Abstract
Pls1 tetraspanins were shown for some pathogenic fungi to be essential for appressorium-mediated penetration into their host plants. We show here that Podospora anserina, a saprobic fungus lacking appressorium, contains PaPls1, a gene orthologous to known PLS1 genes. Inactivation of PaPls1 demonstrates that this gene is specifically required for the germination of ascospores in P. anserina. These ascospores are heavily melanized cells that germinate under inducing conditions through a specific pore. On the contrary, MgPLS1, which fully complements a ΔPaPls1 ascospore germination defect, has no role in the germination of Magnaporthe grisea nonmelanized ascospores but is required for the formation of the penetration peg at the pore of its melanized appressorium. P. anserina mutants with mutation of PaNox2, which encodes the NADPH oxidase of the NOX2 family, display the same ascospore-specific germination defect as the ΔPaPls1 mutant. Both mutant phenotypes are suppressed by the inhibition of melanin biosynthesis, suggesting that they are involved in the same cellular process required for the germination of P. anserina melanized ascospores. The analysis of the distribution of PLS1 and NOX2 genes in fungal genomes shows that they are either both present or both absent. These results indicate that the germination of P. anserina ascospores and the formation of the M. grisea appressorium penetration peg use the same molecular machinery that includes Pls1 and Nox2. This machinery is specifically required for the emergence of polarized hyphae from reinforced structures such as appressoria and ascospores. Its recurrent recruitment during fungal evolution may account for some of the morphogenetic convergence observed in fungi.
Convergent evolution of trophic strategies and morphogenetic processes is a hallmark of fungi (27). For example, many fungi belonging to different fungal clades are phytopathogenic, whereas their close relatives are saprobic. These plant parasitic fungi may penetrate into their host plants using a specialized structure called the appressorium. Appressoria are attached to the surface of plants and redirect fungal growth into the host tissues underneath through the formation of a penetration peg. Due to their importance in pathogenicity, appressoria are studied in fungi such as Magnaporthe grisea, a devastating pathogen of rice (25, 54), Botrytis cinerea, a wide-host-range plant pathogen (19), and Colletotrichum lindemuthianum, an important pathogen of beans (13). These three fungi belong to two different clades of ascomycetes. M. grisea and C. lindemuthianum are in the Sordariomycetes and B. cinerea in Leotiomycetes. A significant number of species related to these three fungi are saprobic. Which of the parasitic or saprobic lifestyles displayed by these organisms arise by convergent evolution is thus crucial to understand their evolution. An answer to this question is to establish whether appressoria are homoplasious or homologous in Sordariomycetes and Leotiomycetes and whether appressorium-specific processes may also be found in saprobic fungi.
Appressoria of M. grisea, C. lindemuthianum, and B. cinerea, have different morphologies, cellular organizations, and ontologies. In M. grisea and C. lindemuthianum, appressoria are differentiated at the tip of germ tubes issued from asexual spores on contact with a plant surface. These appressoria are dome-shaped single cells that are heavily melanized. It has been shown that collapse of the spore and transfer of its content to the appressorium are necessary for functionality in M. grisea (49). In B. cinerea, appressoria are also differentiated at the tip of germ tubes issued from asexual spores on contact with a plant surface. However, this structure is also called a pseudo-appressorium because it is not an individualized cell. Indeed, B. cinerea pseudo-appressoria are still connected to the spore from which they are derived without the formation of a septum. Furthermore, they are not heavily melanized, although they display a reinforcement of their cell wall (19). This observation suggests that these two types of appressoria have evolved convergently: i.e., appressoria from Leotiomycetes and Sordariomycetes are homoplasious. Alternatively, they could be homologous structures despite these morphogenetic differences. In support of the homology hypothesis, it has been shown that appressoria from these three fungi require a functional tetraspanin gene to infect host plants (11, 19, 51). Indeed Mgpls1−, Clpls1−, and Bcpls1− deletion mutants are nonpathogenic. They are all specifically impaired in the production of the penetration peg, while their appressoria appear normal. These data suggest an evolutionary conservation of appressorium physiology among these fungi and therefore homology of appressoria instead of homoplasy. However, the recurrent use of the Pls1 tetraspanins during fungal evolution for a function specific to appressoria cannot be ruled out.
Tetraspanins are eukaryotic small integral membrane proteins identified in animals and fungi (26, 50). At least 33 distinct tetraspanins were identified in humans, 37 in Drosophila melanogaster, and 20 in Caenorhabditis elegans, but only 3 in fungi: Pls1 and Tps2 in basidiomycetes and Pls1 and Tps3 in ascomycetes (26, 31). In M. grisea, Pls1 is expressed in the appressoria and localizes either at the plasma membrane or in vacuoles (11), while it is expressed in both appressoria and vegetative mycelium in C. lindemuthianum (51) and in conidia, germ tubes, and appressoria during host penetration in B. cinerea (19). Although these proteins display limited sequence similarities, they share conserved secondary structures including four transmembrane domains and a cysteine based pattern in their EC2 large extracellular loop (42, 48). In animals, tetraspanins act as molecular adaptors involved in the formation of protein complexes localized in tetraspanin-enriched microdomains of membranes (23), called the “tetraspanin web” (7). Animal tetraspanins are involved in different biological processes including sperm-egg fusion, entry of parasites or viruses into host cells, cell-cell interactions in central nervous system or immune system (B-cell-T-cell immunological synapse), cell adhesion, motility, polarity, and trafficking of membrane proteins (23, 33).
Surprisingly, additional Pls1 orthologues were identified in saprobic fungi devoid of appressorium such as Neurospora crassa (20), Coprinopsis cinerea (26), Chaetomium globosum, Podospora anserina, Trichoderma reesei, and Phanerochete chrysosporium (31). To date, the role of Pls1 tetraspanins in these nonpathogenic fungi is unknown. In this report, we used the model ascomycete Podospora anserina, a coprophilous fungus closely related to N. crassa, to study the role of Pls1 in a saprobic fungus devoid of appressorium. The phenotype of the PaPls1 deletion mutant revealed a crucial role for this gene in germination of ascospores, which are heavily melanized in this species. Interestingly, the germination of nonmelanized M. grisea ascospores is unaffected in the MgPLS1 deletion mutant. These data suggest that the germination of P. anserina melanized ascospores and the formation of the penetration peg in M. grisea melanized appressoria share homologous determinants. Our data thus provide new insights indicating that the ability to generate a peg through a pore of a reinforced structure (including appressoria and ascospores) may be gained by convergent evolution through the recruitment of a highly specialized homologous machinery, providing a strong argument for the recurrent convergent evolution of morphogenetic processes in fungi.
MATERIALS AND METHODS
Strains and culture conditions.
