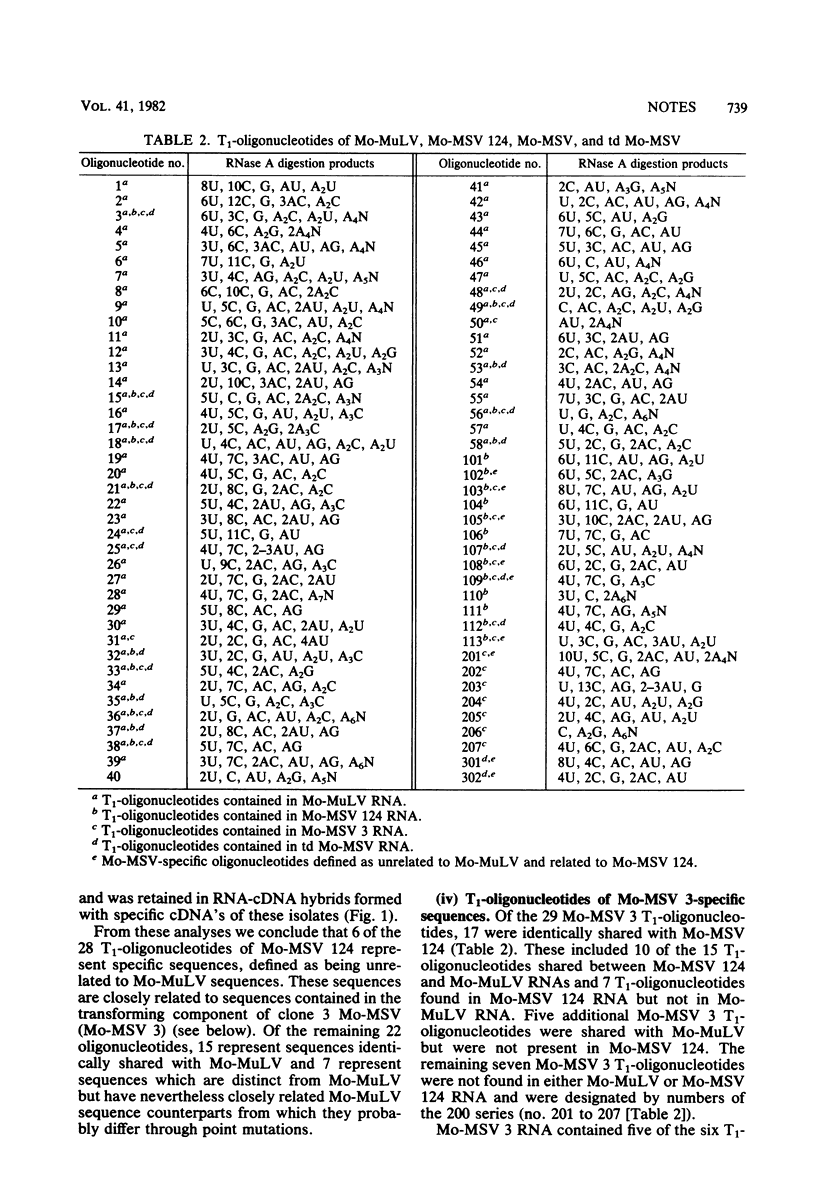
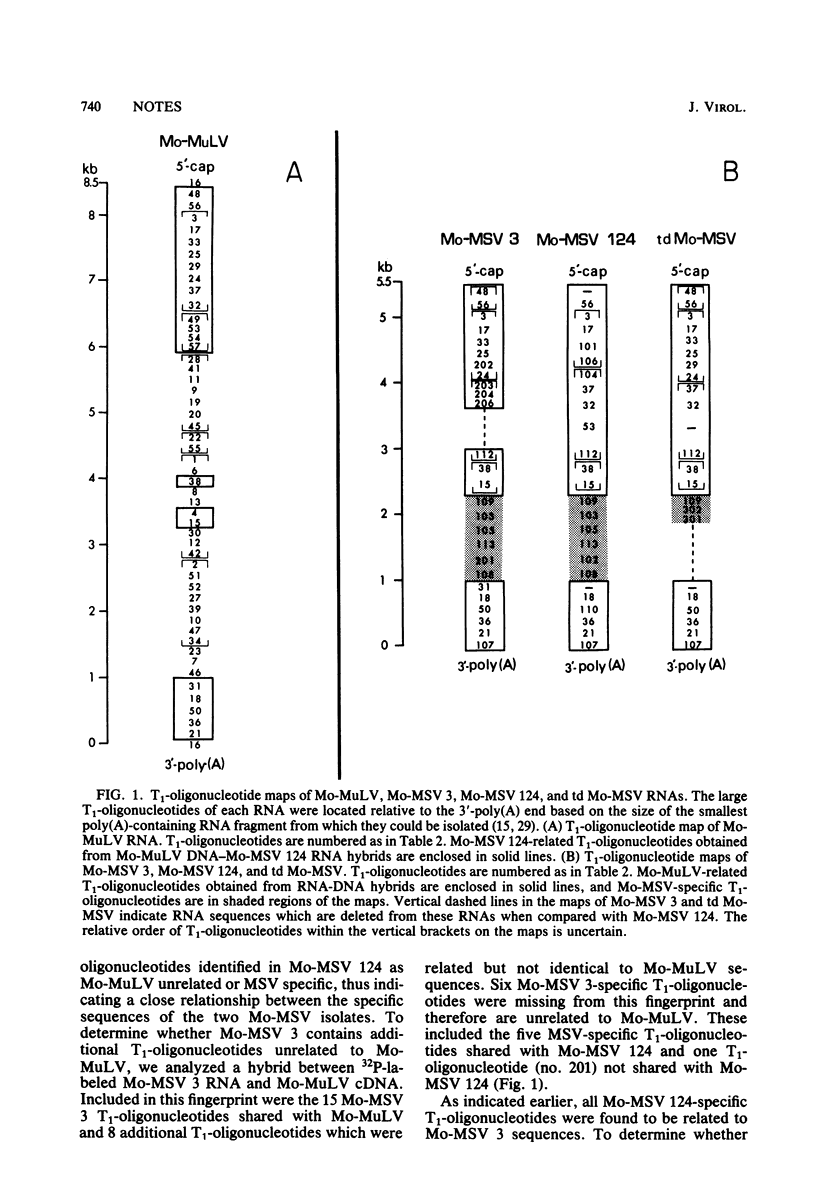
Abstract
A transformation-defective (td) deletion mutant of Moloney murine sarcoma virus (td Mo-MSV) and a transforming component termed Mo-MSV 3 were cloned from a stock of clone 3 Mo-MSV. To define the defect of the transforming function, the RNA of td Mo-MSV was compared with those of Mo-MSV 3 and of another transforming variant termed Mo-MSV 124 and with helper Moloney murine leukemia virus (Mo-MuLV). The RNA monomers of td Mo-MSV and Mo-MSV 3 comigrated on polyacrylamide gels and were estimated to be 4.8 kilobases (kb) in length. In agreement with previous analyses, the RNA of Mo-MSV 124 measured 5.5 kb and that of Mo-MuLV measured 8.5 kb. The interrelationships among the viral RNAs were studied by fingerprinting and mapping of RNase T1-resistant oligonucleotides (T1-oligonucleotides) and by identification of T1-oligonucleotides present in hybrids formed by a given viral RNA with cDNA's made from another virus. The nontransforming td Mo-MSV RNA lacked most of the Mo-MSV-specific sequence, i.e., the four 3′-proximal T1-oligonucleotides of the six T1-oligonucleotides that are shared by the Mo-MSV-specific sequences of Mo-MSV 3 and Mo-MSV 124. The remaining two Mo-MSV-specific oligonucleotides identified td Mo-MSV as a deletion mutant of MSV rather than a deletion mutant of Mo-MuLV. td Mo-MSV and Mo-MSV 124 exhibited similar deletions of gag, pol, and env sequences which were less extensive than those of Mo-MSV 3. Hence, td Mo-MSV is not simply a deletion mutant of Mo-MSV 3. In addition to their MSV-specific sequences, all three MSV variants, including td Mo-MSV, shared the terminal sequences probably encoding the proviral long terminal repeat, which differed from their counterpart in Mo-MuLV. This may indirectly contribute to the oncogenic potential of MSV. A comparison of td Mo-MSV sequences with either Mo-MSV 124 or Mo-MSV 3 indicated directly, in a fashion similar to the deletion analyses which defined the src gene of avian sarcoma viruses, that Mo-MuLV-unrelated sequences of Mo-MSV are necessary for transformation. A definition of transformation-specific sequences of Mo-MSV by deletion analysis confirmed and extended previous analyses which have identified Mo-MuLV-unrelated sequences in Mo-MSV RNA and other studies which have described transformation of mouse 3T3 fibroblasts upon transfection with DNAs containing the Mo-MSV-specific sequence.