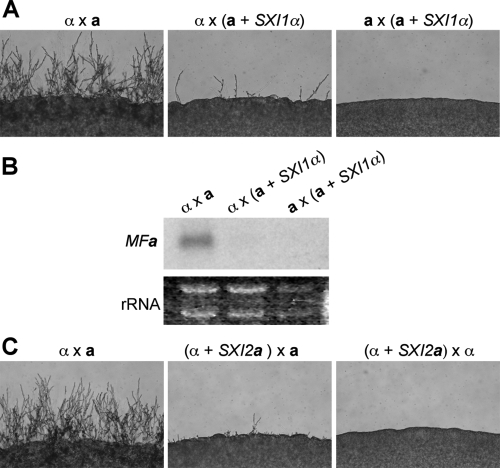
On the other hand, the progeny that are recombinant showed a sterile phenotype. As expected, the strains that carry the sxi1α deletion were sterile when crossed to a or α cells. Progeny with both the wild-type MATa allele and the SXI1α gene at the ura5 locus exhibited a strong mating defect when cocultured with α cells and were sterile when cocultured with a cells. Compared to wild-type a cells, cells with an additional SXI1α gene unlinked to MAT produced much less hyphae when crossed to a wild-type α strain for 48 h (Fig. 3A). This result suggested that the presence of a functional Sxi1α/Sxi2a heterodimer in an a cell inhibited the mating response, which is analogous to the action of the a1/α2 repressor in Saccharomyces cerevisiae (10). A similar finding was also observed in U. maydis: cells harboring compatible homeodomain proteins are attenuated in fusion (20). During a normal mating process, the Sxi1α/Sxi2a heterodimer forms after cell-cell fusion and orchestrates gene expression to maintain the dikaryotic state. Although no systemic studies have been conducted to identify the downstream targets of the Sxi1α/Sxi2a heterodimer, the pheromone genes MFa and MFα are known to be repressed by the Sxi1α/Sxi2a heterodimer; cells lacking either Sxi1α or Sxi2a exhibit elevated pheromone gene expression during mating (12, 13). Therefore, we hypothesized that the reduced fertility seen in strains that express both homeodomain proteins was likely due to a decreased expression of the pheromone genes.
FIG. 3.
Strains that express both Sxi1α and Sxi2a have a mating defect. (A) The wild-type a and a SXI1α strains were crossed to the serotype D a and a tester strains JEC20 and JEC21 on MS medium, incubated for 2 days in the dark at 25°C, and photographed at ×100 magnification. (B) Northern analysis demonstrated that the expression of the pheromone gene is not induced during mating of the a SXI1α strain. a and a SXI1α strains were crossed to serotype A wild-type a and α strains H99 and KN99a. RNA was extracted from the mating mixtures and probed with a PCR fragment of the MFa gene. (C) The wild-type α and α SXI2a strains were crossed to JEC20 and JEC21 on MS medium, incubated for 2 days in the dark at 25°C, and photographed at ×100 magnification.
To test this hypothesis, RNA was extracted from cells cultured under mating conditions and Northern analysis was conducted. In the wild-type a × α cross, the expression of the MFa pheromone genes was significantly induced; in contrast, no such induction was detected if the a partner carried an additional SXI1α gene (Fig. 3B). The decreased expression of pheromone genes correlated well with the reduced mating observed, demonstrating that this is part of the reason, if not the sole reason, why a cells carrying an additional SXI1α gene exhibit reduced fertility. In the case of crossing a cells to a cells with SXI1α, no mating response was observed. This was expected because no pheromone communication between these two cell types could be established even though Sxi1α is present in one of the partners (Fig. 3A).
Crossing an “a” strain to a wild-type α strain—the reciprocal tripolar cross.
To determine whether mating type influences the outcome of a tripolar mating, we also conducted a reciprocal “a” × α cross (Fig. 2). Progeny were isolated by spore dissection and analyzed. Independent segregation of the MAT locus and the transgenic SXI2a gene at the ura5 locus were again observed. Among the 37 progeny isolated, 7 were α strains, 6 were “a” strains, 10 were sxi2a strains, and the remaining 14 were α strains carrying the SXI2a gene at the ura5 locus (Table 3). Similar to the findings seen in the “α” × a cross, three out of the four types of the progeny were fertile (α and “a”) or sterile (sxi2a), as predicted. The last type of isolate, which were cells bearing the ectopic SXI2a gene, also exhibited mating defects compared to wild-type α cells, indicating that the presence of a functional Sxi1α/Sxi2a heterodimer in α cells decreases the efficiency of mating (Fig. 3C). Therefore, we conclude that having both homeodomain proteins in one cell, whether a or α, inhibits mating.