The P. anserina S (big S) strain (39) used for this study has its genome sequence available at http://podospora.igmors.u-psud.fr (17). The pks1-193 (formerly 193) P. anserina big S strain carries a mutation in a polyketide synthase gene required for melanin biosynthesis. It displays nonpigmented mycelia, perithecia, and ascospores (12, 38). Standard culture conditions, media, and genetic methods were described previously (40). Menadione was added at 10−4 to 10−6 M, t-butyl-hydroxyperoxide at 10−5 to 10−7 M, and H2O2 at 0.25, 0.05, and 0.01%. P. anserina is pseudohomothallic, producing homokaryotic and heterokaryotic mycelia from small and large ascospores, respectively.
An s (small s) Δmus51::phleoR strain was kindly provided by C. Sellem. It has undergone deletion of its mus51 gene, encoding the Ku70 protein involved in nonhomologous end-joining DNA repair, and has an increased frequency of targeted gene replacement (16). The Δmus51::phleoR mutation was introduced in the S strain by 10 successive backcrosses. The phleomycin marker of the Δmus51::phleoR big S mutant was replaced by the su8-1 marker using homologous recombination, and Δmus51::su8-1 big S strains of both mating types were constructed. These Δmus51::su8-1 strains also displayed a high frequency of targeted gene replacement (see below). Δmus51::su8-1 strains were crossed with pks1-193 mutants of the opposite mating type to recover pks1-193 Δmus51::su8-1 mat+ and pks1-193 Δmus51::su8-1 mat− progeny.
DNA manipulation and deletion of P. anserina PaPls1 by targeted gene replacement.
DNA was extracted as described previously (32), and standard protocols were followed for DNA manipulation (2). The upstream (LB [700 bp]) and downstream (RB [750 bp]) regions of PaPls1 were obtained by PCR amplification using P. anserina big S strain genomic DNA as a template and primer pairs Pls1-L1/Pls1-L2-SfiIa and Pls1-R1-SfiIb/Pls1-R2, respectively (see Table S1 in the supplemental material). The hygromycin B resistance cassette (hph) was obtained by digesting the plasmid pFV8 (52) with SfiI (hph/SfiI). The amplified 700-bp LB and 750-bp RB fragments from the PaPls1 locus were digested with SfiI and ligated with T4 DNA ligase (Roche) to an hph/SfiI fragment. The amplification of the 2.8-kb PaPls1 replacement cassette was obtained using primers Pls1-L3 and Pls1-R3 and the previous ligation product as a template. This final PCR fragment was used to transform P. anserina protoplasts from pks1-193 Δmus51::su8-1 mat+ and pks1-193 Δmus51::su8-1 mat− strains as described previously (9). Three hygromycin B-resistant transformants (hygR) were obtained, one mat− and two mat+. These primary transformants were crossed to a wild-type strain, and progeny with pigmented ascospores lacking pks1-193 or pks1-193 Δmus51::su8-1 mutations were recovered. ΔPaPls1::hph mutants were distinguished from Δmus51::su8-1 ΔPaPls1::hph mutants by the ability of su8-1 to suppress of the lack of pigmentation of pks1-193 ascospores in additional crosses with a pks1-193 mutant. Progeny lacking pks1-193 and Δmus51::su8-1 were obtained, and the replacements at the PaPls1 locus were assessed by Southern hybridization as depicted in Fig. S1 in the supplemental material. Two PaPls1 deletion mutants, one mat+ and one mat−, were selected for further studies.
Complementation of PaPls1 deletion mutants with wild-type PaPls1 and phenotypic analyses.
Plasmid pCM421-8.5 carries the M. grisea MgPLS1 wild-type allele (11). Green fluorescent protein (GFP) was fused to the C terminus of MgPLS1 under the control of MgPls1 promoter and terminator. NcoI and XbaI sites were introduced by PCR just before the Stop codon of MgPls1 by using a 937-bp subclone from pCM421-8.5 as a template. pEGFP (Clontech, Ozyme) was used to obtain a PCR product containing an NcoI site at the start of the GFP open reading frame (ORF) and a XbaI site at the end of the GFP ORF. This PCR product was fused just before the Stop codon of the modified MgPls1-937-bp NruI subclone and reintroduced as an NruI fragment into pCM421-8.5 to obtain the plasmid pCM421-8.5-MgPls1-GFP. This plasmid and the pBC-phleo plasmid conferring resistance to phleomycin (43) were used to cotransform the ΔPaPls1 mat− mutant.
P. anserina life span and crippled growth phenotypes were assessed as previously described (45, 46). Hyphal interference, peroxide accumulation, and cell death at mycelial confrontation zones was assayed as described previously (44). Perithecium formation, fertility in crosses, and ascospore germination were assessed as described previously (36). M. grisea crosses were performed as described previously (47). Four Mgpls1::hph progenies from the cross between the original punchless mutant (Mgpls1::hph) (11) and a wild-type M. grisea strain (M4) of the opposite mating type were used to perform two wild-type × Mgpls1::hph and two Mgpls1::hph × Mgpls1::hph crosses. Eighteen days after mycelium confrontation, three perithecia from each cross were deposited on water agar (4.5% agar) and opened with a sterile scalpel under a stereoscopic binocular to liberate asci. After 16 h on water agar at 26°C, ascospore germination was observed.
Wounded barley leaf fragments (cv. Plaisant) were inoculated with 50-μl droplets of spore suspension (3.105 spores/ml) from M. grisea wild-type strain P1.2 or the ΔMgpls1 P1.2 mutant as described previously (18). Tricyclazole stock solution (Ehrenstorfer GmbH, Augsburg, Germany) was mixed with spores at a final concentration of 20 μg/ml before inoculation.
Phylogenetic analysis.
Protein sequences were aligned using ClustalX 1.8 and transferred to GenDoc for visualization. This alignment was used to construct a phylogenetic tree using the maximum likelihood method (PHYML software) (21, 22) and transferred to Mega3.1 for visualization (30). Bootstrap values are expressed as percentages of 100 replicates.
RESULTS
Identification of the Pls1 tetraspanin-encoding gene from P. anserina.
