Abstract
Cell wall integrity is crucial for fungal growth, survival, and pathogenesis. Responses to environmental stresses are mediated by the highly conserved Pkc1 protein and its downstream components. In this study, we demonstrate that both oxidative and nitrosative stresses activate the PKC1 cell integrity pathway in wild-type cells, as measured by phosphorylation of Mpk1, the terminal protein in the PKC1 phosphorylation cascade. Furthermore, deletion of PKC1 shows that this gene is essential for defense against both oxidative and nitrosative stresses; however, other genes involved directly in the PKC1 pathway are dispensable for protection against these stresses. This suggests that Pkc1 may have multiple and alternative functions other than activating the mitogen-activated protein kinase cascade from a “top-down” approach. Deletion of PKC1 also causes osmotic instability, temperature sensitivity, severe sensitivity to cell wall-inhibiting agents, and alterations in capsule and melanin. Furthermore, the vital cell wall components chitin and its deacetylated form chitosan appear to be mislocalized in a pkc1Δ strain, although this mutant contains wild-type levels of both of these polymers. These data indicate that loss of Pkc1 has pleiotropic effects because it is central to many functions either dependent on or independent of PKC1 pathway activation. Notably, this is the first time that Pkc1 has been implicated in protection against nitrosative stress in any organism.
Cryptococcus neoformans is a pathogenic yeast that causes pulmonary infections and meningoencephalitis in immunocompromised persons, primarily those infected with human immunodeficiency virus (for a review, see reference 7). Individuals with AIDS are particularly vulnerable to opportunistic fungal infections and cryptococcosis. It has been estimated that approximately 10% of patients with AIDS in the United States and roughly 30% of human immunodeficiency virus-infected people in portions of Africa will contract cryptococcosis, and if left untreated, the disease is fatal. C. neoformans is ubiquitous and is found worldwide in the soil.
Fungal cells, including C. neoformans, are continuously exposed to a wide variety of environmental stresses in both their natural habitats and their resident hosts. These stresses include changes in temperature, osmolarity, pH, nutrients, and exposure to reactive oxygen and nitrogen molecules. They subsequently sense and react to changes in their environment through a complex network of signal transduction pathways, thus adapting to their surroundings and ensuring repair to cellular damage. Identification and characterization of components that comprise signaling pathways have been a major focus of research in the past two decades. Understanding how these various pathways function and interact is important, not only because of the desire to understand basic principles but also because deregulation of these pathways is a major cause of a variety of cancers.
Responses to environmental stresses are mediated by mitogen-activated protein kinase (MAPK) phosphorylation cascades in eukaryotic cells. The MAPKs themselves are highly conserved among eukaryotes and have been characterized extensively for Saccharomyces cerevisiae (reviewed by Heinisch et al. [21]) and to a lesser extent for C. neoformans (16, 29). The components of the PKC1-MAPK signaling pathway are vital to maintaining integrity of the cell. The core components directly involved in this kinase cascade (reviewed by Levin [31]) include (i) the small GTP binding protein Rho1p; (ii) Pck1p; (iii) the serine/threonine MAPK kinase kinase Bck1p; (iv) a pair of functionally redundant threonine/tyrosine MAPK kinases, Mkk1p/Mkk2p; and (v) the MAPK Slt2p. There is evidence that the pathway is activated primarily in a linear manner. Mutants with deletions of members of the pathway often have similar, but not necessarily identical, phenotypes. Deletion of PKC1 or BCK1 abolishes phosphorylation of Slt2p (43, 45), and the constitutively active alleles BCK1-20 and MKK1DD activate downstream members of the pathway (20, 30). However, there is mounting evidence that the inputs to the pathway may not always be strictly linear and that the outputs of the pathway may differ. Activation of this cell integrity pathway via phosphorylation of Slt2p, the terminal kinase in the PKC1 pathway of S. cerevisiae, has been shown to occur preferentially in response to different stresses at alternative points in the cell integrity pathway and not always in a “top-down” manner (20). It has also been shown that phosphorylation of Slt2p, but not components further downstream that are needed for other responses, is required for cell cycle checkpoint signaling (19).
Based on homology with S. cerevisiae and functional analysis (16, 29), the components directly involved in this kinase cascade in C. neoformans include Pkc1, Bck1, Mkk2, and Mpk1, whose S. cerevisiae homolog is Slt2p. Deleterious phenotypes are associated with deletions of genes in this pathway (16, 29). However, a direct comparison of a bck1Δ strain to an mkk2Δ strain showed that the mutations caused similar, but not identical, phenotypes, suggesting that there may be lateral input or differential output from some stresses (16). In C. neoformans, both the sensors that detect cell stresses and initiate activation of the MAPK pathway and the downstream transcription factors affected by phosphorylation are yet to be identified.
The PKC1 gene was originally identified in S. cerevisiae (reviewed by Levin [31]) and is a close homolog of the human protein kinase C gene. Deletion of this gene in S. cerevisiae was found to be lethal in some genetic backgrounds and partially suppressed by osmotic stabilization in others. The existence of more severe phenotypes for PKC1 deletion strains than for strains with deletions of other components in the PKC1 pathway suggests that Pkc1p serves as a central regulator with multiple outputs that affect other pathways and possibly connect different signaling pathways with each other (32). In C. neoformans, PKC1 has been demonstrated to be activated by diacylglycerol (DAG) (23). In addition, the DAG C1 binding domain of Pkc1 is required for the proper localization of laccase in the cryptococcal cell wall and for the formation of melanin (22). The studies presented here further characterize crucial functions of PKC1 in C. neoformans, predominantly demonstrating that activation of the cell integrity pathway occurs in response to both oxidative and nitrosative stresses. Furthermore, the generation of a PKC1 conditional deletion strain has allowed us to establish critical roles for this gene in protection against both oxidative and nitrosative stresses as well as functions in cell wall formation and defense against various external stresses, including alterations in temperature and osmolarity.
MATERIALS AND METHODS
Fungal strains and media.
KN99α and KN99a, well-characterized congenic strains of C. neoformans serotype A, were used as the wild-type strains (39). The PKC1 deletion strain was generated in KN99a (39); cku80Δ KN99α, which displays enhanced homologous recombination of transforming DNA (17); and the Mpk1 Flag-tagged strain (this study). H99 and the mpk1Δ strain in the serotype A background were kindly provided by Joseph Heitman at Duke University (28). The PKC1 pathway mutants and reconstituted bck1Δ, bck1Δ::BCK1, mkk2Δ, and mkk2Δ + MKK2 strains were generated in the H99 background (16). The cap59Δ and lac1Δ lac2Δ controls were generated in the KN99 background (2). All strains were grown on rich medium (YPD [1% yeast extract, 2% Bacto peptone, and 2% dextrose]). Solid media contained 2% Bacto agar. All PKC1 deletion strains were grown on YPD supplemented with 1 M sorbitol. Selective YPD media contained 200 units/ml hygromycin (Calbiochem, San Diego), 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany), or 200 μg/ml Geneticin (Gibco). For induction of capsule formation, strains were grown on DME (13.4 g/liter Dulbecco's modified Eagle's medium [Sigma, D-5648], 25 mM MOPS [morpholinepropanesulfonic acid] [pH 7.0], 1.8% agar) supplemented with 1 M sorbitol. For the determination of melanin production, strains were grown on l-3,4-dihydroxyphenylalanine (l-DOPA) medium (13 mM glycine, 15 mM glucose, 29.4 mM KH2PO4, 10 mM MgSO4·7H2O, 3 μM thiamine, 5 μM d-biotin, 2% agar, and 1 mM L-DOPA, pH 5.5) supplemented with 1 M sorbitol. For nitrosative stress sensitivity determination, serial dilutions of cells were plated on YNB medium (pH 4.0) (6.7 g/liter Difco yeast nitrogen base without amino acids, 2% glucose, 2% agar, 25-ml/liter 1 M succinate [made pH 4.0 with HCl]) supplemented with 1 M sorbitol. For oxidative stress sensitivity determination, serial dilutions of cells were plated on YNB medium (pH 4.0) supplemented with 1 M sorbitol and also on YNB medium (pH 7.0) (6.7 g/liter Difco yeast nitrogen base without amino acids, 2% glucose, 2% agar, 50-ml/liter 1 M MOPS [made pH 7.0 with NaOH]) supplemented with 1 M sorbitol. All strains were maintained at 30°C unless otherwise indicated.
Generation of Mpk1 fusion proteins.
Four independent strains were created in both the KN99a and KN99α backgrounds, in which the Mpk1 protein was tagged at the carboxy terminus with either Flag (GATTACAAGGATGACGACGATAAG)- or V5 (GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG)-encoding sequences using overlap PCR. A DNA fragment containing the 3′ end of MPK1 genomic sequence followed by the Flag tag immediately preceding the stop codon TAG (native to Mpk1) and the 5′ end of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) terminator was amplified using primers Mpk1-tag 14 and Mpk1-Flag tag RC, yielding a product 609 bp in length. A 4,310-bp fragment comprised of the GAPDH terminator joined to the G418 resistance cassette and a portion of the MPK1 genomic sequence immediately downstream of the MPK1 stop codon was amplified from another construct using primers Flag-tag/GAPDH and Mpk1-tag 11. Nested primers Mpk1-tag 8 and Mpk1-tag 13 were used to generate the full-length Flag-tagged Mpk1 (containing the GAPDH terminator joined to the G418 resistance cassette). The presence of the Flag tag was determined by PCR using one primer that bound specifically to the tag sequence (Flag-RC) and another primer that bound upstream of the region of intended homologous recombination within the MPK1 gene (Mpk1-tag 14). Homologous integration at the MPK1 locus was further verified by a 5′ PCR screen (using primers Mpk1-tag 14 and Cn-actin7, which binds within the G418 resistance cassette), a 3′ PCR screen (using primers Mpk1-tag 11 and NMT-3000, which binds within the G418 resistance cassette), and a full-length PCR screen (using primers Mpk1-tag 11 and Mpk1-tag 14, both of which lie outside the region of homologous recombination). An identical strategy was utilized to create the V5-tagged construct, except that primer Mpk1-V5 tag RC was used in place of Mpk1-Flag tag RC, V5-tag/GAPDH was used in place of Flag-tag/GAPDH, and V5-RC was used in place of Flag-RC for screening. In order to rule out ectopic integration of the Flag-tagged construct, Southern blot analysis was performed. Genomic DNA from isolates found to be correct by PCR screening was digested with either BglII or XhoI, and blots were probed with an 801-bp fragment of the G418 gene to confirm the presence of only one integrated construct. The Mpk1 Flag-tagged strain in the KN99α background was subsequently used for studies presented in this paper.