TABLE 3.
Segregation analysis of progeny derived from an α (H99) × “a” (JF289) tripolar cross
| Genotype | Phenotype | No. |
|---|---|---|
| α | Fertile | 7 |
| “a” | Fertile | 6 |
| sxi2a | Sterile | 10 |
| α SXI2a | Sterile | 14 |
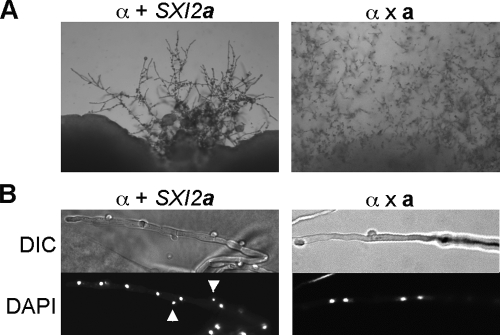
Cells with an active Sxi1α/Sxi2a heterodimer are self-filamentous.
Serotype A haploid cells with an active Sxi1α/Sxi2a heterodimer are mating impaired, and cultures become self-filamentous after prolonged incubation on mating medium (Fig. 4A). DAPI (4′,6′-diamidino-2-phenylindole) staining showed that these self-filamentous hyphae were mononucleate and had unfused clamp cells, which is in contrast to the dikaryons observed in a wild-type a × α cross (Fig. 4B). Furthermore, basidia with very few basidiospores were observed. These phenotypes are in contrast to those in previous studies, which demonstrated that expressing Sxi1α in a cells or expressing Sxi2a in α cells resulted in robust filamentation and spore formation in divergent serotype D strains (12, 13, 15). There are two factors that might contribute. First, there are intrinsic differences in the ability to undergo filamentous growth between the two serotypes: serotype D strains are known to exhibit more-robust hyphal growth, as evident from faster progression of sexual morphogenesis and their ability to undergo fruiting in the absence of a mating partner. Second, the promoter used to drive expression of the homeodomain transcription factors might also affect this phenotype. In this study, SXI1α or SXI2a are driven by their endogenous promoters, while in previous studies, they were expressed from the constitutive GPD1 gene promoter (12, 13). Based on our observations, we conclude that expressing both Sxi1α and Sxi2a can render both a and α cells self-filamentous in serotype A strains. Nonetheless, the presence of both homeodomain proteins appears insufficient to efficiently complete the sexual cycle.
FIG. 4.
Cells with an active Sxi1α/Sxi2a heterodimer are self-filamentous. (A) The left panel shows the self-filamentous phenotype of the α SXI2a strain on V8 medium (pH 5) after 3 weeks of incubation. The right panel shows mating filaments produced in a wild-type a × α cross after 3 weeks. (B) DAPI (4′,6′-diamidino-2-phenylindole) staining shows that the filaments produced by the α SXI2a strain are monokaryons (left), in contrast to the dikaryons (right) observed in a wild-type a × α cross. Arrowheads indicate two nuclei present in clamp cells in α SXI2a hyphae. DIC, differential interference contrast.
Crossing an “a” strain to an “α” strain—a tetrapolar cross.
After characterizing the basic properties of the two reciprocal tripolar crosses, we next examined the mating behavior of a tetrapolar cross (Fig. 2). “a” and “α” strains were cocultured on mating-inducing media, and basidiospores were randomly isolated by micromanipulation. Among the 48 progeny analyzed, 11 “a” and 14 “α” parental types were found. In addition, 22 recombinants equally divided into two classes (sxi1α SXI2a or sxi2a SXI1α) were isolated (Table 4). One progeny was identified to be an “a”/“α” diploid strain that contained genetic information from both parents.
TABLE 4.
Segregation analysis of progeny derived from an “α” (JF306) × “a” (JF289) tetrapolar crossa
| Genotype | Phenotype | No. |
|---|---|---|
| “α” | Fertile | 14 |
| “a” | Fertile | 11 |
| sxi1α SXI2a | Fertile when crossed with sxi2aSXI1α | 11 |
| sxi2aSXI1α | Fertile when crossed with sxi1α SXI2a | 11 |
One progeny isolated was an “a”/“α” diploid.