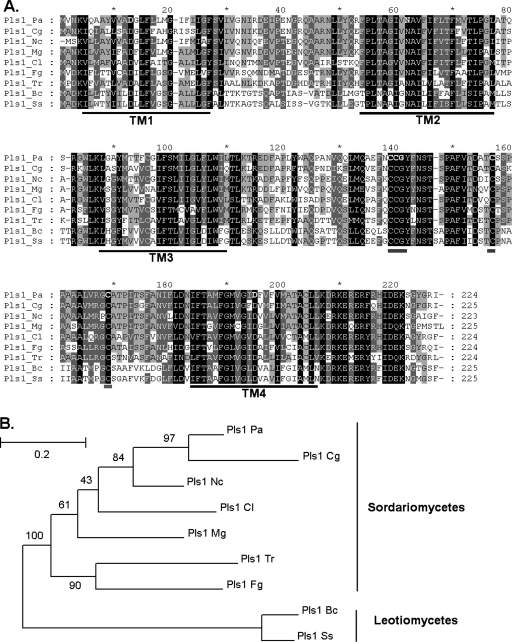
A single MgPls1 hit (Pa_1_19270; E value, e−76) was identified in the P. anserina predicted proteome (http://podospora.igmors.u-psud.fr) (17) using BlastP (1) and designated as PaPls1. The PaPls1 protein sequence displayed 65% identity and 83% similarity to MgPls1 (Fig. 1A). PaPls1 is located on P. anserina chromosome 1 close to the mating-type locus in a region devoid of recombination. Eighteen PaPls1 expressed sequence tags (ESTs) were identified (P. anserina EST database) (17), including 13 ESTs from mycelia, 4 from developing fructifications (perithecia), and 1 from germinating ascospores. The recovery of PaPls1 ESTs obtained from different tissues suggests that this gene is transcribed throughout the life cycle of the fungus. These ESTs showed that the exon/intron boundaries predicted in Pa_1_19270 were correct. PaPls1 displays two exons of 402 bp and 273 bp, interrupted by an intron of 76 bp. MgPLS1 has an additional intron (20).
FIG. 1.
Alignment and phylogenetic tree of Sordariomycetes and Leotiomycetes Pls1 tetraspanins. (A) Pls1 proteins from Sordariomycetes and Leotiomycetes display numerous conserved motifs in transmembrane domains (TM1 to -4 [underlined in black]), in ECL2 including a cysteine pattern (underlined in gray), and in the C-terminus tail. Protein sequences were aligned using ClustalX 1.8. Conserved amino acids are boxed in black (identical) and gray (similar). (B) PaPls1 clusters with CgPls1 and NcPls1, as expected, since the corresponding fungal species are closely related. The phylogenetic tree was constructed using the previous alignment (A) and a maximum likelihood method (PHYML) with BcPls1 and ScPls1 as outgroups. Bootstrap values are expressed as percentage of 100 replicates. Abbreviations for Sordariomycetes: Pls1_Pa, Podospora anserina Pa_1_19270; Pls1_Cg, Chaetomium globosum CHGG_06472.1; Pls1_Nc, Neurospora crassa AJ504996; Pls1_Cl, Colletotrichum lindemuthianum AJ504995; Pls1_Mg, Magnaporthe grisea AX058239; Pls1_Fg, Fusarium graminearum FG08695.1; Pls1_Tr, Trichoderma reesei jgi Trire2 4514 fgenesh1_pm.C_scaffold_13000033. Abbreviations for Leotiomycetes: Pls1_Bc, Botrytis cinerea AJ504994; Pls1_Ss, Sclerotinia sclerotiorum SS1G_05586.1.
Analysis of the PaPls1 protein sequence with TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) revealed four transmembrane segments (TM1 to -4) that delimit a small extracellular loop (EC1), a small intracellular loop (IC), a large extracellular loop (EC2), and a short cytoplasmic tail similar in size and locations (Fig. 1A) as Pls1 tetraspanins (20). PaPls1 TMs have charged/polar amino acids conserved among fungal tetraspanins, and PaPls1 ECL2 displayed the same cysteine-based pattern as Pls1 tetraspanins (CCG-X14-C-X10-C [Fig. 1A]). Finally, the C-terminal tail of PaPls1 is almost identical (>70% identity [Fig. 1A]) to the conserved C-terminal tail sequence of Pls1 tetraspanins (20). This analysis shows that PaPls1 has all the structural hallmarks of Pls1 tetraspanins (20, 31, 50), suggesting that it corresponds to a functional protein.
Fungal Pls1 tetraspanins identified in the ascomycetes B. cinerea, C. lindemuthianum, N. crassa, and Gibberella zeae (20, 26, 50) were aligned with PaPls1. The CgPls1 tetraspanin was identified in the genome of Chaetomium globosum (CHGG_06472.1 at http://www.broad.mit.edu/annotation/genome/chaetomium_globosum/Home.html), and its protein sequence was aligned with those of Pls1 tetraspanins. This alignment was used to construct a phylogenetic tree using the maximum likelihood method (PHYML) (22). This phylogenetic tree (Fig. 1B) shows that PaPls1 is highly similar to CgPls1, as expected since C. globosum is a fungal species closely related to P. anserina (27). The position of the single PaPls1 intron that is conserved among PLS1 genes from pyrenomycetes (20) and the topology of the phylogenetic tree suggest that Pls1 tetraspanins are encoded by a family of orthologous genes.
Inactivation of PaPls1 leads to a strong defect in ascospore germination.
To investigate the role of PaPls1 in the P. anserina life cycle, we constructed a deletion mutant (ΔPaPls1) by targeted gene replacement. As PaPls1 is closely linked to the mating-type locus in a region devoid of recombination, we constructed ΔPaPls1 deletion mutants in pks1-193 Δmus51::su8-1 strains with different mating types. These ΔPaPls1 mutants of opposite mating types are needed to study the role of PaPls1 in the sexual cycle that requires crosses between ΔPaPls1 mutants. Transformants carrying only the ΔPaPls1 deletion were obtained by crossing and characterized by Southern hybridization (see Fig. S1 in the supplemental material). This analysis revealed that the PaPls1 gene was replaced by the hph resistance cassette in different transformants designated as ΔPaPls1 mat+ and ΔPaPls1 mat−, respectively. ΔPaPls1 mutants are identical to the wild type for vegetative phenotypes, including growth rate, mycelium morphology, life span (37), ability to display crippled growth cell degeneration (45), and hyphal interference (44). ΔPaPls1 mutants were also tested for their ability to perform cell fusion after hyphal anastomosis. ΔPaPls1 mat+ lys2-1 and ΔPaPls1 mat+ leu1-1 mutants unable to grow on minimal medium were constructed by crossing the ΔPaPls1 mat+ strain with auxotrophic mutants. Mycelia from both double mutants were mixed and deposited onto M2 minimal medium. Mycelia able to grow on minimal medium were observed as soon as 24 h after plating, at the same rate as those formed when mixing mycelia from lys2-1 and leu1-1 strains. These results demonstrate that hyphae from ΔPaPls1 mat+ lys2-1 and ΔPaPls mat+ leu1-1 strains are fusing and forming heterokayotic hyphae growing on minimal medium, as observed for the wild type.