Western blot analysis.
Cultures were grown overnight in YNB (pH 4.0) (see “Fungal strains and media” above) or in YPD containing 1 M sorbitol for the pkc1Δ strain at 25°C with shaking and then diluted to an optical density at 650 nm (OD650) of 0.2 in 100 ml of the same medium. The cells were grown with shaking for an additional 2.5 h and then induced with 10 μg/ml calcofluor white (Sigma, F-3543; stock solution of 15 mg/ml made in H2O and titrated into solution with NaOH) for 2 h. The cells were harvested at 1,800 rpm and 10°C for 10 min and then washed once in ice-cold sterile milliQ water. The pellets were frozen on dry ice and subsequently at −80°C. The cells were thawed on ice, and 0.2 ml of 1× lysis buffer (containing 10 mM Tris-HCl [pH 7.5], 0.4 mM MgCl2, 2% glycerol, 1× protease inhibitor cocktail [Roche, no. 11836170001], and 10 μl/ml each phosphatase inhibitor cocktail 1 and cocktail 2 [Sigma, P-2850 and P-5726, respectively]). The cells were beaten for 5.5 min at 4°C with approximately one-third to one-half the supernatant volume of 0.5-mm zirconia/silica beads (BioSpec Products, Inc., no. 11079105z) on a Disruptor Genie (Scientific Industries). Lysates were spun down in a microcentrifuge at 14,000 rpm and 4°C for 20 min, and the supernatant was collected. Protein concentrations were determined using Quick Start Bradford dye reagent (Bio-Rad, no. 500-0205). For each sample, 50 μg of total protein was loaded on 8% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and proteins were transferred to nitrocellulose. For phosphorylated Mpk1 determination, a 1:2,500 dilution of phospho-p44/42 MAPK (Thr202/Tyr204) rabbit polyclonal antibody (Cell Signaling Technology, no. 9101) was used. Total Mpk1 protein independent of phosphorylation was detected using a 1:5,000 dilution of DYKDDDDK (Flag) tag rabbit polyclonal antibody (Cell Signaling Technology, no. 2368). All primary antibodies were diluted in TBST (25 mM Tris-HCl [pH 8.1], 145 mM NaCl, 0.1% Tween 20) plus 5% bovine serum albumin and allowed to bind overnight at 4°C. Secondary antibodies used were goat anti-rabbit immunoglobulin G peroxidase conjugated (Sigma, A-6154) and were diluted 1:1,000 in TBST. Proteins were detected using the ECL Plus Western blotting detection system (GE Healthcare, no. RPN2132).
Activation of the PKC1 pathway by oxidative and nitrosative stresses and Western blot analysis.
The Mpk1 Flag-tagged strain (KN99α background) was grown overnight in YNB (pH 4.0, with succinate) at 25°C, diluted the next morning to an OD650 of 0.2, allowed to grow for 2.5 h, and then either uninduced for 60 min (midpoint of aliquots) or treated with 0.5 mM diamide for 30, 60, 90, and 120 min or with 1 mM H2O2 or 1 mM NaNO2 for 30, 60, 90, 120, and 150 min. The bck1Δ, mkk2Δ, mpk1Δ, and H99 control strains were grown and induced in YNB (pH 4.0) similarly. The pkc1Δ strain (and Mpk1-Flag control strain for that experiment) was grown in YPD containing 1 M sorbitol, diluted to an OD650 of 0.05, allowed to grow for 3 h, and then induced with either 1 mM NaNO2 for 30 min or 1 mM H2O2 for 60 min. Cells were harvested and then lysed as described above, and 50 μg of total protein for each sample was loaded and analyzed by SDS-polyacrylamide gel electrophoresis as before (see “Western blot analysis” above). Nitrocellulose membranes were probed initially with a 1:2,500 dilution of phospho-p44/42 MAPK (Thr202/Tyr204) rabbit polyclonal antibody (Cell Signaling Technology, no. 9101) and then stripped and reprobed with a 1:5,000 dilution of DYKDDDDK (Flag) Tag rabbit polyclonal antibody (Cell Signaling Technology, no. 2368). Secondary antibodies used were goat anti-rabbit immunoglobulin G peroxidase conjugated (Sigma, A-6154) and were diluted 1:1,000 in TBST. Proteins were detected using the ECL Plus Western blotting detection system (GE Healthcare, no. RPN2132).
Drug resistance markers.
Drug resistance markers for the PKC1 deletion constructs were amplified using primers 3-PKC1 and 6-PKC1. A 1,927-bp region of the plasmid pHL001 was amplified to create a nourseothricin resistance (NAT) cassette (33), which was used in the construction of pkc1Δ. Descriptions of all primers used in this work can be found in Table S1 in the supplemental material.
Generation of pkc1Δ.
The pkc1Δ construct was made by fusing together the 5′ region of PKC1 including 896 bp of genomic sequence (made by PCR using primers 7-PKC1 and 4-PKC1), the 1,884-bp NAT cassette (made from the plasmid pHL001-STM212 utilizing primers 3-PKC1 and 6-PKC1) (33, 38), and the 3′ region of PKC1 including 908 bp of sequence (made by PCR using primers 5-PKC1 and 8-PKC1). The PKC1 deletion construct consisting of 3,401 bp was generated utilizing overlap PCR and resulted in the fusion of the 5′ region of PKC1, the NAT resistance cassette, and the 3′ region of PKC1 in a single PCR with primers 1-PKC1 and 2-PKC1 (11). This fusion resulted in the deletion of 3,721 bp of the PKC1 gene. This linear construct was biolistically transformed into C. neoformans KN99a (39), the KN99α cku80Δ strain (17), and the Mpk1-Flag strain (KN99α background, this study).
Transformation of C. neoformans.
C. neoformans strains KN99α, KN99a, KN99α cku80Δ, and Mpk1-Flag were transformed using biolistic techniques (25, 42). Cells were grown to late log phase in YPD, concentrated, and plated onto YPD agar supplemented with 1 M sorbitol for transformation. The cells were bombarded with 0.6-μm gold particles (Bio-Rad) coated with DNA according to the manufacturer's recommendations and allowed to sit for 4 to 6 h on nonselective medium following bombardment. They were then recovered by adding 0.8 ml phosphate-buffered saline (PBS) to the plate and scraping the cells into suspension with a spreader. The cells were transferred to selective YPD plates supplemented with 1 M sorbitol containing 100 μg/ml nourseothricin and incubated at room temperature. Transformants were observed within 3 to 5 days. Subsequent experiments were performed with pkc1Δ isolates generated both in the KN99a background and in the Mpk1-Flag background.
Complementation of pkc1Δ.
The pkc1Δ strain in the KN99a background was transformed by electroporation (14). An overnight culture of the pkc1Δ strain was grown in 5 ml YPD containing 1 M sorbitol at 30°C with shaking. The following morning, 2 ml of overnight culture were diluted into 50 ml of fresh YPD containing 1 M sorbitol with shaking at 30°C for 6 h. The cells were spun at 1,800 × g for 10 min, washed in 50 ml electroporation buffer (0.27 M sucrose, 10 mM Tris-HCl [pH 7.5], 1 mM MgCl2) containing 1 mM of freshly added dithiothreitol at room temperature, and spun again at 2,200 × g for 5 min. The cells were resuspended in 150 μl of electroporation buffer without dithiothreitol. Either 0.5 or 1 μg of a linear 5.2-kb construct containing the wild-type PKC1 gene generated by PCR using primers 1-PKC1 and 2-PKC1 from KN99α was added to 75 μl of pkc1Δ cells and mixed gently by pipetting. The mixture was transferred to a 0.2-cm-gap cuvette (Bio-Rad) and electroporated at settings of 470 V, 50 μF, and 1,000 Ω with a time constant of ∼25 ms. One milliliter of YPD containing 1 M sorbitol was added immediately, transferred to a sterile tube, and incubated at room temperature overnight. The following morning, the cells were spun at 3,000 × g for 5 min., and all but 100 μl of the supernatant was removed. The cells were plated on YPD containing no sorbitol and incubated at 37°C for 3 days as a double selection for transformants.
Analysis of transformants.
All transformants were allowed to grow for 3 to 5 days on selective medium (either on plates containing nourseothricin at 30°C or on YPD without sorbitol at 37°C for PKC1 complementation). Colonies were then passaged five times on nonselective medium to remove unstable transformants (either on YPD plates containing no nourseothricin or on YPD plus 1 M sorbitol at 30°C for PKC1 complementation) and then replated on selective medium to determine stable antibiotic resistance or ability to grow without osmotic stabilization at 37°C in the case of PKC1 complementation. Only transformants that grew well under selective pressure were counted as stable transformants and considered for further study.
PCR screening and Southern blot analysis of transformants.