Mating assays were then conducted to determine the sex-identity of each type of the progeny. The “a”/“α” diploid strain is self-fertile, providing additional evidence that the ectopically expressed homeodomain proteins are functional. In addition, the “a” and “α” progeny unequivocally behaved as a and α cells in mating assays, as expected.
The sex-identity of the recombinant progeny is less predictable because these strains lack the endogenous homeodomain protein and instead express the homeodomain gene of the opposite sex. Although a key sex-determining gene has been exchanged, the remaining information at the MAT locus, including the pheromone and pheromone receptors, remains of the original mating type. To address this question, the sxi1α SXI2a and sxi2a SXI1α strains were crossed to wild-type a and α cells to determine their sexual identity.
The results of mating assays indicated that both the sxi1α SXI2a and the sxi2a SXI1α strains were sterile when crossed to either serotype D wild-type a or α tester strains (Fig. 5A). In the (sxi1α SXI2a) × α cross, no mating structures, including hyphae, basidia, or spores, were observed. When the same strain was crossed to wild-type a cells, scarce hyphae were randomly distributed on the edges of the mating colonies. Furthermore, the morphology of these hyphae was abnormal and readily distinguishable from hyphae produced by wild-type mating. No basidiospores were observed, indicating that hyphae were unable to complete sexual development. These results indicate that expressing SXI2a in the sxi1α mutant does not alter the sex-identity of the cells; in addition, it suggests that a compatible pheromone and pheromone receptor are still required for mating even when the two partners have compatible homeodomain proteins. Similar findings were observed in the (sxi2a SXI1α) × α or the (sxi2a SXI1α) × a crosses (Fig. 5A).
FIG. 5.
The sxi1α SXI2a and the sxi2a SXI1α strains are sterile when crossed to a or α cells but are interfertile. (A) Strains of the indicated genotype were crossed to JEC20 and JEC21 on MS medium and photographed after 5 days of incubation. Aberrant filaments but no basidiospores were observed. (B) Two strains of the indicated genotypes were cocultured on MS medium and incubated for 5 days in the dark at 25°C. Abundant hyphae and basidiospore chains were seen at the edge of the mating colony.
Surprisingly, we found that when a sxi1α SXI2a strain was crossed to the serotype A wild-type a or “a” cells, a few basidiospores were observed at the edges of mating colonies, although the amount was much less abundant than that generated from a wild-type cross (see Fig. S1 in the supplemental material). This result is unexpected because, in this cross, a functional heterodimer is absent in the cells. It is known that homeodomain proteins generally can be divided into two categories, HD1 and HD2, which have distinct functions: HD1 has a nuclear localization signal, whereas HD2 does not. The main function of HD1 (Sxi1α) is to translocate the homeodomain protein complex into the nucleus, where the HD2 (Sxi2a) protein can bind DNA to regulate gene expression, according to classic studies of the corresponding proteins in Coprinopsis cinerea conducted by Casselton and colleagues (32). This unusual mating behavior is likely attributable to modest overexpression of the SXI2a gene, as we found that three copies of the SXI2a transgene were inserted into the ura5 locus in strain JF289 (data not shown).
Although the sxi1α SXI2a and the sxi2a SXI1α strains are sterile when crossed to both a and α cells, we hypothesized that these two strains should be interfertile when crossed to each other. In this setting, the two mating partners have compatible pheromone and pheromone receptor systems, which trigger pheromone signaling that leads to mating responses, including cell-cell fusion. After the two cells fuse, Sxi1α and Sxi2a, although now encoded by the “a” and “α” nucleus, respectively, still have access to each other to form a functional heterodimer to govern the expression of downstream targets. To test this, the sxi1α SXI2a and the sxi2a SXI1α strains were cocultured on MS medium for two weeks. Abundant hyphae, basidia, and chains of basidiospores were observed, indicating that the two parental strains were mating compatible (Fig. 5B). Furthermore, viable progeny (basidiospores) were isolated from this cross. The genotypes and the number of the progeny isolated are listed in Table 5. Among the 19 progeny isolated, 9 were parental type and the other 10 were recombinant, demonstrating that the basidiospores generated from the (sxi1α SXI2a) × (sxi2a SXI1α) cross were viable and that independent chromosomal assortment occurred during meiosis. In summary, the “a” by “α” tetrapolar cross produces progeny with four different mating types, and any given progeny is only fertile with 1/4 of its siblings: the sine qua non of a tetrapolar mating system.