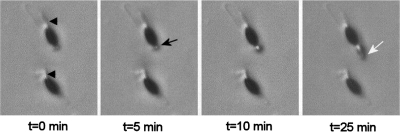
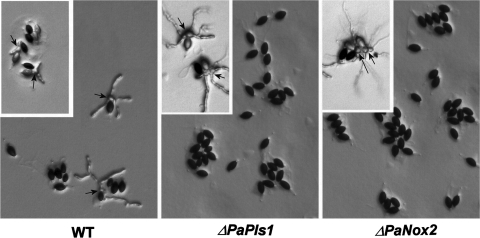
Crosses between ΔPaPls1 mat+ and ΔPaPls1 mat− mutants led to normal sexual fructifications (perithecia) with mature ascospores ejected as in wild-type crosses. However, only 1 out of 104 ΔPaPls1 ascospores germinated on the germination-inducing G medium, unlike wild-type ascospores, which germinated at 100% on this medium. On culture media unable to induce germination, such as M2 minimal medium, ΔPaPls1 ascospores germinated at a frequency identical to that of the wild type (10−4). The ΔPaPls1 defect was ascospore autonomous since ascospores from wild-type progeny obtained in wild-type × ΔPaPls1 crosses germinated normally, whereas those from ΔPaPls1 progeny did not. PaPls1+/ΔPaPls1 dikaryotic ascospores had a wild-type germination rate, showing that the ΔPaPls1 defect was recessive. In P. anserina, two cells form the ascospore, a large heavily melanized one and a small hyaline one, called the primary appendage. On G medium, wild-type ascospore germination occurs at a germ pore located at the anterior side of the ascospore (i.e., at the opposite end of the primary appendage) (3, 35). The first stage of germination is the extrusion of a spherical structure from the pore that reaches up to one-third of the size of the ascospore (Fig. 2), called the “germination peg.” This extrusion starts a few hours after the deposition of ascospores on the inducing medium (usually between 4 and 10 h, depending on the batch of germination medium) and is completed in 10 to 15 min (Fig. 2). Immediately after this extrusion, new polarized hyphae originate from the germination peg within a few minutes and develop into mycelia. In the ΔPaPls1 mutants, the ascospore germination process is stopped at an early stage since germination pegs are not formed (Fig. 3).
FIG. 2.
Time course of the germination of a wild-type P. anserina ascospore on inducing medium. P. anserina ascospores are bicellular, with one large heavily melanized cell and a small hyaline one, the primary appendage (arrowheads). The germination starts with the extrusion of a peg (black arrow) at the pole opposite to the primary appendage, where the germ pore is located (3). The white arrow points to a newly formed hypha with polarized growth issued from the spherical germination peg. This hypha appears 20 min after the germination peg.
FIG. 3.
Lack of P. anserina ascospore germination in ΔPaPls1 and ΔPaNox2 mutants. Germination of melanized (main panels) and unmelanized (inserts) ascospores on inducing medium. Ascospores from the main panels originate from crosses between wild type and wild type, ΔPaPls1 and ΔPaPls1, and ΔPaNox2 and ΔPaNox2 strains of opposite mating types. Ascospores from the inserts originate from crosses between the wild type and pks1-193 (WT), ΔPaPls1 and pks1-193 ΔPaPls1 (ΔPaPls1), and ΔPaNox2 and ΔPaNox2 pks1-193 (ΔPaNox2) strains. Arrows point to germination pegs. In the main panels, melanized ascospores from ΔPaPls1 and ΔPaNox2 mutants are unable to germinate, while wild-type ascospores do germinate. In the inserts, ΔPaPls1 and ΔPaNox2 melanized ascospores did not germinate, while several ΔPaPls1 and ΔPaNox2 unmelanized ascospores did germinate, demonstrating that the inhibition of melanin biosynthesis suppresses ΔPaPls1 and ΔPaNox2 defects. In the wild-type × pks1-193 cross (WT insert), only the nonmelanized ascospores have germinated when the picture was taken, indicating that ascospores lacking melanin germinate before pigmented ones.
M. grisea MgPLS1 can functionally replace PaPls1.
The ΔPaPls1 mat− strain was cotransformed with a vector carrying the PaPls1 wild-type allele and the pBC-phleo plasmid conferring resistance to phleomycin (43). Eight out of 45 transformants resistant to phleomycin produced ascospores that germinated with 100% efficiency when crossed with a ΔPaPls1 mat+ strain. In the control experiment, transformation was done with an empty vector. All of the 100 transformants obtained produced nongerminating ascospores. This result demonstrates that the ascospore germination defect of ΔPaPls1 is due to the inactivation of PaPls1. The ΔPaPls1 mat− mutant was also cotransformed with pBC-phleo plasmid and pCM421-8.5-MgPls1-GFP plasmid that expressed an MgPLS1-GFP fusion protein. The transformants resistant to phleomycin were crossed with the ΔPaPls1 mat+ mutant, and the resulting ascospores were assessed for germination on inducing medium. Two phleomycin-resistant transformants among 48 displayed a 100% ascospore germination frequency on inducing medium similar to that of the wild type. Both expressed a fluorescent protein, demonstrating that MgPLS-GFP complements the ΔPaPls1 mutation. Therefore, MgPLS1 is a functional orthologue of PaPls1.
The M. grisea ΔMgpls1 mutant has a normal ascospore germination efficiency.
Since ΔPaPls1 mutants have a strong defect in ascospore germination, we assessed the germination efficiency of asexual spores and ascospores from an M. grisea MgPLS1 deletion mutant (ΔMgpls1). Asexual spores from the ΔMgpls1 mutant germinated normally. The ΜgPLS1 original insertion mutant (Mgpls1::hph), which displays the same pathogenicity defect as the ΔMgpls1 mutant, was previously crossed to wild-type M. grisea strain M4 (11). Their progeny were analyzed for their resistance to hygromycin B (Mgpls1::hph), pathogenicity, and mating type by performing novel Mgpls1::hph × wild-type crosses (11). The recovery of nonpathogenic progeny resistant to hygromycin (Mgpls1::hph genotype) at a frequency (45%) expected for single-gene segregation, suggests that this mutation has no effect on ascospore germination. To confirm this hypothesis, ΔMgpls1 progeny of opposite mating types were crossed, yielding fertile perithecia that produced ascospores with a normal germination rate of 90%. These results show that the inactivation of MgPLS1 has no effect on ascospore germination in M. grisea.
Phenotypes of PaPls1 and PaNox2 deletion mutants are identical.
The mutant with deletion of the PaNox2 gene, which encodes an NADPH oxidase (36), is the only known P. anserina mutant that has the same specific recessive and autonomous defect in ascospore germination as the ΔPaPls1 mutant. NADPH oxidases are membrane enzymes involved in the production of superoxide in a wide range of eukaryotes for signaling or defense purposes (4). Like PaPls1, PaNox2 is expressed throughout the life cycle since nine ESTs for this genes are present in the database (17): six from mycelia and three from perithecia. Ascospores of the ΔPaNox2 mutant are stopped at an early stage of their germination process, since no germination peg is formed as observed for ΔPaPls1 ascospores (Fig. 3). Addition to the germination medium of compounds generating reactive oxygen species such as menadione (a quinone-generating superoxide in the cells), t-butyl-hydroxyperoxide (an inorganic peroxide) or hydrogen peroxide failed to restore ΔPaPls1 ascospore germination. The same result was already observed for ΔPaNox2 ascospores (36).