Only pkc1Δ isolates that were positive for the NAT resistance marker after passage on nonselective medium were chosen for PCR screening (see Fig. S1 in the supplemental material). For pkc1Δ as well as pkc1Δ::PKC1constructs, a three-primer PCR screen was used to prove homologous integration on both the 5′ (primers 7-PKC1, 9-PKC1, and NAT-1) and 3′ (primers 8-PKC1, 10-PKC1, and Cn-actin7) ends of all of the constructs (described by Nelson et al. (37) and Gerik et al. (16)). A full-length screen was also used to confirm the presence of the entire construct (either the PKC1 deletion cassette or the wild-type gene) using primers 7-PKC1 and 8-PKC1. In order to rule out ectopic integration of the pkc1Δ deletion cassette, Southern blot analysis was performed. Genomic DNAs from deletion isolates correct by PCR screening were digested with either NcoI or XhoI, and blots were probed with a 556-bp fragment of NAT to confirm the presence of only one integrated construct (see Fig. S1B in the supplemental material). For complementation of the mutant, only isolates that were able to grow on YPD without sorbitol at 37°C after passage on YPD plus 1 M sorbitol at room temperature were chosen for PCR screening. All isolates were screened for both the presence of the wild-type gene and the original deletion cassette, with wild-type and parental deletion strains serving as controls. Only complemented isolates that had the wild-type PKC1 gene integrated back into the original locus were used in subsequent experiments (see Fig. S1 in the supplemental material).
Osmotic and temperature sensitivity of pkc1Δ.
Deletion strains were streaked onto YPD plates with and without 1 M sorbitol and grown at 25°C, 30°C, and 37°C for 5 days. Alternatively, strains were grown in 2 ml of YPD plus 1 M sorbitol overnight at 30°C with shaking. The next morning, they were diluted to an OD650 of 1.0 in PBS plus 1 M sorbitol. Tenfold serial dilutions were made in PBS plus 1 M sorbitol, and 5 μl of these was plated on YPD plates containing 1 M sorbitol and incubated for up to 7 days at 30°C, 37°C, and 39°C.
Analysis of nitrosative and oxidative stress responses.
KN99a, pkc1Δ, and pkc1Δ::PKC1 strains were grown overnight at 30°C in YPD containing 1 M sorbitol. The following day, cells were counted using a hemocytometer; 1 × 107 cells were diluted into 200 μl of PBS containing 1 M sorbitol in a 96-well plate, and 10-fold serial dilutions were made in the same buffer. Five microliters of each dilution was plated on both YPD (pH 4.0 and pH 7.0) and YNB (pH 4.0 and pH 7.0) containing 1 mM NaNO2 (pH 4.0 only), 0.5 mM or 1 mM H2O2, or 1 mM diamide. Plates were incubated at 30°C for up to 7 days. The PKC1 pathway mutants and reconstituted bck1Δ, bck1Δ::BCK1, mkk2Δ, mkk2Δ + MKK2 (16), and mpk1Δ (29) strains and the parent strain H99 were grown overnight at 30°C in YPD. The following day, each strain was diluted to an OD650 of 1.0 in PBS, and 10-fold serial dilutions were made in the same buffer. Five microliters of each dilution was plated on YNB (pH 4.0) and YNB (pH 4.0) containing1 mM NaNO2, 0.5 mM or 1 mM H2O2, or 1 mM diamide. The same strains were also plated on YNB (pH 4.0) plates containing 1 M sorbitol with and without nitrosative and oxidative stressors for comparison to the pkc1Δ mutant. All plates were made fresh the same day. Plates were incubated at 30°C for up to 3 days.
Cell wall inhibitor assays.
Strains were grown in 2 ml of YPD plus 1 M sorbitol overnight at 30°C with shaking. The next morning, they were diluted to an OD650 of 1.0 in PBS plus 1 M sorbitol. Tenfold serial dilutions were made in PBS plus 1 M sorbitol and 5 μl of these was plated on YPD plates containing 1 M sorbitol without and with 0.5, 1.0, and 1.5 mg/ml calcofluor white (Fluorescent Brightner 28; Sigma, F-3543); 0.2, 0.5, and 1.0 mg/ml caffeine (Sigma, C-0750); 0.01, 0.03, and 0.06% SDS (AnalaR Biochemical, BDH Chemicals Ltd., Poole, England); 0.5% Congo red (Sigma, C-6767); and 1.5 M NaCl. Plates were incubated for up to 7 days at 30°C.
Capsule analysis.
Cells grown on YPD plates containing 1 M sorbitol for 2 days were plated on DME plates (13.4 g/liter Dulbecco's modified Eagle's medium [Sigma, D-5648], 25 mM MOPS [pH 7.0], 1.8% agar) containing 1 M sorbitol and incubated vertically in the presence of 5% CO2 for 5 days at 30°C. Individual isolates of the pkc1Δ, wild-type, pkc1Δ::PKC1 complemented, and capsule-deficient cap59Δ strains were resuspended in a 1:4 India ink-H2O solution and observed on day 5 at a magnification of ×1,000 (Olympus AHBT3 microscope).
Cryptocrit analysis.
The KN99a, pkc1Δ, pkc1Δ::PKC1, and cap59Δ strains were grown on DME agar plates containing 1 M sorbitol the presence of 5% CO2 for 5 days at 30°C to induce capsule formation. Cells were scraped off of the plates into microcentrifuge tubes containing 1 ml PBS plus 1 M sorbitol, spun for 1 minute in a microcentrifuge at 5,000 rpm, resuspended in 0.5 ml PBS plus 1 M sorbitol, counted with a hemocytometer, and then diluted to 109 cells/ml in PBS plus 1 M sorbitol containing 10% formalin. The formalin killed the Cryptococcus without altering the capsule (18). Forty microliters of each strain, containing 4 ×107 cells, was used to measure the packed cell volume, or cryptocrit, as previously described by Alspaugh et al. (1). Briefly, the cells were added to heparinized Microhematocrit capillary tubes (Fisher, 02-668-66) and spun for 2 minutes in a Microhematocrit centrifuge. The cryptocrit was measured as the length of the packed-cell phase divided by the length of the total suspension in the capillary tube. Each strain was examined in triplicate and the values averaged to obtain a percentage. The average diameter of cells excluding the capsule for each strain was determined by measuring 50 cells each under a magnification of ×1,000, and pellet volumes for each strain were also determined and averaged.
Melanin formation.
Freshly growing strains from 2-day-old YPD plates containing 1 M sorbitol were streaked onto l-DOPA plates (8) containing 1 M sorbitol and grown at 30°C for 2 to 7 days. Cells were also grown overnight in YPD containing 1 M sorbitol and diluted to an OD650 of 1.0. Five 10-fold serial dilutions were made in PBS plus 1 M sorbitol, and 5 μl of each dilution was plated onto l-DOPA plates containing 1 M sorbitol and incubated at 30°C for 2 to 10 days.
Microscopy.
All strains were grown for 3 days in YPD medium containing 1 M sorbitol at 25°C. Five hundred microliters was centrifuged at 3,000 rpm for 3 min in a microcentrifuge, washed once with 0.5 ml McIlvaine's buffer (pH 6.0), and then resuspended in 0.5 ml of the same buffer (34). For eosin Y staining, 30 μl of a 5-mg/ml eosin Y (Sigma, no. 119830) stock solution was added to each tube and shaken gently for 30 to 40 min at room temperature. For pontamine staining, the cells were washed in 1× PBS and then resuspended in 180 μl PBS; 20 μl of a 1:1,000 dilution of pontamine in PBS solution (Fast Scarlet 4B; Bayer Corp.) (24) was added to each tube and shaken gently for 30 to 40 min at room temperature. The eosin Y-stained cells were washed one time in 0.5 ml McIlvaine's buffer (pH 6.0), and the pontamine-stained cells were washed with 0.5 ml PBS, and then pelleted at 3,000 rpm for 2 min. The cells were resuspended in 0.3 ml McIlvaine's buffer (pH 6.0) (for eosin Y) or in 0.3 ml PBS (for pontamine), and samples were viewed at a magnification of ×1,000 by bright-field and fluorescence illumination (excitation wavelengths were set at 548 nm for pontamine and 488 nm for eosin Y). To determine viability of eosin Y and pontamine stained cells, cells were also stained simultaneously with eosin Y or pontamine and a 1:1 dilution of trypan blue (Sigma [no. T-8154], 0.4% solution) for the last 10 min of staining, washed, and viewed microscopically as described above. For confocal microscopy, cells were stained as described above and coverslipped with 0.17-mm-thick glass coverslips. Imaging was carried out using an Olympus FV-1000 MPE confocal microscope. A plan-apochromat 60× NA 1.42 oil immersion objective was used to collect all images. Excitation wavelengths were set at 488 nm and 548 nm for eosin Y and pontamine imaging, respectively, and images were collected with 10 μs of dwell time and resolution settings of 1,024 by 1,024.
Determination of chitin and chitosan contents in pkc1Δ cells.
Cells were cultured in liquid YPD supplemented with 1 M sorbitol, using a starting OD650 of 0.05 and shaking at 27°C for 3 to 4 days. To measure the chitin and chitosan contents as well as the dry weight of cells, samples of each culture were divided into three 0.5- to 1.0-ml aliquots in tared 2-ml microcentrifuge tubes. Cells were collected by centrifugation at 14,000 rpm for 2 min, the medium was removed, and the tubes were spun again at 14,000 rpm for 1 min to remove residual medium. The wet weight of each cell pellet was measured. Cells of one aliquot were suspended with 1 ml of YPD and the centrifugation repeated to remove residual sorbitol; the cell pellet was dried at 37°C for 3 days, weighed, and then discarded. Dry weights of the other two aliquots were calculated from the dry weight/wet weight ratio of this aliquot of cells.
To measure chitin plus chitosan, one of the remaining aliquots of cells was treated with acetic anhydride to convert chitosan to chitin, and the other was left untreated to measure chitin only. Both were alkaline extracted and then treated with chitinase to completely digest chitin to N-acetylglucosamine, which was then measured using the Morgan-Elson assay (6). The difference between the two measurements estimated the amount of chitosan. The procedures followed those described in detail by Baker et al. (2).