TABLE 5.
Segregation analysis of progeny derived from a (sxi1α SXI2a) × (sxi2a SXI1α) tetrapolar cross
| Genotype | Phenotype | No. |
|---|---|---|
| sxi1α SXI2a | Fertile when crossed with sxi2aSXI1α | 3 |
| sxi2aSXI1α | Fertile when crossed with sxi1α SXI2a | 6 |
| “α” | Fertile | 2 |
| “a” | Fertile | 8 |
Finally, χ2 tests were performed to assess the distribution of F1 progeny in all genetic crosses analyzed in this study (Tables 2 to 5). In each case, the chi-square test indicated that the goodness of fit was satisfactory for the expected 1:1:1:1 Mendelian segregation for two unlinked markers (with χ2 values of 3.17, 4.19, 0.57, and 4.79, respectively).
Exchanging Sxi1α and Sxi2a does not affect mitochondrial inheritance.
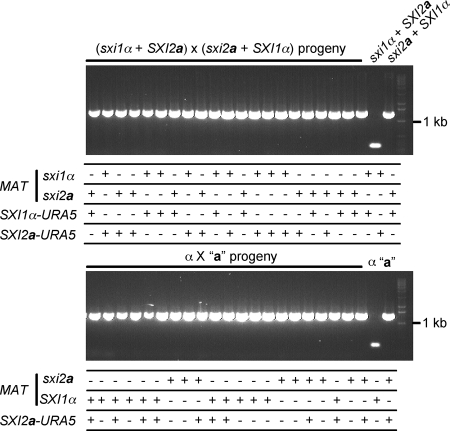
Uniparental mitochondrial inheritance is regulated by Sxi1α and Sxi2a in C. neoformans, but the underlying molecular mechanism remains unclear (36). Because the sxi1α SXI2a mutant strain can cross to the sxi2a SXI1α mutant and generate viable progeny, we addressed whether the uniparental mitochondrial inheritance would be affected in this cross, where the two parents carried the reciprocal Sxi1/2 HD protein of the opposite mating type. In particular, we hypothesized that α cells harboring the SXI2a gene might become the mitochondrial donor. To test this hypothesis, we followed the mitochondrial inheritance pattern in a sxi2a SXI1α (YPH227) × sxi1α SXI2a (YPH716) cross, in which the two strains have different mitochondrial DNA sequences that can be distinguished by different mitochondrial COX1 alleles (34). As shown in Fig. 6, all progeny isolated still inherited mitochondria from the sxi2a SXI1α parent, which contains the MATa allele but lacks Sxi2a. This demonstrates that exchanging the homeodomain proteins does not interfere with uniparental mitochondrial inheritance, suggesting that another a- or α-specific gene is responsible for uniparental mitochondrial inheritance.
FIG. 6.
Exchanging the homeodomain proteins does not alter uniparental mitochondrial inheritance. Progeny were isolated from the cross sxi1α SXI2a (YPH716, mitochondria type I) × sxi2a SXI1α (YPH227, mitochondria type II) or α (H99, mitochondria type I) × “a” (JF289 mitochondria type II) and typed for mitochondrial inheritance. All progeny inherited the type II mitochondria from the a parent. Nuclear markers were also scored by drug resistance, PCR and mating assays. “+” indicates the sxi1α or the sxi2a mutants, which are NEO or NAT resistant, and also the presence of the SXI1α -URA5 or SXI2a-URA5 transgenes, identified by PCR analysis. Mating assays were conducted to score the wild-type SXI1α allele in the MAT locus.
DISCUSSION
Within the mushroom fungi (homobasidiomycetes), it is estimated that 10% are homothallic (self-fertile), 25 to 35% are bipolar, and 55 to 65% are tetrapolar (30). The tetrapolar mating system is unique in basidiomycetes and has never been seen in the ascomycetes or zygomycetes. It is common to see mating system transitions in closely related basidiomycete species; several genera, including Coprinopsis and Ustilago, have been found to encompass both bipolar and tetrapolar species and transitions from bipolar to tetrapolar and from tetrapolar to bipolar appear to have occurred.