We have previously shown that ascospores from the double mutant carrying the pks1-193 and the ΔPaNox2 mutations germinate at a high frequency (50%) on both inducing and noninducing media, like ascospores from the pks1-193 mutant (36). These experiments show that pks1-193 mutation suppresses the germination defect of ΔPaNox2 ascospores. The pks1-193 mutants carry a recessive null mutation in a P. anserina gene encoding the polyketide synthase involved in the first step of 1,8-dihydroxynaphthalene (DHN) melanin biosynthesis (12). These mutants are unable to synthesize DHN melanin and display unmelanized hyphae, perithecia, and ascospores. Crosses between pks1-193 ΔPaPls1 mat+ and pks1-193 ΔPaPls1 mat− strains were performed, and the ascospores produced germinated at high frequency (50%) on both inducing and noninducing media. This result shows that the absence of melanin suppressed the ΔPaPls1 ascospore germination defect. Unmelanized pks1-193, pks1-193 ΔPaNox2, or pks1-193 ΔPaPls1 ascospores germinate rapidly after deposition onto the inducing medium. Their germination peg, which is often larger, is produced earlier than in wild-type melanized ascospores (usually 2 to 5 h after being deposited on the petri plates), although it develops in 10 to 15 min as in the wild type (Fig. 3). As in wild-type ascospores, the germination peg from these mutants switches to polarized growth and differentiated filamentous hyphae.
In M. grisea, the inhibition of the DHN melanin biosynthesis, either genetically in melanin-deficient mutants or chemically using inhibitors such as tricyclazole, leads to nonmelanized appressoria unable to penetrate into intact host tissues (10, 14, 24). However, M. grisea melanin-deficient mutants affected in the Alb, Rsy, and Buf genes cause lesions similar to the wild type when inoculated on wounded leaves, presumably as a consequence of their ability to penetrate through wounds. The pathogenicity defect of the ΔMgpls1 mutant differs strongly from that of melanin-deficient mutants as ΔMgpls1 mutants are unable to penetrate both unwounded and wounded leaves (11). ΔMgpls1 spores treated with tricyclazole differentiated unmelanized appressoria but were unable to infect intact or wounded barley leaves (no lesions [data not shown]). Therefore, the inhibition of melanin biosynthesis does not suppress the penetration and colonization defects of ΔMgpls1.
Co-occurrence of PLS1 and NOX2 genes in fungal genomes.
To further support the hypothesis that both PLS1 and NOX2 genes are involved in the same pathway, we have searched in available sequences of fungal genomes for the co-occurrence of these two genes. As seen in Table S2 in the supplemental material, whereas NOX2 was found in Batrachochytrium dendrobatidis, a Chytridiomycota, PLS1 seems to be present only in higher fungi (i.e., the Dikarya). Strikingly, in higher fungi, NOX2 and PLS1 are either both present, as in P. anserina, M. grisea, and N. crassa, or both absent, as in Aspergillus or Mycosphaerella. The absence of PLS1/NOX2 likely results from losses in different clades of ascomycete and basidiomycete species. The absence of these two genes seems to rely on independent events since they are not located on the same chromosomal locus in fungi. The loss of PLS1/NOX2 has occurred repeatedly in both ascomycete and basidiomycete species, especially in lineages with species living mostly as yeasts, such as Taphrinomycotina and Saccharomycotina yeasts and Sporobolomyces roseus. Fungal species that have both PLS1 and NOX2 differentiate either melanized appressoria or melanized ascospores germinating through a pore, while fungi lacking PLS1/NOX2, such as Aspergillus, Ustilago maydis, and Cryptococcus neoformans, mostly behave as yeast cells or form melanized filamentous hyphae (53, 56), without being able to differentiate reinforced structures germinating through a pore. These data strongly suggest that they have been maintained during evolution as a consequence of their involvement in the same cellular function.
DISCUSSION
Phenotype of P. anserina ΔPaPls1 mutants.
P. anserina is a fungus that lives as a saprobe on herbivore dung, does not differentiate appressoria, and does not attack plants. Nevertheless, it possesses an expressed gene coding for a tetraspanin of the Pls1 family. Thanks to its ease of handling, this fungal species is well suited to study the role of Pls1 tetraspanin in general cellular processes other than pathogenicity on plants. Most animal tetraspanins are involved in cell fusion (e.g., Cd9) (23) or in cell-cell interactions (e.g., Cd81) (33). Therefore, it was important to assess if PaPls1 has a role in either sexual or vegetative cell fusion. Our results show that PaPls1 is not required for sexual and vegetative cell fusion (anastomosis), as indicated by the behavior of PaPls1 null mutants in cell fusion during sexual crosses or during the formation of vegetative heterokaryons. ΔPaPls1 mutants also exhibited normal hyphal interference, indicating that these mutants interact normally with other fungi and were similar to the wild type with regard to mycelial growth on different media, senescence, and crippled growth cell degeneration (29, 37, 45). Following fertilization, PaPls1 null mutants have a normal sexual process, as they matured fructifications like the wild type. However, we showed that PaPls1 has an important role in the life cycle of P. anserina, as it is required for the germination of melanized ascospores. This defect is the only phenotype we observed in PaPls1 deletion mutants.
Roles of Pls1 tetraspanins in P. anserina and M. grisea.
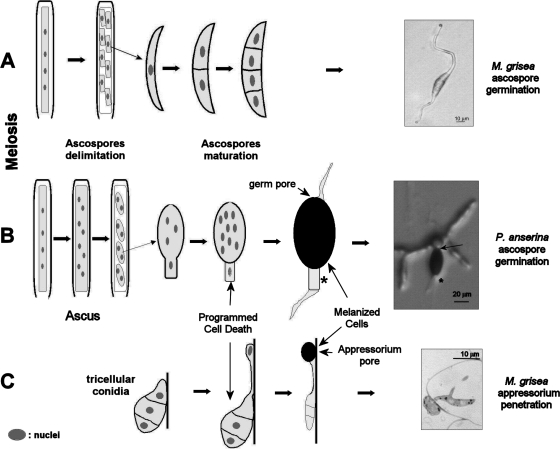
MgPLS1 and PaPls1 genes are functional orthologues, since MgPLS1 fully complements the germination defect of the ΔPaPls1 null mutant. Yet, MgPLS1 is required for the formation of the penetration peg originating at the pore of M. grisea melanized appressorium (11), while it is required for ascospore germination in P. anserina. We have verified that asexual spores and sexual ascospores from the M. grisea Mgpls1 deletion mutant germinate as efficiently as the wild type, as this phenotype was not reported previously (11). Therefore, the phenotypes in both fungal species strongly differ. While P. anserina does not differentiate appressoria, both species produce ascospores. However, it should be noted that ascospores from these related fungal species are quite different (Fig. 4). M. grisea ascospores are tetracellular nonmelanized structures (28, 55), while P. anserina ascospores are bicellular structures with only one cell being heavily melanized (3). In M. grisea, ascospore germination is similar to vegetative spore germination, with germ tubes frequently originating without latency at opposite tips of a single ascospore on any media, including water agar (Fig. 4). On the contrary, P. anserina ascospores germinate only on specific inducing media at a germ pore (3) (Fig. 4) and germination starts by the extrusion of a spherical isotropic cell, the germination peg, which further differentiates into polarized hyphae (Fig. 2). Our results thus suggest that Pls1 tetraspanins are involved in a cellular process specifically required for the germination of the melanized ascospores from P. anserina that is not needed for the germination of the nonmelanized ascospores from M. grisea. The P. anserina ascospore structure and mode of germination may be dictated by the necessity for these cells, unlike those of M. grisea, to be resting and melanized in order to withstand their passage through the digestive tract of herbivores. The relief from this resting stage requires a trigger mimicked by the specific nutritional conditions of the germination medium. One can hypothesize that, in nature, this trigger would be the passage through a digestive tract.