RESULTS
The PKC1 cell integrity pathway is activated in response to both oxidative and nitrosative stresses.
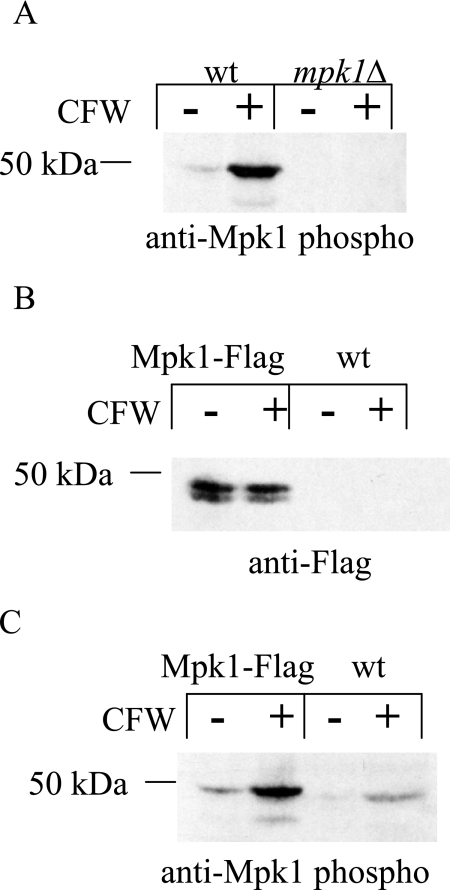
C. neoformans presumably encounters both reactive oxygen and nitrogen intermediates in its primary ecological niche as well as in alveolar macrophages of mammalian hosts (reviewed by Brown et al. [5]). In S. cerevisiae, the PKC1 cell integrity pathway is activated upon exposure to oxidative stress, and it has been shown that PKC1 and its upstream components are essential mediators of this effect (45). Moreover, it was demonstrated that Slt2p, the yeast homolog of Mpk1, was phosphorylated in response to induction with both diamide and peroxide but not as a result of nitrosative stress (45). Therefore, we hypothesized that activation of the PKC1 pathway was crucial for protection against reactive oxygen species and wanted to determine whether it was also essential for protection against nitrosative stress in C. neoformans wild-type cells. The terminal protein in the PKC1 pathway, Mpk1, was activated in response to the cell wall-perturbing agent calcofluor white (Fig. 1A) (29). Mpk1 became phosphorylated, i.e., was activated, and the commercially available phospho-p44/p42 antibody recognized the dually phosphorylated form of this 50-kDa protein, thereby providing a means for measuring PKC1 pathway activation. Furthermore, the 50-kDa band was absent in a strain lacking MPK1 under both induced and uninduced conditions, showing that the phospho-antibody specifically recognized Mpk1 (Fig. 1A).
FIG. 1.
FLAG-tagged Mpk1 allows for the detection of total Mpk1, the terminal kinase in the PKC1 pathway, independent of phosphorylation. (A) Western blot analysis was done using the anti-Mpk1 phospho-antibody to probe a blot containing total protein from wild-type or mpk1Δ lysates, which were made from cells that were either uninduced or induced with calcofluor white (CFW). The amount of the 50-kDa band increased in the wild-type strain in response to CFW, and the absence of the band in the mpk1Δ lysate indicated that the band in the wild-type lysate was Mpk1. (B) The gene construct encoding the Mpk1 protein fused to the Flag epitope at the carboxy terminus replaced the wild-type MPK1 locus in KN99α. The panel shows Western blot analysis using an antibody against the Flag tag and lysates from the Mpk1-Flag tagged strain and the wild-type strain either induced or uninduced with CFW. Bands of the appropriate size were detected in the Mpk1-Flag strain but not the wild-type strain, suggesting that the protein detected was the Mpk1-Flag fusion. (C) The same blot from panel B probed with an antibody specific for the phosphorylated form of Mpk1. Phosphorylation of both the fusion protein and the wild-type protein was induced by CFW.
In this study, we demonstrate that oxidative and nitrosative stressors also induce Mpk1 phosphorylation in C. neoformans. To test this, we first needed to develop the tools to recognize total Mpk1 protein independent of phosphorylation. A fusion protein was created using either the Flag or V5 tag at the carboxy terminus of the Mpk1 protein in both the KN99a and KN99α backgrounds immediately preceding the stop codon. Deletion of MPK1 was shown to have severe, deleterious phenotypes (29), but the strain with the wild-type MPK1 gene replaced with the fusion protein had no detectable phenotypes (data not shown). The Mpk1 Flag-tagged fusion construct in the KN99α background was used in subsequent experiments to determine activation of the PKC1 pathway.
Using an antibody to the Flag epitope, a band of the appropriate size was detected in the Mpk1-Flag strain in both the induced and uninduced lanes but was absent in the wild-type strain lacking the Mpk1-Flag fusion protein, demonstrating that the band was specific for Flag-tagged Mpk1 (Fig. 1B). Upon induction of the PKC1 pathway with 10 μg/ml calcofluor white, Mpk1 phosphorylation was induced in a strain containing the Flag-tagged construct as well as in the wild-type strain containing endogenous Mpk1 as detected using the anti-phospho-Mpk1 antibody (Fig. 1C). This result suggested that the Mpk Flag-tagged fusion protein was phosphorylated in response to a cell wall-damaging agent, similar to the response seen in the wild-type strain.
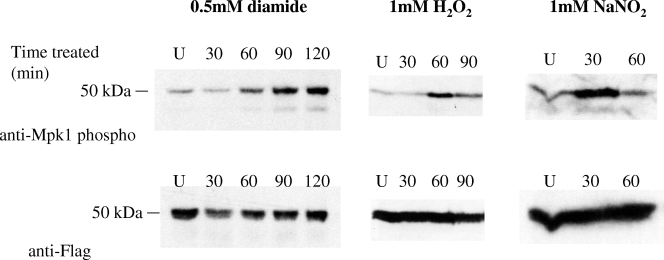
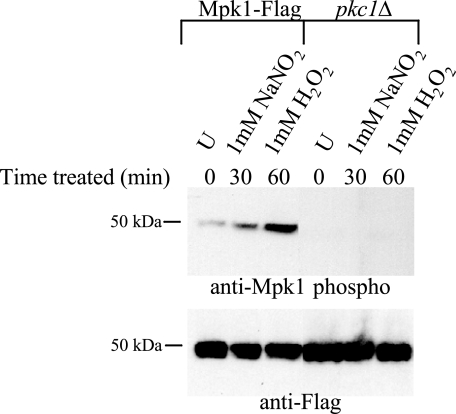
To test whether oxidative and nitrosative stressors induced Mpk1 phosphorylation, the Mpk1 Flag-tagged strain was treated with diamide, H2O2, or NaNO2 (Fig. 2). Upon addition of diamide, phosphorylated Mpk1 increased for at least 2 hours. With the addition of another oxidative stressor, H2O2, Mpk1 phosphorylation was more transient, peaking at approximately 1 hour and then decreasing after 90 min. For the nitrosative stressor NaNO2, the response was more rapid than that with H2O2, peaking at 30 min and lessening after 1 hour. Thus, the PKC1 pathway was induced upon exposure to both oxidative and nitrosative stresses. Each of these blots was stripped and reprobed with anti-Flag antibody to demonstrate that although the PKC1 pathway is activated (as a measure of phosphorylated Mpk1), the total amount of Mpk1 did not increase over time.
FIG. 2.
Mpk1 is induced by both oxidative and nitrosative stresses. The Mpk1-Flag tagged strain (KN99α background) was grown overnight in YNB (pH 4.0) at 25°C and diluted the next morning to an OD650 of 0.2. The cells were grown for an additional 2.5 h and then treated with 0.5 mM diamide for 30, 60, 90, and 120 min; with 1 mM H2O2 for 30, 60, and 90 min; or with 1 mM NaNO2 for 30 and 60 min. The uninduced control (U) for each was harvested at 60 min. Blots containing 50 μg of total protein from cell lysates were analyzed by probing sequentially with anti-Mpk1 phospho-antibody and then anti-Flag antibody after stripping the membranes.
PKC1 is not essential, and the pkc1Δ mutant is a conditional mutant dependent upon osmotic stabilization.
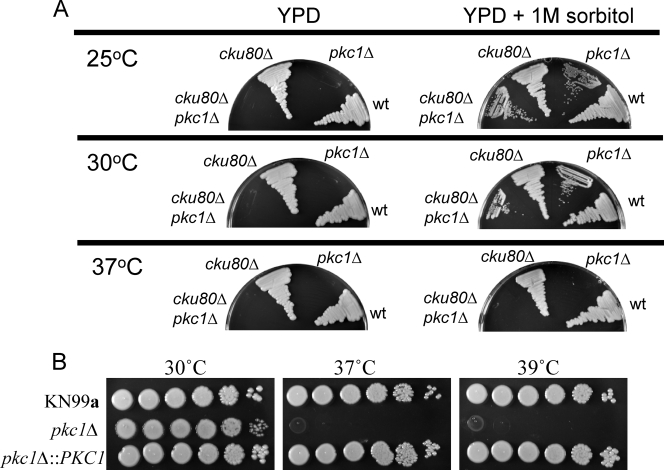
To further examine the role of PKC1 in various stress responses and maintenance of cell integrity, conditionally lethal PKC1 deletion strains were independently generated in two genetic backgrounds of C. neoformans. A linear deletion construct containing the NAT resistance cassette (33) in place of the PKC1 gene, flanked by approximately 900 bp of upstream and downstream sequence for homologous recombination, was used to transform both the wild-type strain KN99a (39) and the cku80Δ mutant, a strain with a decreased rate of nonhomologous end joining and therefore increased homologous recombination (17). Following transformation, cells were plated on YPD both with and without 1 M sorbitol (both containing NAT). No colonies grew on plates lacking an osmotic stabilizer; stable isolates containing the PKC1 deletion cassette and NAT resistance marker were obtained only on plates containing 1 M sorbitol. Isolates harboring the PKC1 deletion were confirmed by PCR (see Fig. S1 in the supplemental material), and the presence of a single insertion was verified by Southern blotting using a portion of the NAT gene as a probe (see Fig. S1 in the supplemental material). Figure 3A demonstrates that a deletion of PKC1 in two different strains causes an inability to grow in the absence of an osmotic stabilizer at 25°C, 30°C, or 37°C. Thus, deletion of PKC1 results in conditional lethality and is dependent upon osmotic stabilization.