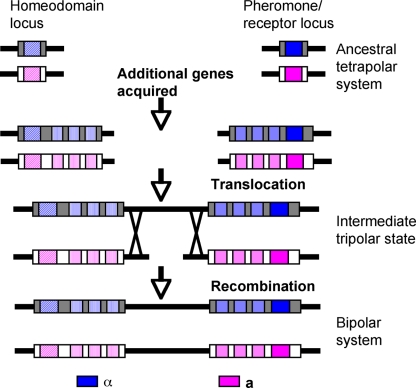
In this study, experimental evidence is provided to support the previously proposed model that the bipolar species C. neoformans descends from an ancestral tetrapolar fungus (7, 8). In this model, two ancestral unlinked MAT loci, one encoding the homeodomain proteins and the other encoding the pheromone and pheromone receptors, first expanded to form two larger gene clusters via the acquisition of sex-related genes. Next, the two loci were fused via a chromosomal translocation event in one mating type while the two loci remained unfused in the other mating type. During this transitional stage an unusual mating system operated among the population that we have termed the “tripolar” system to reflect the presence of linked and unlinked sex determinants in the mating partners. Finally, the tripolar system collapsed to a bipolar one via recombination (Fig. 7). By genetically engineering strains in which the homeodomain genes were physically unlinked to MAT, we demonstrated that a tetrapolar mating system can operate in C. neoformans. The engineered strains with two unlinked MAT loci were able to complete the sexual cycle and produce viable, fertile progeny. Thus, these results provide experimental evidence validating that the ancestor of C. neoformans might have harbored a tetrapolar mating system.
FIG. 7.
A simplified model for the evolution of MAT in C. neoformans. The ancestral tetrapolar MAT loci encode homeodomain protein genes and the pheromones and pheromone receptors. Major evolutionary steps included gene acquisition, translocation, and the collapse of the tripolar system to a bipolar one.
Inbreeding versus outcrossing lifestyle.
The two key considerations in comparing the impacts of different fungal mating systems on genetic exchange are the relative rates of inbreeding among progeny of defined genetic crosses and the relative rates of outcrossing between progeny of a genetic cross with isolates from the broader general population. From the viewpoint of population genetics, one major difference between a tetrapolar and a bipolar mating system is the degree of inbreeding allowed. In a bipolar cross, only two mating types are present among the progeny generated; therefore, there is a 50% chance that any two siblings are sexually compatible. In contrast, progeny with four different mating types are generated from a tetrapolar cross, and each mating type is only compatible with one of the other three kinds, restricting the chances of inbreeding to 25% (Table 6).
TABLE 6.
One-fourth of the progeny pairings derived from a tetrapolar mating are interfertile
| Genotype | Crossa
|
|||
|---|---|---|---|---|
| “α” | “a” | sxi1α SXI2a | sxi2aSXI1α | |
| “α” | − | + | − | − |
| “a” | + | − | −/+ | − |
| sxi1α SXI2a | − | −/+ | − | + |
| sxi2aSXI1α | − | − | + | − |
+, fertile; −, sterile; −/+, defective mating reaction with only a few basidiospore chains observed.
With respect to outcrossing, it is not the pattern of sexuality (bipolar versus tetrapolar) that governs the relative level but rather the number of alleles that are present at the MAT locus. Multiallelic mating systems, whether they are bipolar or tetrapolar, promote outbreeding. For representative species such as S. cerevisiae or C. neoformans, which are bipolar with two alleles or idiomorphs, the relative frequencies of inbreeding and outcrossing are both 50% in populations in which the mating type is balanced (Table 7). For representative tetrapolar multiallelic species, such as C. cinerea or Schizophyllum commune, the frequency of inbreeding is 25% and the frequency of outcrossing is ∼99% (Table 7). Of note, in the bipolar multiallelic species C. disseminatus, the frequency of inbreeding is 50% yet the frequency of outcrossing is ∼99%, and for the tetrapolar species U. maydis in which the pheromone/pheromone receptor locus has only two alleles, the frequency of inbreeding is restricted to 25% but the frequency of outcrossing is only 50% (Table 7). Thus, there are clear impacts on mating patterns via transitions between both bipolar and tetrapolar mating systems and via expansions and contractions in the numbers of alleles that reside at these sex-determining loci. As C. neoformans evolved from a tetrapolar multiallelic ancestor into a bipolar biallelic species, outcrossing potential would have been restricted and inbreeding potential increased, restricting genetic exchange in the population by promoting mating between more closely related isolates. Table 7 summarizes the inbreeding versus outcrossing frequencies for various fungal mating systems and species and some examples which remain to be discovered.