FIG. 4.
Ontogeny of ascospore and appressorium in P. anserina and M. grisea. (A) Formation of ascospores in M. grisea results in four-cell hyaline ascospores that germinate spontaneously (55). Ascospores were observed 16 h after deposition onto water agar (4.5% agar) at 26°C. The picture shows the germination of a wild-type ascospore that occurred at two opposite tips of the spore. (B) Formation of ascospore in P. anserina results in two-celled ascospores (3). One cell undergoes enlargement and melanization, while the other undergoes programmed cell death to form the primary appendage (*). Ascospores were observed 4 h after deposition onto germination medium at 27°C. Germination proceeds only on a special medium rich in ammonium acetate through the germ pore (arrow), which is located opposite of the dead cell. The picture shows the germination of a typical wild-type ascospore with its germination peg. (C) Appressoria are formed at the tip of the germ tube issued from the germination of a three-cell spore produced by asexual reproduction on contact with an hydrophobic hard surface (8). The single-cell appressorium strongly adheres to the plant surface and penetrates its host through the formation of a penetration peg at a basal pore as depicted in the picture.
Comparison of PaPls1 and PaNox2 mutant defects.
In P. anserina, the NADPH oxidase 2 (PaNox2) is essential for ascospore germination (36). Like PaPls1, PaNox2 is expressed throughout the life cycle. Nevertheless, the phenotypes of the ΔPaPls1 and ΔPaNox2 deletion mutants are similar and restricted to the same early stage of the germination of melanized ascospores. A possibility is that both genes are required during vegetative growth only when P. anserina encounters special conditions in nature not present on petri plates. Attempts to assay the superoxide burst in ΔPaPls1 ascospores, as previously described for the wild type and ΔPaNox2 mutant (36), were unsuccessful, as this assay proved highly variable (P. Silar, unpublished data). Therefore, it was not possible to test whether PaPls1 is necessary to generate a superoxide burst in ascospores or not. Similarly, the identities of phenotypes exhibited by the two mutants prevented us from performing epistasis analysis. Further experiments are thus needed to know whether PaPls1 and PaNox2 are either involved in the same pathway or in different pathways controlling the same morphogenetic process. Recent studies of orthologues of PaNox2 in M. grisea (MgNOX2) and B. cinerea (BcNOX2) have shown that these genes are also required for appressorium-mediated penetration (15, 41). The phenotypes of NOX2 and PLS1 null mutants are similar in these two fungi (penetration-deficient appressoria), supporting our hypothesis that they are involved in the same specific cellular process. This hypothesis is reinforced by the fact that both PLS1 and NOX2 genes have been conserved in the same species during fungal evolution.
ΔPaPls1 and ΔPaNox2 ascospore germination defect is suppressed by the inhibition of melanin biosynthesis.
The germination of nonmelanized ascospores from the pks1-193 mutant occurs on both inducing and noninducing media (36). Then, on germination medium, it carries on as in wild-type melanized ascospores. The only observed differences between the pks1-193 mutant and the wild type are that the emergence of the germination peg happens earlier and that this cellular extrusion reaches a larger size in nonmelanized ascospores. Therefore, the absence of melanin mainly facilitates germination of ascospores, either by triggering the germination process or relieving its inhibition, but does not change the morphogenetic process itself. We have shown that the melanin deficiency of the P. anserina pks1-193 mutant suppressed the ΔPaPls1 ascospore germination defect, as observed for ΔPaNox2 (36). These results further confirm that PaPls1 and PaNox2 are involved in a cellular process specifically required for the germination of melanized ascospores. They could be either required for the triggering of ascospore germination on inducing medium or required after this trigger for proper germination. One simple hypothesis compatible with these observations is that PaPls1 and PaNox2 are both involved in the same pathway responsible for the weakening of the germ pore in melanized ascospores, which is not needed in nonmelanized ascospores. For example, this pathway could control the inhibition of the deposition of the melanin layer at the pore or promote the degradation of the pore cell wall. According to this hypothesis, ΔPaPls1 and ΔPaNox2 melanized ascospores could have a pore cell wall that cannot be degraded, preventing the extrusion of the germination peg.
The fact that the defect in appressorium-mediated penetration of the M. grisea MgPLS1 null mutant is not suppressed by the absence of melanin is not too surprising. Indeed during P. anserina ascospore germination, a physical barrier does not impair the extrusion of the peg, at least on agar media, allowing the germination peg to be easily extruded from the nonmelanized ascospore. On the contrary, in M. grisea, appressoria are firmly attached to the plant surface and the penetration peg has to breach a strong physical barrier to “germinate.” This is impossible in nonmelanized appressoria and likely explains the lack of suppression of the Mgpls1− mutant defect by melanin inhibition. Melanin-deficient mutants are still able to attack wounded leaves by an unknown mechanism. This was not observed in melanin-deficient MgPLS1 null mutants that are unable to attack wounded leaves.
Comparison of P. anserina ascospore germination and M. grisea appressorium-mediated penetration.
Since the only defects observed in P. anserina ΔPaPls1 and ΔPaNox2 mutants as well as M. grisea ΔMgpls1 and ΔMgnox2 mutants are, respectively, the lack of ascospore germination and the absence of formation of the penetration peg in mature appressoria, we tentatively suggest that these two apparently different morphogenetic processes have recruited homologous genes (Fig. 4). Many additional lines of evidence support this hypothesis. First, P. anserina ascospores and M. grisea appressoria both “germinate” through the differentiation of, respectively, a germination peg and a penetration peg after an induction. Second, P. anserina ascospores and M. grisea appressoria are heavily melanized cells. In both species, the melanin is deposited between the membrane and the cell wall as a dense continuous layer (3, 14, 24). Third, both structures harbor a pore (i.e., a nonmelanized region with a very thin cell wall). In M. grisea, the pore is located at the base of the appressorium in direct contact with the plant surface. In P. anserina, the germ pore is located at one tip of the ascospore at the opposite end of the primary appendage (3, 35). In M. grisea, melanin acts as a semipermeable membrane retaining osmolytes required for the generation of its high internal turgor pressure (14, 24). This turgor pressure is required for the perforation of plant cuticle and cell wall by the penetration peg. It is not known if P. anserina ascospores generate a turgor pressure. However, P. anserina nonmelanized ascospores are very fragile and frequently burst upon contact with a needle or an agar plate, suggesting that these cells may have some internal turgor.