FIG. 3.
Deletion of PKC1 requires osmotic stabilization, is temperature sensitive, and is complemented by reintroduction of the PKC1 gene. (A) Strains with PKC1 deletions in two genetic backgrounds, KN99a or cku80Δ (KN99α), were streaked onto YPD plates with or without 1 M sorbitol and grown at 25, 30, or 37°C for 5 days. The pkc1Δ strains failed to grow in the absence of sorbitol at any temperature and in the presence of sorbitol at 37°C. The parental strains (KN99a or cku80Δ in the KN99α background) were also plated as controls. wt, wild type. (B) Temperature sensitivity of pkc1Δ is rescued in the complemented strain. The pkc1Δ strain in the KN99a background (KN99a) and the complemented pkc1Δ::PKC1 strain were grown overnight in YPD plus 1 M sorbitol and then diluted to an OD650 of 1.0. Tenfold serial dilutions were plated onto YPD plates containing 1 M sorbitol and grown for 5 days at 30, 37, or 39°C.
PKC1 deletion strains are temperature sensitive.
High temperature is a stress that C. neoformans presumably encounters both in a mammalian host and in its ecological niche. To test whether deletion of PKC1 rendered strains sensitive to physiologic temperature important for virulence in mammalian hosts, pkc1Δ strains were tested for viability at both 37°C and 39°C. The pkc1Δ strains with both the wild-type KN99a and the cku80Δ backgrounds grew slightly slower than the parental strains when either streaked or serially diluted on YPD containing 1 M sorbitol. Additionally, the colony size of the deletion mutants was smaller than that of the parent strains, and the colonies consistently exhibited a glossier, more yellow appearance than the wild-type strains. Growth of the PKC1 deletion strains in both the KN99a and cku80Δ backgrounds was completely abolished when cells were grown on YPD plates containing 1 M sorbitol at 37°C (Fig. 3A). To test whether these phenotypes were due to deletion of PKC1, a complemented strain was constructed (see Fig. S1C in the supplemental material). Both the requirement for osmotic stabilization and the temperature sensitivity of the pkc1Δ strain were rescued in the complemented pkc1Δ::PKC1 strain at both 37° and 39°C (data not shown and Fig. 3B). The requirement for osmotic support and severe temperature sensitivity distinguishes Pkc1 from its downstream components, which include Bck1, Mkk2, and Mpk1. Deletions of genes encoding these downstream components cause slow growth on cell wall inhibitors and some, but not severe, temperature sensitivity but do not result in conditional lethality in the absence of sorbitol (16, 29).
PKC1 is essential for protection against both nitrosative and oxidative stresses.
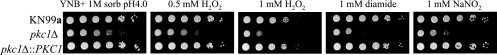
Given that the PKC1 pathway is activated in response to both oxidative and nitrosative stress as determined by Mpk1 phosphorylation (Fig. 2), it was probable that PKC1 would be crucial for protection against one or both of these stresses. Therefore, we hypothesized that a deletion would compromise the cells’ ability to overcome both oxidative and nitrosative challenges. Serial dilutions of the KN99a, pkc1Δ, and pkc1Δ::PKC1 strains were plated on YNB medium (pH 4.0) containing 1 M sorbitol in the presence or absence of the oxidative stressor diamide or H2O2 or the nitrosative stressor NaNO2. After three days growth at 30°C, the pkc1Δ strain exhibited sensitivities to all reagents tested (Fig. 4). It also grew slightly less well on YNB (pH 4.0) containing no stressor than did either the wild-type or complemented strain. The deletion strain was an order of magnitude more sensitive to 0.5 mM H2O2 and 2 orders of magnitude more sensitive to 1 mM H2O2 than either KN99a or the pkc1Δ::PKC1 strain. A deletion of PKC1 also rendered the cells 2 to 3 orders of magnitude more sensitive to 1 mM diamide and 2 orders of magnitude more sensitive to 1 mM NaNO2 than either the wild-type or complemented strain. Thus, PKC1 is critical for protection against both oxidative and nitrosative stresses.
FIG. 4.
PKC1 is important for protection against both oxidative and nitrosative stresses. The wild-type KN99a, the pkc1Δ, and the complemented pck1Δ::PKC1 strains were grown overnight in YPD containing 1 M sorbitol at 30°C and then diluted to 5 × 107 cells/ml in PBS containing 1 M sorbitol. Five-microliter portions of 10-fold serial dilutions were plated onto YNB plates containing 1 M sorbitol plus the indicated oxidative or nitrosative stressor and grown for 3 days at 30°C. All plates were at pH 4.0.
PKC1 is necessary for induction of Mpk1 phosphorylation during both nitrosative and oxidative stresses.
In order to determine whether the PKC1 pathway was activated by nitrosative and oxidative stress when PKC1 was deleted, a pkc1Δ strain was created from the Mpk1 Flag-tagged strain. Western blot analysis was performed to observe Mpk1 phosphorylation, and in contrast to the case for the wild type, no phosphorylation was detected for the pkc1Δ strain when this strain was induced with either 1 mM NaNO2 or 1 mM H2O2 (Fig. 5) or 10 μg/ml calcofluor white (data not shown), using the anti-phospho-Mpk1 antibody. This blot was stripped and reprobed with anti-Flag antibody to demonstrate that, importantly, the total amount of Mpk1 did change compared to that in the wild type.
FIG. 5.
PKC1 is necessary for Mpk1 phosphorylation in response to both nitrosative and oxidative stresses. The pkc1Δ strain in the Mpk1 Flag-tagged background and the Mpk1-Flag control strain were grown overnight in YPD plus 1 M sorbitol at 25°C and diluted the next morning to an OD650 of 0.05. The cells were grown for an additional 3 h and then treated with 1 mM NaNO2 for 30 min or 1 mM H2O2 for 60 min. The uninduced control (U) for each was harvested at 10 min. Blots containing 50 μg of total protein from cell lysates were analyzed by probing sequentially with anti-Mpk1 phospho-antibody then anti-Flag antibody after stripping the membrane.
Genes downstream of PKC1 in the cell integrity pathway are dispensable for resistance to oxidative and nitrosative stresses.
To determine whether additional components of the PKC1 pathway were required for protection against oxidative and nitrosative stresses, mutants with deletions of the three other known MAPK genes in the C. neoformans PKC1 pathway were tested for sensitivity to diamide, H2O2, and NaNO2. These genes are BCK1 and MKK2 (16) and MPK1 (29). Interestingly, the bck1Δ, mkk2Δ, and mpk1Δ deletion mutants demonstrated no sensitivities to either of the oxidative stressors (diamide and H2O2) or to the nitrosative stressor (NaNO2) up to concentrations of 1 mM on YNB with or without 1 M sorbitol at pH 4.0 (data not shown). Furthermore, upon analysis by Western blotting, no Mpk1 phosphorylation was detected in the bck1Δ, mkk2Δ, and mpk1Δ strains when they were induced with 1 mM NaNO2 or 1 mM H2O2 (data not shown). This showed that although the PKC1 pathway was activated by oxidative and nitrosative stressors in a wild-type strain, the downstream components of the pathway were dispensable for resistance.
Deletion of PKC1 results in severe cell integrity phenotypes.
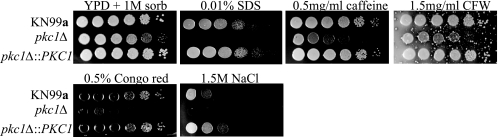
Identified PKC1 pathway components have been shown to be essential for resistance to a variety of cell stresses in C. neoformans (16, 29). Therefore, to further examine the role of the PKC1gene in the maintenance of cell integrity, we chose to subject the pkc1Δ mutant to a variety of cell wall-perturbing agents. Pkc1 and its downstream components involved in the MAPK cascade comprise a highly conserved major cell integrity pathway and are essential for proper biosynthesis and maintenance of the cell wall. In order to ascertain the ability of pkc1Δ to survive in the presence of cell wall-inhibiting agents, serial dilutions were plated on YPD plates containing 1 M sorbitol in addition to SDS, NaCl, caffeine, calcofluor white, or Congo red (Fig. 6) (16). The pkc1Δ strain failed to grow when subjected to either 1.5 M NaCl or 0.01% SDS. The PKC1 deletion strain also demonstrated a sensitivity to 0.5% Congo red that was 3 orders of magnitude greater than that of the wild type. Additionally, the pkc1Δ mutant exhibited increased sensitivity to both 1.5 mg/ml calcofluor white and 0.5 mg/ml caffeine, by approximately 2 orders of magnitude compared to wild type, after growth at 30°C for 3 days. Not surprisingly, these data demonstrate that deletion of PKC1 rendered cells more susceptible to cell wall-perturbing agents.
FIG. 6.
PKC1 is vital for protection against cell wall stress. Cultures of KN99a and the pkc1Δ and complemented pck1Δ::PKC1 strains were grown overnight in YPD containing 1 M sorbitol at 30°C and then diluted to an OD650 of 1.0. Five-microliter portions of 10-fold serial dilutions were plated onto YPD plates containing 1 M sorbitol plus the indicated inhibitor and grown for 3 days at 30°C. CFW, calcofluor white.
Capsule is aberrant in a pkc1Δ mutant.