TABLE 7.
Chances of inbreeding and outcrossing in different fungal mating systems
| Mating system | Representative species | No. of alleles in the population (n)a | Chance of inbreeding (%) | Chance of outcrossing (%)b |
|---|---|---|---|---|
| Bipolar-biallelic | C. neoformans | 2 | 50 | 50 |
| Bipolar-multiallelic | C. disseminatus | ∼123 | 50 | 99.2 |
| Tetrapolar-bi/biallelic | Remains to be discovered | 2 (PRL), 2 (HPL) | 25 | 50 |
| Tetrapolar-bi/multiallelic | U. maydis | 2 (PRL), >25 (HPL) | 25 | 50 |
| Tetrapolar-multi/biallelic | Remains to be discovered | >2 (PRL), 2 (HPL) | 25 | 50 |
| Tetrapolar-multi/multiallelic | S. commune | ∼81 (PRL), ∼288 (HPL) | 25 | 98.8 |
| C. cinerea | >200 (PRL), >200 (HPL) | 25 | 99.5 |
PRL, pheromone/receptor locus; HPL, homeodomain protein locus.
n − 1/n. In tetrapolar mating systems, the smaller number between the pheromone/receptor alleles and homeodomain protein alleles determine the chance of outcrossing.
Examining the impact on inbreeding in our experiments revealed several interesting findings. In a tripolar cross, in which one parent has two unlinked MAT loci while the other has one contiguous functional MAT locus, only 1/8 (12.5%) of the progeny pairings are interfertile (Table 8). The tripolar state is hypothesized to be an intermediate state during the transition from tetrapolar to bipolar because the fusion of the two ancestral MAT loci via chromosomal translocation is likely to first occur with one mating type. Therefore, this transitional stage occurs in a population with two different MAT configurations: some have the fused, single MAT locus and the others have two unfused MAT loci. The fact that only 12.5% of the progeny pairings produced by a tripolar cross are mating compatible suggests that in this transition state, the organism shifted to a life style in which inbreeding was even more restricted. Moreover, in a tripolar cross, 50% of the progeny are not only unable to mate with their siblings but also are unable to mate with cells of any other mating type, as experimentally demonstrated. These progeny either lack a homeodomain protein or express an additional homeodomain protein of the opposite mating type, leading to the formation of a functional heterodimer prior to cell fusion (Table 8). In both cases, these cells have significantly reduced fertility and fail to engage in sexual reproduction. In contrast, all progeny generated from a bipolar or a tetrapolar cross are genetically fertile and are able to identify potential mating partners with a compatible MAT configuration.
TABLE 8.
One-eighth of the progeny pairings derived from a tripolar mating are interfertile
| Genotype | Crossa
|
|||
|---|---|---|---|---|
| “α” | a | sxi2a | α SXI2a | |
| “α” | − | + | − | − |
| a | + | − | − | − |
| sxi2a | − | − | − | − |
| α SXI2a | − | − | − | − |
+, fertile; −, sterile.
The hypothesis that the tripolar state is a transitional state under strong selection pressure is supported experimentally, as only 12.5% of the progeny pairings are interfertile while 50% of the progeny are sterile. This would be a considerable disadvantage and therefore might have directly or indirectly facilitated the transition from the tripolar system to a bipolar one. It is known that a significant portion of isolates of some species closely related to C. neoformans are sterile; thus, it is possible that some isolates of these species could exist in the transitional tripolar state. For example, it was recently reported that of 33 isolates of the new species Kwoniella mangroviensis identified in the Florida Everglades, 26 (∼80%) were sterile in genetic crosses (33). A further approach to explore and support this inference might be to examine naturally occurring sterile isolates to ascertain whether any might represent this hypothesized tripolar intermediate state. One caveat is that if the selective pressure is sufficient, this state may no longer be extant. While we favor the hypothesis that the tripolar state is unstable and deleterious, we acknowledge that it is conceivable that it might also prove to be beneficial under some environmental conditions or in some populations. For example, a chromosomal translocation could possibly contribute to enhance fitness, or linking the homeodomain and the pheromone/receptor loci may coordinate gene expression and also enhance fitness, which could be related or unrelated to mating.
Conversions between tetrapolar and bipolar mating systems.