Additionally, besides these shared morphological characteristics, P. anserina ascospore germination and M. grisea appressorium peg formation rely on similar physiological processes such as the catabolism of lipids. M. grisea Pth2 peroxisomal carnitine acetyltransferase (5) and Pex6 protein involved in peroxisome assembly (5) are required for the degradation of lipids stored in the appressorium, and the mutants inactivated for these genes cannot build up sufficient turgor pressure to efficiently penetrate the host plant. In P. anserina, peroxisome-deficient mutants are also quantitatively impaired in ascospore germination, likely through a defect in lipid catabolism (6). Another striking feature of P. anserina ascospore formation is the fact that the ascospore starts as a bicellular structure. Soon, one cell dies and its cytoplasmic content disappears (3). This is reminiscent of the collapse of the spore associated with the formation of M. grisea appressorium (49). Additionally, autophagy genes such as ATG8 (49) and ATG1 (34) are required for appressorial lipid catabolism, turgor pressure buildup, and appressorium-mediated penetration in M. grisea. In P. anserina, similar autophagy mutants display a low ascospore germination rate (C. Clavé, personal communication).
C. lindemuthianum appressoria are less studied than those of M. grisea. Their ontogeny, morphology, and physiology suggest that they are homologous to M. grisea appressoria. B. cinerea appressoria derive from the swelling of a germ tube tip and are less melanized. One common determinant among these apparently different appressoria is the differentiation of a penetration peg, whose formation and penetration of host tissues are controlled by PLS1 and NOX2. Melanized ascospores that germinate through a pore and appressoria that differentiate a penetration peg are produced by a large number of fungi, likely through convergent evolution, as they are scattered throughout the fungal kingdom. The presence of PLS1/NOX2 in these fungal species may result from repetitive convergent selection of the same machinery used for the “germination” of apparently very different cells. The ecological factors involved in the selection of such complex structures and their associated “germination” processes may be an advantage for saprobes to resist herbivore digestive tracts or harsh conditions and for pathogens to enter rapidly into host plants by breaching the plant cuticle and cell wall. The recurrent but discontinuous use of the same homologous machinery in unrelated species may have facilitated this convergent evolution observed either at the level of spore morphology (presence versus absence of melanin and germination pores) or through lifestyle-associated morphogenetic processes (presence versus absence of melanized appressoria) in Eumycota, as observed for other morphogenetic characters in the fungal tree of life (27).
Supplementary Material
Acknowledgments
We thank Sylvie François, Magali Prijeant, and Joëlle Milazzo for expert technical assistance. We also thank Dominique Job for critical reading of the manuscript.
Philippe Silar's laboratory was supported by funding from CNRS/Université de Paris Sud (France). Karine Lambou was supported by a fellowship from Bayer CropScience (Lyon, France).
Footnotes
Published ahead of print on 29 August 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 3.Beckett, A., R. Barton, and I. M. Wilson. 1968. Fine structure of the wall and appendage formation in ascospores of Podospora anserina. J. Gen. Microbiol. 5389-94. [DOI] [PubMed] [Google Scholar]
- 4.Bedard, K., and K. H. Krause. 2007. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87245-313. [DOI] [PubMed] [Google Scholar]
- 5.Bhambra, G. K., Z. Y. Wang, D. M. Soanes, G. E. Wakley, and N. J. Talbot. 2006. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 6146-60. [DOI] [PubMed] [Google Scholar]
- 6.Boisnard, S., G. Ruprich-Robert, M. Picard, and V. Berteaux-Lecellier. 2004. Peroxisomes, p. 61-78. In R. Bramble and G. A. Marzluf (ed.), The Mycota, vol. III. Springer, Berlin, Germany. [Google Scholar]
- 7.Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell Mol. Life Sci. 581189-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourett, T. M., and R. J. Howard. 1990. In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68329-342. [Google Scholar]
- 9.Brygoo, Y., and R. Debuchy. 1985. Transformation by integration in Podospora anserina. I. Methodology and phenomenology. Mol. Gen. Genet. 200128-131. [Google Scholar]
- 10.Chumley, F. G., and B. Valent. 1990. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol. Plant-Microbe Interact. 3135-143. [Google Scholar]
- 11.Clergeot, P. H., M. Gourgues, J. Cots, F. Laurans, M. P. Latorse, R. Pepin, D. Tharreau, J. L. Notteghem, and M. H. Lebrun. 2001. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 986963-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppin, E., and P. Silar. 2007. Identification of PaPKS1, a polyketide synthase involved in melanin formation and its use as a genetic tool in Podospora anserina. Mycol. Res. 111901-908. [DOI] [PubMed] [Google Scholar]
- 13.Deising, H. B., S. Werner, and M. Wernitz. 2000. The role of fungal appressoria in plant infection. Microbes Infect. 21631-1641. [DOI] [PubMed] [Google Scholar]
- 14.de Jong, J. C., B. J. McCormack, N. Smirnoff, and N. J. Talbot. 1997. Glycerol generates turgor in rice blast. Nature 389244. [Google Scholar]
- 15.Egan, M. J., Z. Y. Wang, M. A. Jones, N. Smirnoff, and N. J. Talbot. 2007. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 10411772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Khoury, R., C. H. Sellem, E. Coppin, A. Boivin, M. F. Maas, R. Debuchy, and A. Sainsard-Chanet. 2008. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53249-258. [DOI] [PubMed] [Google Scholar]
- 17.Espagne, E., O. Lespinet, F. Malagnac, C. Da Silva, J. Jaillon, B. M. Porcel, A. Couloux, B. Segurens, J. Poulain, V. Anthouard, S. Grossetete, H. Khalili, E. Coppin, M. Dequard-Chablat, M. Picard, V. Contamine, S. Arnaise, A. Bourdais, V. Berteaux-Lecellier, D. Gautheret, R. P. de Vries, E. Battaglia, P. M. Coutinho, E. G. J. Danchin, B. Henrissat, R. El Khoury, A. Sainsard-Chanet, A. Boivin, B. Pinan-Lucarre, C. H. Sellem, R. Debuchy, P. Wincker, J. Weissenbach, and P. Silar. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fudal, I., J. Collemare, H. U. Böhnert, D. Melayah, and M.-H. Lebrun. 2007. Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium-mediated penetration. Eukaryot. Cell 6546-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourgues, M., A. Brunet-Simon, M. H. Lebrun, and C. Levis. 2004. The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol. Microbiol. 51619-629. [DOI] [PubMed] [Google Scholar]