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson P., Goldfarb M. P., Weinberg R. A. A defined subgenomic fragment of in vitro synthesized Moloney sarcoma virus DNA can induce cell transformation upon transfection. Cell. 1979 Jan;16(1):63–75. doi: 10.1016/0092-8674(79)90188-0. [DOI] [PubMed] [Google Scholar]
- Ball J., McCarter J. A., Sunderland S. M. Evidence for helper independent murine sarcoma virus. I. Segregation of replication-defective and transformation-defective viruses. Virology. 1973 Nov;56(1):268–284. doi: 10.1016/0042-6822(73)90305-x. [DOI] [PubMed] [Google Scholar]
- Bassin R. H., Tuttle N., Fischinger P. J. Isolation of murine sarcoma virus-transformed mouse cells which are negative for leukemia virus from agar suspension cultures. Int J Cancer. 1970 Jul 15;6(1):95–107. doi: 10.1002/ijc.2910060114. [DOI] [PubMed] [Google Scholar]
- Blair D. G., Oskarsson M., Wood T. G., McClements W. L., Fischinger P. J., Vande Woude G. G. Activation of the transforming potential of a normal cell sequence: a molecular model for oncogenesis. Science. 1981 May 22;212(4497):941–943. doi: 10.1126/science.7233190. [DOI] [PubMed] [Google Scholar]
- Canaani E., Aaronson S. A. Isolation and characterization of naturally occuring deletion mutants of Moloney murine sarcoma virus. Virology. 1980 Sep;105(2):456–466. doi: 10.1016/0042-6822(80)90046-x. [DOI] [PubMed] [Google Scholar]
- Canaani E., Robbins K. C., Aaronson S. A. The transforming gene of Moloney murine sarcoma virus. Nature. 1979 Nov 22;282(5737):378–383. doi: 10.1038/282378a0. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Verma I. M., Duesberg P. H., Davidson N. Heteroduplex analysis of the RNA of clone 3 Moloney murine sarcoma virus. J Virol. 1979 Dec;32(3):1028–1032. doi: 10.1128/jvi.32.3.1028-1032.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M., Hageman T. C., Maxam A. M., Haseltine W. A. Structure of the genome of Moloney murine leukemia virus: a terminally redundant sequence. Cell. 1978 Apr;13(4):761–773. doi: 10.1016/0092-8674(78)90226-x. [DOI] [PubMed] [Google Scholar]
- Dina D., Beemon K., Duesberg P. The 30S Moloney sarcoma virus RNA contains leukemia virus nucleotide sequences. Cell. 1976 Oct;9(2):299–309. doi: 10.1016/0092-8674(76)90120-3. [DOI] [PubMed] [Google Scholar]