In the fungal kingdom, tetrapolar mating systems have thus far been observed only in the Basidiomycota phylum and isolates in other phyla are bipolar. Therefore, it is thought that the bipolar mating system is ancestral and the tetrapolar mating system evolved within the basidiomycetes. However, basidiomycetes contain species with both tetrapolar and bipolar mating systems. The bipolar system appears to have repeatedly and independently evolved from tetrapolar mating systems, leading to a wide distribution of bipolar species among different clades of basidiomycetes. From an evolutionary perspective, it is thus thought that the tetrapolar system had a more-ancient origin within the phylum. As first proposed by Raper, simple genetic changes may lead to such transitions (30). In the first model, a chromosomal translocation that fuses the pheromone/receptor and homeodomain loci into a nonrecombining region could create bipolarity. The structures of the MAT loci in U. hordei, M. globosa, and C. neoformans all support this idea. Furthermore, the fact that these species are distantly related, and the differences in gene content within the MAT locus, provide evidence that the fusion of the two loci occurred independently during evolution. In the second model, mutations that lead to self-compatibility in either one of the loci may also enable cells to abandon the self-activating locus for self- and non-self-discrimination. Such mutants have occurred in nature and have been isolated in laboratories (6, 26). Finally in the third model, Raper proposed that the regulatory function of one locus could be gradually assumed by the other. However, no example has been found thus far of this last hypothesis.
Evolution of the tetrapolar mating system.
The tetrapolar mating system has only been observed in the basidiomycete lineage, and studies show that in other major lineages, sex is governed by a single bipolar MAT locus. How then did the tetrapolar mating system first evolve? We propose that in an ancestral bipolar basidiomycete, MAT encoded the homeodomain proteins, and the pheromone and pheromone receptor genes were unlinked to the homeodomain MAT locus. The pheromone and pheromone receptor genes evolved to be linked to each other and self-activating. Diversification of alleles led to at least two pairs of self-activated receptor-pheromone gene pairs. Finally, recombination occurred within the pheromone and pheromone receptor alleles and led to the separation of the compatible pheromone and receptor pair. This separation event then forced successful recognition as only possible between two non-self individuals, and thus, the pheromone/receptor was incorporated to function as one of two unlinked sex determinants (see Fig. S2 in the supplemental material).
In summary, this study provides experimental evidence to support and extend our previous model of the evolution of the MAT locus in C. neoformans. We showed that by unlinking the two major self/non-self sex determinants, C. neoformans can complete the sexual cycle with a tetrapolar mating system. Furthermore, the tripolar transitional state was also experimentally mimicked and demonstrated to possibly be detrimental to the population from an evolutionary perspective based on a further-restricted inbreeding potential (12.5%) and a large population (50%) of sterile progeny. The disadvantage of being tripolar might have accelerated the transition from a tripolar mating system to a bipolar one. Our studies also illustrate how a bipolar mating system could give rise to a tetrapolar mating system, which may mirror the events by which the ancestral tetrapolar mating system first arose in the basidiomycete phylum. Finally, our studies provide evidence that sex-determining regions expand by translocation and linkage, similar to models for early steps in sex chromosome evolution.
Supplementary Material
Acknowledgments
We thank Keisha Findley, Alex Idnurm, Soo Chan Lee, Banu Metin, and Tom Petes for critical reading of the manuscript and Anna Floyd for technical assistance. We acknowledge use of the C. neoformans serotype A sequencing project (Duke University/Broad Institute).