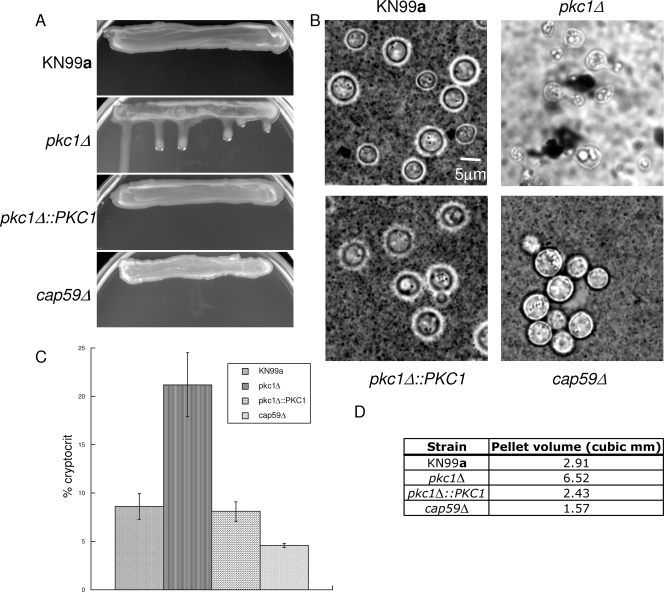
The polysaccharide capsule surrounding C. neoformans is essential for virulence and is anchored to the cell wall via α-1,3-glucan (40). Due to the severely compromised cell wall integrity of the PKC1 deletion strain, we chose to examine its capsule. Furthermore, it was observed that the PKC1 deletion strain, when grown on YPD-1 M sorbitol plates, appeared glossier than the wild-type strain, indicating an alteration in capsule production or attachment (data not shown). In order to analyze capsule formation, pkc1Δ cells were plated on DME plates containing 1 M sorbitol and incubated for 5 days at 30°C in the presence of 5% CO2. After 5 days on DME, the deletion strain was shinier and runnier than the wild type, suggesting an increase in capsule production in this strain (Fig. 7A).
FIG. 7.
Deletion of PKC1 causes altered capsule on capsule induction medium. (A) Strain KN99a, a strain with pkc1Δ in the KN99a background, the pkc1Δ::PKC1 strain, and the capsule-deficient cap59Δ strain in the KN99 background were streaked onto DME plates containing 1 M sorbitol and allowed to grow vertically for 5 days at 30°C in the presence of 5% CO2. The pkc1Δ strain was shinier and runnier than the parental or the complemented strain, and the strain lacking CAP59 appeared duller and drier. (B) India ink staining of the pkc1Δ strain is abnormal compared to that of the wild type. Strains were grown on DME plus 1 M sorbitol plates for 5 days at 30°C in the presence of 5% CO2. Individual isolates were resuspended in India ink-dH2O at a 1:4 ratio and viewed at a magnification of ×1,000. A representative isolate for each strain is shown. (C) Strains were grown on DME plus 1 M sorbitol plates for 5 days at 30°C in the presence of 5% CO2, and then the packed cell volume divided by total cell suspension (cryptocrit) was calculated for the KN99a, pkc1Δ, pkc1Δ::PKC1, and cap59Δ strains and shown as a percentage. All strains were analyzed in triplicate. Error bars indicate standard deviations. (D) The pellet cell volumes for each strain were calculated. These were determined from the average diameters, excluding capsule, of 50 cells each of the KN99a, pkc1Δ, pkc1Δ::PKC1, and cap59Δ strains (which were calculated to be 6.61, 4.57, 5.88, and 5.41 μm, respectively). These data suggest that although pkc1Δ cells are smaller than wild type, the increase in cryptocrit indicates that they produce more capsule than KN99a cells.
Individual isolates of the pkc1Δ, pkc1Δ::PKC1, wild-type, and cap59Δ acapsular strain were suspended in a 1:4 India ink-H2O solution and observed microscopically after 5 days of growth on DME medium containing 1 M sorbitol in the presence of 5% CO2. Capsule is normally visualized as a halo around the cells due to exclusion of the dye. Interestingly, India ink staining of the pkc1Δ strain did not show a large amount of capsule surrounding the cells, but it appeared that the ink bound to large patches surrounding the cells, in comparison to both the wild-type and reconstituted strains (Fig. 7B). The India ink particles seemed to extensively aggregate, to the point of clearing the background. The pkc1Δ cells also exhibited clumping, as is often observed with strains possessing aberrant capsule or a compromised cell wall.
In order to further investigate capsule in the pkc1Δ mutant, the packed cell volume was measured after 5 days of growth on DME containing 1 M sorbitol in the presence of 5% CO2 at 30°C (1). When equal numbers of cells were spun down in microcapillary tubes, the packed cell volume divided by the total cell suspension, or cryptocrit, of the pkc1Δ strain was 21.2%, compared to 8.6% and 8.1% for the wild-type and the PKC1 complemented strains, respectively (Fig. 7C). The portion of the packed cell volume that can be attributed to cells excluding capsule was determined from the average diameters of the cells. Fifty cells each of the KN99a, pkc1Δ, pkc1Δ::PKC1, and cap59Δ strains were measured under the microscope and averaged, and they were calculated to be 6.61, 4.57, 5.88, and 5.41 μm, respectively. The diameter was used to calculate the volume of the cryptocrit containing 4 × 107 cells that is attributable to cell mass, rather than capsule. These data show that the portion of the cryptocrit of the pkc1Δ strain that is attributable to cells is smaller than that for the wild type. Thus, given that equal numbers of cells for each strain were analyzed and that the pkc1Δ cells were smaller than those of both the wild-type and complemented strains (data not shown and Fig. 7D), the majority of cryptocrit for the PKC1 deletion strain was due to capsule. The percentage of cryptocrit for the cap59Δ strain was 4.6%, thus strengthening the idea that most of the packed cell volume for the pkc1Δ strain was due to capsule.
Deletion of PKC1 results in decreased melanin production.
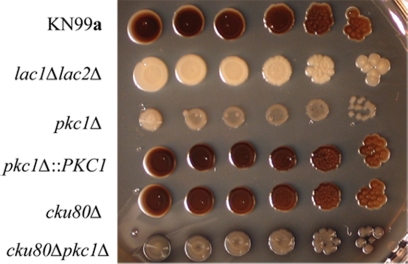
Deletion of the C1 domain of Pkc1 in C. neoformans has been shown to reduce production of the pigment melanin, an important virulence factor, and to decrease the activity of laccase, the enzyme required for melanin synthesis (22) and that is localized to the cell wall (47). Therefore, the ability of pkc1Δ strains to produce melanin was examined by plating serial dilutions of the strains on l-DOPA medium containing 1 M sorbitol. The plates were incubated for 21 days at 30°C. Similar to the C1 domain deletion strain, the pkc1Δ strains in both the KN99a and cku80Δ backgrounds produced some pigment. In the KN99 background, the level of pigmentation was intermediate between those the parental strain and the lac1Δlac2Δ deletion strain, which produces no melanin (Fig. 8).
FIG. 8.
Deletion of PKC1 results in decreased melanin production. Deletion strains with both the KN99a and cku80Δ (KN99α) backgrounds were grown overnight in YPD containing 1 M sorbitol and then diluted to an OD650 of 1.0. Five-microliter portions of 10-fold serial dilutions were plated onto l-DOPA plates containing 1 M sorbitol and allowed to grow for 21 days at 30°C. A lac1Δ lac2Δ strain in the KN99 background was plated as a negative control; it produces no melanin.
Chitin and chitosan staining are altered in pkc1Δ, but both chitin and chitosan content approach wild-type levels.
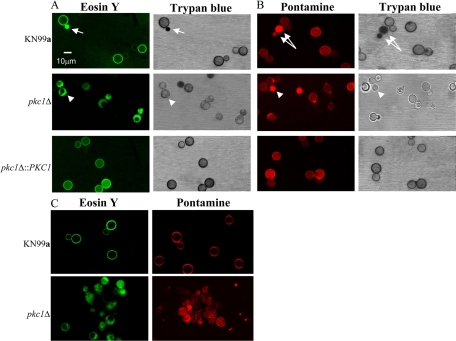
Chitin and its deactylated form, chitosan, have been demonstrated to be important components of the C. neoformans cell wall (2, 3). The PKC1 deletion strain exhibited extremely compromised cell wall integrity, so we wanted to determine whether those defects could be due to altered polysaccharide composition, specifically, aberrations in chitin and/or chitosan content or localization. To determine if the distribution of chitin and chitosan was altered, the cells were stained with the dye pontamine (Fast Scarlet 4B), which is thought to stain specifically for chitin (24), and with eosin Y, which stains chitosan but not chitin (2, 9). The pkc1Δ strain exhibited altered pontamine and eosin Y staining compared to the wild type, and complementation of the deletion restored normal staining (Fig. 9A and B). The wild-type and complemented strains revealed uniform, ring-like staining and showed binding of both dyes, presumably to the cell wall. The pkc1Δ strain, however, demonstrated some bright, diffuse staining with pontamine and revealed increased staining at the bud neck. Furthermore, it showed a polarized staining pattern with eosin Y, often staining more brightly on one half of the cell than the other, and it appeared that large vacuoles were present in some of the pkc1Δ cells and that these excluded eosin Y but not pontamine.
FIG. 9.
Eosin Y and pontamine staining of the pkc1Δ strain are aberrant compared to wild type. The KN99a, pkc1Δ, and pkc1Δ::PKC1 strains were grown for 72 h in YPD containing 1 M sorbitol at 25°C. Cells were stained with eosin Y in addition to Trypan blue (A) and with pontamine in addition to trypan blue (B) as described in Materials and Methods. Eosin Y and pontamine staining were visualized using fluorescence microscopy (excitation wavelengths of 488 nm and 548 nm, respectively), and trypan blue staining of the same fields was visualized using light microscopy. Eosin Y and pontamine stainings are seen as uniform rings surrounding the cells containing a wild-type PKC1 gene and as more polarized and diffuse staining, respectively, for the pkc1Δ cells. Arrows represent dead wild-type cells (calculated to be ∼0.5% of the population [data not shown]) that have taken up eosin Y or pontamine as well as trypan blue. Arrowheads indicate mutant cells that have taken up either eosin Y or pontamine but not trypan blue. (C) Confocal images from the midsection of the Z series (see Fig. S2 in the supplemental material) for each strain. For all images, excitation wavelengths were set at 488 nm for eosin Y and 548 nm for pontamine.