- 20.Gourgues, M., P. H. Clergeot, C. Veneault, J. Cots, S. Sibuet, A. Brunet-Simon, C. Levis, T. Langin, and M. H. Lebrun. 2002. A new class of tetraspanins in fungi. Biochem. Biophys. Res. Commun. 2971197-1204. [DOI] [PubMed] [Google Scholar]
- 21.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 22.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemler, M. E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6801-811. [DOI] [PubMed] [Google Scholar]
- 24.Howard, R. J., and M. A. Ferrari. 1989. Role of melanin in appressorium function. Exp. Mycol. 13403-418. [Google Scholar]
- 25.Howard, R. J., and B. Valent. 1996. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50491-512. [DOI] [PubMed] [Google Scholar]
- 26.Huang, S., S. Yuan, M. Dong, J. Su, C. Yu, Y. Shen, X. Xie, Y. Yu, X. Yu, S. Chen, S. Zhang, P. Pontarotti, and A. Xu. 2005. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86674-684. [DOI] [PubMed] [Google Scholar]
- 27.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G. H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schussler, J. E. Longcore, K. O'Donnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443818-822. [DOI] [PubMed] [Google Scholar]
- 28.Kato, H., and T. Yamaguchi. 1982. The perfect state of Pyricularia oryzae Cav. from rice plants in culture. Ann. Phytopathol. Soc. Jpn. 48607-612. [Google Scholar]
- 29.Kicka, S., C. Bonnet, A. K. Sobering, L. P. Ganesan, and P. Silar. 2006. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA 10313445-13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 31.Lambou, K., D. Tharreau, A. Kohler, C. Sirven, M. Marguerettaz, C. Barbisan, A. C. Sexton, E. M. Kellner, F. Martin, B. J. Howlett, M. J. Orbach, and M. H. Lebrun. 2008. Fungi have three tetraspanin families with distinct functions. BMC Genomics 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecellier, G., and P. Silar. 1994. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 25122-123. [DOI] [PubMed] [Google Scholar]
- 33.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5136-148. [DOI] [PubMed] [Google Scholar]
- 34.Liu, X.-H., J.-P. Lu, L. Zhang, B. Dong, H. Min, and F.-C. Lin. 2007. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 6997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mai, S. H. 1976. Morphological studies in Podospora anserina. Am. J. Bot. 63821-825. [Google Scholar]
- 36.Malagnac, F., H. Lalucque, G. Lepere, and P. Silar. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41982-997. [DOI] [PubMed] [Google Scholar]
- 37.Marcou, D. 1961. Notion de longévité et nature cytoplasmique du déterminant de la sénescence chez quelques champignons. Ann. Sci. Nat. Bot. Sér. 12 2653-764. [Google Scholar]
- 38.Picard, M. 1971. Genetic evidence for a polycistronic unit of transcription in the complex locus “14” in Podospora anserina. I. Genetic and complementation maps. Mol. Gen. Genet. 11135-50. [DOI] [PubMed] [Google Scholar]
- 39.Rizet, G. 1952. Les phénomènes de barrage chez Podospora anserina. I. Analyse génétique des barrages entre souches S and s. Rev. Cytol. Biol. Veg. 1351-92. [Google Scholar]
- 40.Rizet, G., and C. Engelmann. 1949. Contribution à l'étude génétique d'un Ascomycète tétrasporé: Podospora anserina (Ces.) Rehm. Rev. Cytol. Biol. Veg. 11201-304. [Google Scholar]
- 41.Segmüller, N., L. Kokkelink, S. Giesbert, D. Odinius, J. van Kan, and P. Tudzynski. 2008. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant-Microbe Interact. 21808-819. [DOI] [PubMed] [Google Scholar]
- 42.Seigneuret, M., A. Delaguillaumie, C. Lagaudriere-Gesbert, and H. Conjeaud. 2001. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 27640055-40064. [DOI] [PubMed] [Google Scholar]
- 43.Silar, P. 1995. Two new easy-to-use vectors for transformations. Fungal Genet. Newsl. 4273. [Google Scholar]
- 44.Silar, P. 2005. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol. Res. 109137-149. [DOI] [PubMed] [Google Scholar]
- 45.Silar, P., V. Haedens, M. Rossignol, and H. Lalucque. 1999. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics 15187-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silar, P., and M. Picard. 1994. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J. Mol. Biol. 235231-236. [DOI] [PubMed] [Google Scholar]
- 47.Silué, D., D. Tharreau, N. J. Talbot, P.-H. Clergeot, J.-L. Notteghem, and M.-H. Lebrun. 1998. Identification and characterization of apf1− in a non-pathogenic mutant of the rice blast fungus Magnaporthe grisea which is unable to differentiate appressoria. Physiol. Mol. Plant Pathol. 53239-251. [Google Scholar]
- 48.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2003. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28106-112. [DOI] [PubMed] [Google Scholar]
- 49.Veneault-Fourrey, C., M. Barooah, M. Egan, G. Wakley, and N. J. Talbot. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312580-583. [DOI] [PubMed] [Google Scholar]
- 50.Veneault-Fourrey, C., K. Lambou, and M. H. Lebrun. 2006. Fungal Pls1 tetraspanins as key factors of penetration into host plants: a role in re-establishing polarized growth in the appressorium? FEMS Microbiol. Lett. 256179-184. [DOI] [PubMed] [Google Scholar]
- 51.Veneault-Fourrey, C., D. Parisot, M. Gourgues, R. Lauge, M. H. Lebrun, and T. Langin. 2005. The tetraspanin gene ClPLS1 is essential for appressorium-mediated penetration of the fungal pathogen Colletotrichum lindemuthianum. Fungal Genet. Biol. 42306-318. [DOI] [PubMed] [Google Scholar]
- 52.Villalba, F., J. Collemare, P. Landraud, K. Lambou, V. Brozek, B. Cirer, D. Morin, C. Bruel, R. Beffa, and M. H. Lebrun. 2008. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet. Biol. 4568-75. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., P. Aisen, and A. Casadevall. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 633131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, J. R., X. Zhao, and R. A. Dean. 2007. From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv. Genet. 57175-218. [DOI] [PubMed] [Google Scholar]
- 55.Yaegashi, H., and T. T. Hebert. 1976. Perithecial development and nuclear behavior in Pyricularia. Phytopathology 66122-126. [Google Scholar]
- 56.Youngchim, S., R. Morris-Jones, R. J. Hay, and A. J. Hamilton. 2004. Production of melanin by Aspergillus fumigatus. J. Med. Microbiol. 53175-181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.