This work was supported by NIH R01 grant AI50113.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bakkeren, G., G. Jiang, R. L. Warren, Y. Butterfield, H. Shin, R. Chiu, R. Linning, J. Schein, N. Lee, G. Hu, D. M. Kupfer, Y. Tang, B. A. Roe, S. Jones, M. Marra, and J. W. Kronstad. 2006. Mating factor linkage and genome evolution in basidiomycetous pathogens of cereals. Fungal Genet. Biol. 43655-666. [DOI] [PubMed] [Google Scholar]
- 2.Bakkeren, G., J. Kämper, and J. Schirawski. 2008. Sex in smut fungi: structure, function and evolution of mating type complexes. Fungal Genet. Biol. doi: 10.1016/j.fgb.2008.1004.1005. [DOI] [PubMed]
- 3.Bakkeren, G., and J. W. Kronstad. 1994. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. USA 917085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capel, B. 1998. Sex in the 90s: SRY and the switch to the male pathway. Annu. Rev. Physiol. 60497-523. [DOI] [PubMed] [Google Scholar]
- 5.Charlesworth, B. 1991. The evolution of sex chromosomes. Science 2511030-1033. [DOI] [PubMed] [Google Scholar]
- 6.Fowler, T. J., M. F. Mitton, L. J. Vaillancourt, and C. A. Raper. 2001. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 1581491-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler, F. S. Dietrich, and J. Heitman. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, J. A., Y. P. Hsueh, K. M. Findley, and J. Heitman. 2007. Evolution of the mating-type locus: the basidiomycetes, p. 19-34. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 9.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 21036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herskowitz, I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342749-757. [DOI] [PubMed] [Google Scholar]
- 11.Hsueh, Y. P., A. Idnurm, and J. Heitman. 2006. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull, C. M., M.-J. Boily, and J. Heitman. 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hull, C. M., R. C. Davidson, and J. Heitman. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1alpha. Genes Dev. 163046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idnurm, A., Y. S. Bahn, K. Nielsen, X. Lin, J. A. Fraser, and J. Heitman. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3753-764. [DOI] [PubMed] [Google Scholar]
- 15.Idnurm, A., and J. Heitman. 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Idnurm, A., F. J. Walton, A. Floyd, and J. Heitman. 2008. Identification of the sex genes in an early diverged fungus. Nature 451193-196. [DOI] [PubMed] [Google Scholar]
- 17.James, T. Y., P. Srivilai, U. Kues, and R. Vilgalys. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 1721877-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31245-276. [DOI] [PubMed] [Google Scholar]
- 19.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laity, C., L. Giasson, R. Campbell, and J. Kronstad. 1995. Heterozygosity at the b mating-type locus attenuates fusion in Ustilago maydis. Curr. Genet. 27451-459. [DOI] [PubMed] [Google Scholar]
- 21.Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood, and J. W. Kronstad. 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 9615026-15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuba, C., I. Miura, and J. Merila. 2008. Disentangling genetic vs. environmental causes of sex determination in the common frog, Rana temporaria. BMC Genet. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 714831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno, S. 1967. Sex chromosomes and sex-linked genes. Springer-Verlag, New York, NY.
- 26.Olesnicky, N. S., A. J. Brown, S. J. Dowell, and L. A. Casselton. 1999. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 182756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhurst, S. M., and P. M. Meneely. 1994. Sex determination and dosage compensation: lessons from flies and worms. Science 264924-932. [DOI] [PubMed] [Google Scholar]
- 28.Perfect, J. R., N. Ketabchi, G. M. Cox, C. W. Ingram, and C. L. Beiser. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 313305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn, A. E., A. Georges, S. D. Sarre, F. Guarino, T. Ezaz, and J. A. Graves. 2007. Temperature sex reversal implies sex gene dosage in a reptile. Science 316411. [DOI] [PubMed] [Google Scholar]
- 30.Raper, J. 1966. Genetics of sexuality in higher fungi. The Ronald Press, New York, NY.
- 31.Smith, C. A., and A. H. Sinclair. 2004. Sex determination: insights from the chicken. Bioessays 26120-132. [DOI] [PubMed] [Google Scholar]
- 32.Spit, A., R. H. Hyland, E. J. Mellor, and L. A. Casselton. 1998. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc. Natl. Acad. Sci. USA 956228-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Statzell-Tallman, A., C. Belloch, and J. W. Fell. 2008. Kwoniella mangroviensis gen. nov., sp. nov. (Tremellales, Basidiomycota), a teleomorphic yeast from mangrove habitats in the Florida Everglades and Bahamas. FEMS Yeast Res. 8103-113. [DOI] [PubMed] [Google Scholar]
- 34.Toffaletti, D. L., K. Nielsen, F. Dietrich, J. Heitman, and J. R. Perfect. 2004. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr. Genet. 46193-204. [DOI] [PubMed] [Google Scholar]
- 35.Xu, J., C. W. Saunders, P. Hu, R. A. Grant, T. Boekhout, E. E. Kuramae, J. W. Kronstad, Y. M. Deangelis, N. L. Reeder, K. R. Johnstone, M. Leland, A. M. Fieno, W. M. Begley, Y. Sun, M. P. Lacey, T. Chaudhary, T. Keough, L. Chu, R. Sears, B. Yuan, and T. L. Dawson, Jr. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 10418730-18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, Z., C. M. Hull, S. Sun, J. Heitman, and J. Xu. 2007. The mating type-specific homeodomain genes SXI1alpha and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet. 51187-195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.