To ensure that neither the altered pontamine nor polarized eosin Y staining pattern was simply due to compromised cell wall integrity and subsequent cell death (thus causing either stain to diffuse into the cells), the mutant was tested for uptake of trypan blue as a measure of cell membrane integrity (16). Cells were stained concomitantly with either pontamine and trypan blue or eosin Y and trypan blue, and when the same fields were observed by both fluorescence and bright-field microscopy, it was determined that pkc1Δ cells with both altered pontamine and polarized eosin Y staining excluded trypan blue, indicating cell viability (Fig. 9A and B). Inexplicably, not all cells that appeared in the bright field stained with pontamine or eosin Y. For the control KN99a and pkc1Δ::PKC1 strains, cells with uniform, ring-like eosin Y staining also excluded trypan blue, indicating viability (Fig. 9A).
The diminished and more diffuse staining pattern of pontamine and the aberrant, polarized eosin Y staining pattern of the pkc1Δ strain suggested that staining was occurring intracellularly and that either or both chitin and chitosan might be diminished or improperly localized in the mutant. To further investigate this, confocal microscopy was performed and Z series obtained at 1-μm intervals for both the wild-type and reconstituted strains and at 0.25-μm intervals for the pkc1Δ strain (the diameter of the wild-type and complemented strains was determined to be ∼15 μm, and the diameter of the mutant was somewhat smaller) (Fig. 9C; see Fig. S2 in the supplemental material). These data indicated that both pontamine and eosin Y were inside the majority of mutant cells, although in some cells, the stain also appeared to be localized to the cell wall, particularly with pontamine.
Due to the aberrant staining patterns observed with both pontamine and eosin Y that indicated altered chitin and chitosan, respectively, in the PKC1 deletion mutant, we chose to measure the levels of both of these important cell wall components in the pkc1Δ strain. Surprisingly, the deletion strain contained chitin and chitosan levels that were similar to those in the wild-type strain (data not shown). This strongly suggests that although chitin and chitosan are synthesized at normal levels in the pkc1Δ strain, neither of these components is properly localized to the cell wall.
DISCUSSION
The studies presented here have begun to decipher critical functions of the C. neoformans PKC1 gene and have furthered our understanding of the PKC1 cell integrity pathway in this pathogenic fungus. The generation of strains with conditional deletions in PKC1 has allowed us to begin to assign multiple roles for this gene in protection against a variety of stresses. Not surprisingly, we have shown that C. neoformans PKC1 plays a crucial role in cell integrity, but importantly, we have also demonstrated that it is required for full resistance to both oxidative and nitrosative stresses.
PKC1 deletion strains of C. neoformans are viable only in the presence of the osmotic stabilizer sorbitol. In S. cerevisiae, deletion of PKC1 is lethal in some genetic backgrounds and partially suppressed by osmotic stabilization in others (32), and it remains to be seen if similar results will occur with various cryptococcal species and genetic backgrounds. Not only are the C. neoformans pkc1Δ strains conditionally lethal, they are slower growing, exhibit smaller colony morphology, and are temperature sensitive compared to the wild type when grown with osmotic stabilization (Fig. 3). These phenotypes alone suggest that PKC1 is important for cell integrity in C. neoformans, and this is supported by the sensitivities of the deletion strain to a wide range of cell wall inhibitors (Fig. 6).
Cell wall biogenesis and integrity are essential for viability and are attractive targets for antifungal therapy; therefore, it is vital to understand the composition of the cell wall. Consequently, the cell wall integrity defects of the pkc1Δ strain led us to question what specific deficiencies this strain possessed. We chose to begin this investigation by examining the localization and content of both chitin and chitosan, the deacetylated form of chitin found in vegetatively growing cells and an important component of the cryptococcal cell wall (2, 3). Based on the observation that both pontamine and eosin Y staining of the pkc1Δ strain was aberrant and that both dyes seemed to be located intracellularly (Fig. 9; see Fig. S2 in the supplemental material), the levels of both chitin and chitosan were measured. Surprisingly, neither was significantly different from that in wild-type cells grown under the same conditions. Although it is possible that the cell wall permeability of a pkc1Δ strain is appreciably altered, thus allowing diffusion of pontamine and eosin Y inside the cells and nonspecific staining of cytoplasmic components, the lack of trypan blue uptake and the absence of a defined ring-like structure in mutant cells with abnormal pontamine and eosin Y staining patterns indicated that this was not likely to be the case. However, because the molecular weight of trypan blue is slightly higher than that of either pontamine or eosin Y (960 versus 800 or 700), these results do not completely rule out nonspecific diffusion due to changes solely in membrane or cell wall permeability.
Studies of S. cerevisiae have demonstrated that during cell stress, both chitin and β-glucan syntheses are increased upon phosphorylation of Slt2p (C. neoformans Mpk1 homolog). Upregulation of genes encoding the chitin synthase Chs3p as well as β-1,3-glucan synthases ensues (26, 27, 46). Intriguingly, S. cerevisiae Pkc1p is necessary both for the phosphorylation of Chs3p and for the localization of Chs3p to the plasma membrane in response to heat stress (44). In C. neoformans, Chs3 is the closest homolog to S. cerevisiae Chs3p, and it is the chitin synthase primarily responsible for synthesis of the chitin that is converted to chitosan (3). Therefore, it is possible that in a C. neoformans pkc1Δ strain, Chs3 is not properly localized, given both the pontamine and eosin Y staining patterns of the PKC1 mutant. Another possibility is that Chs3 is trapped or retained inside chitosomes, which are membrane-bound vesicular organelles found in fungi that contain chitin synthase (41). Chitosomes are thought to be intracellular reservoirs for regulated transport of chitin synthase enzymes to the cell wall and potential sites of chitin microfibril synthesis (reviewed by Bartnicki-Garcia [4]). If production of chitin microfibrils (or full-length polymers) occurs intracellularly but these moieties fail to become incorporated in the cell wall, this may help explain in part the wild-type levels of chitin in the pkc1Δ mutant. Staining with eosin Y also indicates that chitosan, in addition to chitin, is mislocalized. Studies by Baker et al. (2) have identified three chitin deacetylases (Cda1, Cda2, and Cda3) in C. neoformans that are responsible for total chitosan production during vegetative growth. These enzymes are proteins that have been localized to the cell wall, most likely through a glycosylphosphatidylinositol anchor remnant (15). It is not known if the chitin is converted to chitosan by the chitin deacetylases while they are covalently attached to the wall or if the deacetylation occurs at an earlier step prior to the chitin deacetylases being linked to the wall. Our eosin Y staining results indicate that the chitosan, as well as chitin, is internal in the pkc1Δ strain. One possible explanation for the mislocalization of both chitin and chitosan is that they are both made in chitosomes, and the chitosomes cannot deliver their cargo to the cell wall in the pkc1Δ mutant. Another possibility is that both chitin synthases and chitin deacetylases are mislocalized and both chitin and chitosan are made internally, rather than at the cell membrane and in the cell wall. In any case, our results suggest that, although wild-type amounts of both chitin and chitosan are being synthesized in the pkc1Δ mutant, these components are not properly localized to the cell wall.
Two major virulence factors of C. neoformans, capsule and melanin, are both cell wall associated. The polysaccharide capsule is anchored to the wall, likely through interactions with α-1,3-glucan (40). On capsule induction medium, the PKC1 deletion strain had a shiny appearance and was runnier than the wild-type strain (Fig. 7). This may indicate that the mutant strain is making more polysaccharide than the wild type, that the capsule composition is altered, or that capsular material is not retained properly by the cell. These last two possibilities are supported by India ink staining. Wild-type strains, when stained with India ink and viewed microscopically, normally exhibit halos surrounding the cells that represent exclusion of ink by the polysaccharide capsule. These halos increase in diameter when cells are grown on capsule induction medium and in strains that overproduce capsule (13). Intriguingly, when the pkc1Δ strain stained with India ink was observed under the microscope, large areas of background that do not contain cells coalesced the ink particles to form large black patches. These patches could be shed capsular material with an altered composition. Interestingly, the background was consistently lighter in color for the pkc1Δ strain than for the other strains that were India ink stained. Furthermore, the possibility that the pkc1Δ strain is making increased capsule was investigated by the determination of cyrptocrit, and this hypothesis was strongly supported by these data.
Melanin is deposited in the cell wall, and the laccase that produces melanin is localized to the wall (47). Melanin appeared to be reduced in the pkc1Δ strain compared to wild type. It has previously been shown by Heung et al. (22) that the DAG C1 domain of Pkc1 is required for the proper localization of laccase in the cryptococcal cell wall and for the formation of melanin. Therefore, it was expected that a deletion of PKC1 would result in decreased pigmentation. Melanin is known to be essential for virulence, although we chose not to determine the ability of the pkc1Δ strain to cause disease in mice because of its severe temperature sensitivity at 37°C. Given the severe defects in the cell wall of the pkc1Δ strain, it is not surprising that there were alterations in both melanin and capsule.