- Dina D., Penhoet E. E. Viral gene expression in murine sarcoma virus(murine leukemia virus)-infected cells. J Virol. 1978 Sep;27(3):768–775. doi: 10.1128/jvi.27.3.768-775.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue D. J., Goldfarb M. P., Sharp P. A., Weinberg R. A. Organization of murine sarcoma virus genomes. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):721–726. doi: 10.1101/sqb.1980.044.01.077. [DOI] [PubMed] [Google Scholar]
- Duesberg P. H. Transforming genes of retroviruses. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):13–29. doi: 10.1101/sqb.1980.044.01.005. [DOI] [PubMed] [Google Scholar]
- Duesberg P. H., Vogt P. K. Differences between the ribonucleic acids of transforming and nontransforming avian tumor viruses. Proc Natl Acad Sci U S A. 1970 Dec;67(4):1673–1680. doi: 10.1073/pnas.67.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. H., Duesberg P. H., Troxler D. H., Scolnick E. M. Spleen focus-forming Friend virus: identification of genomic RNA and its relationship to helper virus RNA. J Virol. 1979 Jul;31(1):133–146. doi: 10.1128/jvi.31.1.133-146.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L., Nunn M., Duesberg P. H., Troxler D., Scolnick E. RNAs of defective and nondefective components of Friend anemia and polycythemia virus strains identified and compared. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):823–835. doi: 10.1101/sqb.1980.044.01.087. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Kleid D. G., Panet A., Rothenberg E., Baltimore D. Ordered transcription of RNA tumor virus genomes. J Mol Biol. 1976 Sep 5;106(1):109–131. doi: 10.1016/0022-2836(76)90303-x. [DOI] [PubMed] [Google Scholar]
- Howk R. S., Troxler D. H., Lowy D., Duesberg P. H., Scolnick E. M. Identification of a 30S RNA with properties of a defective type C virus in murine cells. J Virol. 1978 Jan;25(1):115–123. doi: 10.1128/jvi.25.1.115-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Davidson N. A heteroduplex study of the sequence relationships between the RNAs of M-MSV and M-MLV. Cell. 1977 Mar;10(3):469–477. doi: 10.1016/0092-8674(77)90034-4. [DOI] [PubMed] [Google Scholar]
- Joho R. H., Billeter M. A., Weissmann C. Mapping of biological functions on RNA of avian tumor viruses: location of regions required for transformation and determination of host range. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4772–4776. doi: 10.1073/pnas.72.12.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisel J., Dina D., Duesberg P. Murine sarcoma viruses: the helper-independence reported for a Moloney variant is unconfirmed; distinct strains differ in the size of their RNAs. Virology. 1977 Jan;76(1):295–312. doi: 10.1016/0042-6822(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Maisel J., Klement V., Lai M. M., Ostertag W., Duesberg P. Ribonucleic acid components of murine sarcoma and leukemia viruses. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3536–3540. doi: 10.1073/pnas.70.12.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney J. B. A virus-induced rhabdomyosarcoma of mice. Natl Cancer Inst Monogr. 1966 Sep;22:139–142. [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Aaronson S. A. Complete nucleotide sequence and organization of the Moloney murine sarcoma virus genome. Science. 1981 Oct 23;214(4519):445–450. doi: 10.1126/science.6170110. [DOI] [PubMed] [Google Scholar]
- Rothenberg E., Baltimore D. Increased length of DNA made by virions of murine leukemia virus at limiting magnesium ion concentration. J Virol. 1977 Jan;21(1):168–178. doi: 10.1128/jvi.21.1.168-178.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnic E. M., Goldberg R. J., Parks W. P. A biochemical and genetic analysis of mammalian RNA-containing sarcoma viruses. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):885–895. doi: 10.1101/sqb.1974.039.01.103. [DOI] [PubMed] [Google Scholar]
- Vande Woude G. F., Oskarsson M., McClements W. L., Enquist L. W., Blair D. G., Fischinger P. J., Maizel J. V., Sullivan M. Characterization of integrated Moloney Sarcoma proviruses and flanking host sequences cloned in bacteriophage lambda. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):735–745. doi: 10.1101/sqb.1980.044.01.079. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Beemon K., Vogt P. K. Mapping RNase T1-resistant oligonucleotides of avian tumor virus RNAs: sarcoma-specific oligonucleotides are near the poly(A) end and oligonucleotides common to sarcoma and transformation-defective viruses are at the poly(A) end. J Virol. 1975 Oct;16(4):1051–1070. doi: 10.1128/jvi.16.4.1051-1070.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