The most intriguing result from this analysis of PKC1 is that this gene is vital for both oxidative and nitrosative stress responses. Elegant studies of S. cerevisiae by Vilella et al. (45) showed the involvement of Pkc1p in oxidative stress but could not detect a role for this protein in the nitrosative stress response. Here we demonstrate its importance for the response to oxidative stress and indicate its added importance for protection against nitrosative stress in C. neoformans. Both reactive oxygen and nitrogen species are potentially toxic to the organism, and the cells’ ability to withstand nitrosative stress has been shown to be critical for survival in mammalian hosts (12, 35); additionally, there is mounting evidence that resistance to oxidative stress is also needed for survival in the mammalian host (10, 36). Consistent with the hypersensitivity of the pkc1Δ strain to the two oxidative stressors diamide and hydrogen peroxide and the nitrosative stressor sodium nitrite (Fig. 4), the PKC1 pathway is activated in wild-type strains by these stressors as shown by phosphorylation of Mpk1 (Fig. 2). Cells exposed to diamide, H2O2, and NaNO2 demonstrated slightly different kinetics of Mpk1 phosphorylation, suggesting that there may be distinct sensors for each of these stressors. Moreover, these oxidative and nitrosative stressors could be targeting wall components and thus exerting their effects as a result of cell wall damage.
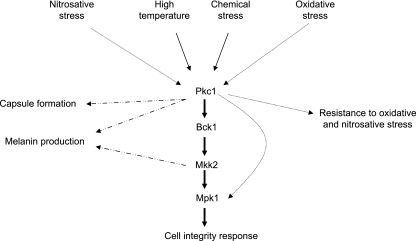
Data from this study and others indicate that PKC1 may have multiple functions in the cell other than activation of the PKC1 pathway with subsequent Mpk1 phosphorylation (Fig. 10). The requirement for the pathway is shown by the severe cell integrity defects exhibited in PKC1 deletion strains (this study) as well as strains with deletions in BCK1, MKK2, and MPK1, the genes that encode downstream MAPKs of Pkc1 (16, 29). However, the pkc1Δ strain has more severe cell integrity defects than the bck1Δ, mkk2Δ, and mpk1Δ strains (16, 29), suggesting an additional role for Pkc1 in cell integrity beyond the activation of the pathway. Furthermore, C. neoformans deletion mutants generated for the MAPKs within this pathway do not have identical phenotypes with each other. For example, melanin production is normal in a bck1Δ strain but delayed in an mkk2Δ strain, suggesting that there could be alternative pathways capable of activating this cell integrity pathway at different points in the phosphorylation cascade (16). Importantly, deletion of PKC1 renders the cells extremely susceptible to oxidative and nitrosative stresses, but BCK1, MKK2, and MPK1 are dispensable for protection against both of those stresses (Fig. 4 and data not shown). The paradox is that Mpk1 is phosphorylated in response to both oxidative and nitrosative stresses, and Pkc1 is required for the phosphorylation of Mpk1 in response to these stresses, but the MAPKs, including Mpk1, that are also needed for phosphorylation are expendable. This lends support to the ideas that although Pkc1 may be activating the pathway in response to reactive nitrogen or oxygen species, this activation is not needed for the protection against these stresses, and that Pkc1 has an alternative output which is needed for the cells’ survival under oxidative or nitrosative stress.
FIG. 10.
Model for PKC1 in signal transduction of various stresses. Solid arrows represent functions in which signaling via the PKC1 pathway and its components is required. Dotted arrows denote functions in which PKC1 is important but pathway components are dispensable. Dashed arrows indicate that a specific gene and function are linked.
The idea that Pkc1 has effects independent of the pathway is also strongly supported by the more severe phenotypes of pkc1Δ than the downstream deletion mutations with all stressors tested. Similarly, Chs3p transport in S. cerevisiae is dependent on Pkc1p and not other members of the pathway when cells react to heat stress (44). The effect of deletions in the downstream kinases on the response to oxidative and nitrosative stresses differs significantly from that of pkc1Δ, indicating that PKC1, at least in response to some stresses, has a distinct mechanism for maintaining cell integrity other than initiating a linear phosphorylation cascade. This reinforces the idea that Pkc1 serves as a central regulator with multiple outputs and that it has additional and critical functions in the cell. Furthermore, this expanded role for PKC1 indicates that the PKC1 pathway is not always activated in a “top-down” manner via PKC1. Collectively, these data demonstrate that loss of Pkc1 has pleiotropic effects because of its central role in many functions, either dependent on or independent of its classical role in PKC1 cell integrity pathway activation.
Supplementary Material
Acknowledgments
We thank Joel Eissenberg for critical reading of the manuscript and helpful suggestions and Tamara Doering for expert capsule advice. We thank Maureen Donlin and Nicole Gilbert for thought-provoking and lively discussions.
This work was supported by National Institute of Health grants RO1AI072195, RO1AI50184, and R01HL088905 to J.K.L.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 175-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, L. G., C. A. Specht, M. J. Donlin, and J. Lodge. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, I. R., C. A. Specht, M. J. Donlin, K. J. Gerik, S. M. Levitz, and J. K. Lodge. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 41902-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartnicki-Garcia, S. 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6957-965. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. M., L. T. Campbell, and J. K. Lodge. 2007. Cryptococcus neoformans, a fungus under stress. Curr. Opin. Microbiol. 10320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall, A., and J. R. Perfect. Cryptococcus neoformans. ASM Press, Washington, DC.
- 8.Chaskes, S., and R. L. Tyndall. 1975. Pigment production by Cryptococcus neoformans from para- and ortho-diphenols: effect of the nitrogen source. J. Clin. Microbiol. 6509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee, S., S. Chatterjee, B. P. Chatterjee, A. R. Das, and A. K. Guha. 2005. Adsorption of a model anionic dye, eosin Y, from aqueous solution by chitosan hydrobeads. J. Colloid Interface Sci. 28830-35. [DOI] [PubMed] [Google Scholar]
- 10.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 1482607-2615. [DOI] [PubMed] [Google Scholar]
- 12.de Jesús-Berríos, M., L. Liu, J. C. Nussbaum, G. M. Cox, J. S. Stamler, and J. Heitman. 2003. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 131963-1968. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 213179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 104538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eigenheer, R. A., Y. Jin Lee, E. Blumwald, B. S. Phinney, and A. Gelli. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 7499-510. [DOI] [PubMed] [Google Scholar]
- 16.Gerik, K. J., M. J. Donlin, C. E. Soto, A. M. Banks, I. R. Banks, M. A. Maligie, C. P. Selitrennikoff, and J. K. Lodge. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58393-408. [DOI] [PubMed] [Google Scholar]
- 17.Goins, C. L., K. J. Gerik, and J. K. Lodge. 2006. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet. Biol. 43531-544. [DOI] [PubMed] [Google Scholar]
- 18.Granger, D. L., J. R. Perfect, and D. T. Durack. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, J. C., E. S. Bardes, Y. Ohya, and D. J. Lew. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3417-420. [DOI] [PubMed] [Google Scholar]
- 20.Harrison, J. C., T. R. Zyla, E. S. Bardes, and D. J. Lew. 2004. Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 2792616-2622. [DOI] [PubMed] [Google Scholar]
- 21.Heinisch, J. J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway is involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32671-680. [DOI] [PubMed] [Google Scholar]
- 22.Heung, L. J., A. E. Kaiser, C. Luberto, and M. Del Poeta. 2005. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J. Biol. Chem. 28028547-28555. [DOI] [PubMed] [Google Scholar]
- 23.Heung, L. J., C. Luberto, A. Plowden, Y. A. Hannun, and M. Del Poeta. 2004. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J. Biol. Chem. 27921144-21153. [DOI] [PubMed] [Google Scholar]
- 24.Hoch, H. C., C. D. Galvani, D. H. Szarowski, and J. N. Turner. 2005. Two new fluorescent dyes applicable for visualization of fungal cell walls. Mycologia 97580-588. [DOI] [PubMed] [Google Scholar]
- 25.Hua, J., J. D. Meyer, and J. K. Lodge. 2000. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab Immunol. 7125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 341049-1057. [DOI] [PubMed] [Google Scholar]
- 27.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 91559-1571. [DOI] [PubMed] [Google Scholar]
- 28.Kojima, K., Y. S. Bahn, and J. Heitman. 2006. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152591-604. [DOI] [PubMed] [Google Scholar]
- 29.Kraus, P. R., D. S. Fox, G. M. Cox, and J. Heitman. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 481377-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K. S., and D. E. Levin. 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin, D. E., F. O. Fields, R. Kunisawa, J. M. Bishop, and J. Thorner. 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62213-224. [DOI] [PubMed] [Google Scholar]
- 33.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39151-154. [DOI] [PubMed] [Google Scholar]
- 34.McIlvaine, T. C. 1921. A buffer solution for colorimetric comparison. J. Biol. Chem. 49183-186. [Google Scholar]
- 35.Missall, T. A., M. E. Pusateri, M. J. Donlin, K. T. Chambers, J. A. Corbett, and J. K. Lodge. 2006. Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: implications for virulence. Eukaryot. Cell 5518-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 511447-1458. [DOI] [PubMed] [Google Scholar]
- 37.Nelson, R. T., B. A. Pryor, and J. K. Lodge. 2003. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet. Biol. 381-9. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, R. T., J. Hua, B. Pryor, and J. K. Lodge. 2001. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics. 157935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 2003 714831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reese, A. J., and T. L. Doering. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 501401-1409. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Herrera, J., and S. Bartnicki-Garcia. 1974. Synthesis of cell wall microfibrils in vitro by a “soluble” chitin synthetase from Mucor rouxii. Science 186357-359. [DOI] [PubMed] [Google Scholar]
- 42.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1751405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres, J., C. J. Di Como, E. Herrero, and M. A. De La Torre-Ruiz. 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 27743495-43504. [DOI] [PubMed] [Google Scholar]
- 44.Valdivia, R. H., and R. Schekman. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 10010287-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilella, F., E. Herrero, J. Torres, and M. A. de la Torre-Ruiz. 2005. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 2809149-9159. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 181013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, X., J. Gibbons, J. Garcia-Rivera, A. Casadevall, and P. R. Williamson. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 695589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.