Abstract
Proteins entering the eukaryotic secretory pathway commonly are glycosylated. Important steps in this posttranslational modification are carried out by mannosyltransferases. In this study, we investigated the putative α-1,2-mannosyltransferase AfMnt1 of the human pathogenic mold Aspergillus fumigatus. AfMnt1 belongs to a family of enzymes that comprises nine members in Saccharomyces cerevisiae but only three in A. fumigatus. A Δafmnt1 mutant is viable and grows normally at 37°C, but its hyphal cell wall appears to be thinner than that of the parental strain. The lack of AfMnt1 leads to a higher sensitivity to calcofluor white and Congo red but not to sodium dodecyl sulfate. The growth of the mutant is abrogated at 48°C but can be restored by osmotic stabilization. The resulting colonies remain white due to a defect in the formation of conidia. Electron and immunofluorescence microscopy further revealed that the observed growth defect of the mutant at 48°C can be attributed to cell wall instability resulting in leakage at the hyphal tips. Using a red fluorescence fusion protein, we localized AfMnt1 in compact, brefeldin A-sensitive organelles that most likely represent fungal Golgi equivalents. The tumor necrosis factor alpha response of murine macrophages to hyphae was not affected by the lack of the afmnt1 gene, but the corresponding mutant was attenuated in a mouse model of infection. This and the increased sensitivity of the Δafmnt1 mutant to azoles, antifungal agents that currently are used to treat Aspergillus infections, suggest that α-1,2-mannosyltransferases are interesting targets for novel antifungal drugs.
The pathogenic mold Aspergillus fumigatus is currently the major cause of airborne fungal infections that menace immunocompromised patients. Its conidia are distributed through the air, and it has been estimated that humans inhale several hundred a day. In the immunocompetent host, invading fungal conidia are eliminated by innate immune cells in the lung, but in cases of severe immunosuppression some A. fumigatus conidia may escape the impaired immune response. After germination, fungal hyphae can infiltrate the surrounding tissue and, in cases of invasive aspergillosis, subsequently spread to different organs (15). Invasive aspergillosis is associated with high mortality rates due to suboptimal diagnostic and therapeutic options. Therefore, there is an urgent need to develop new antifungal agents and to optimize existing therapeutic strategies.
The fungal cell wall represents an excellent drug target, since it is structurally unique, its integrity is a precondition for the survival of the fungus, and it represents an essential barrier for protection against host defense mechanisms (1, 8, 9, 16). The fungal envelope consists of a framework of glycostructures, e.g., chitin and glucans, and a set of cell wall-associated proteins (1). Although at first sight the cell wall appears as a robust and static structure, it is in fact a highly dynamic entity. Infections by fungi always are associated with morphogenetic changes and growth processes, both requiring profound cell wall dynamics. The underlying reorganization of the cell wall is a delicate process in which the fungus has to find the balance between stability and plasticity, an attempt that requires the concerted action of a variety of cell wall proteins.
The addition of N-linked and/or O-linked oligosaccharides is a frequent modification of cell wall proteins. Mannosyltransferases play a crucial role in this process and most likely also are engaged in the generation of other glycoconjugates. Mannosyltransferases are localized in intracellular compartments of the secretory pathway, e.g., the Golgi apparatus or the endoplasmic reticulum (ER). Distinct or multiple disruptions of members of the KTR (for killer toxin resistance) family of mannosyltransferase genes in S. cerevisiae resulted in impaired protein glycosylation (10, 17), and the deletion of the homologous CaMNT1 gene in Candida albicans additionally affected fungal adhesion and virulence (22). At least in C. albicans, mannosyltransferases have been identified as crucial components assuring the maintenance and the robustness of the fungal cell wall (22).
The ScKre2/ScMnt1 α-1,2-mannosyltransferase of S. cerevisiae and the homologous enzymes CaMnt1 and CaMnt2 of C. albicans have been shown to be required for the addition of the second and third mannosyl residue of O-linked carbohydrates (10, 22), and they play a pivotal role in the elaboration of the outer chains of N-linked glycans (17, 27). The importance of α-1,2-mannosyltransferases for the synthesis of O- and N-linked carbohydrates and their possible role in the generation of other glycoconjugates, as well as the fact that humans do not possess any homologous enzymes, make α-1,2-mannosyltransferases promising targets for novel antifungal therapies.
MATERIALS AND METHODS
Strains and culture conditions.
The A. fumigatus wild-type strain D141 and its ΔakuA derivative AfS35 have been described before (14, 31). The procedure used for the isolation of conidia and the composition of the yeast glucose medium (YG) and the Aspergillus minimal medium (AMM) have been described previously (28). The human lung epithelial cell line A549 (ATCC CCL-185) was grown in RPMI 1640 medium supplemented with 5% fetal calf serum.
Sequence analysis and database searches.
Database searches were performed using BlastP and the following databases: GenBank/EMBL/DDBJ, the Central Aspergillus Data Repository (http://www.cadre-genomes.org.uk/Aspergillus_fumigatus/), the Saccharomyces Genome Database (http://www.yeastgenome.org/), and the Candida Genome Database (http://www.candidagenome.org/). Alignments, phylogenetic trees, and bootstrap analyses were generated using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Accession numbers of the protein sequences used are as follows: AfMnt1 (AFUA_5G10760, AFUA_5G12160, and AFUA5G02740), ScKtr1 (P27810/YOR099W), ScKtr2 (P33550/YKR061W), ScKtr3 (P38130/YBR205W), ScKtr4 (P38131/YBR199W), ScKtr5 (P53966/YNL029C), ScKtr6 (P54070/YPL053C), ScKtr7 (P40504/YIL085C), ScKre2 (P27809/YDR483W), ScYur1 (P26725/YJL139C), CaMnt1 (orf19.1665), CaMnt2 (orf19.1663), CaMnt3 (orf19.1010), CaKtr4 (orf19.4475), and CaKtr2 (orf19.4494).
Construction of the Δafmnt1 mutant and the complemented strain.
The generation of A. fumigatus cDNA has been described previously (28). The afmnt1 (Afu5g10760) gene was amplified from cDNA using oligonucleotides mnt1-5′ and mnt1-3′. The oligonucleotides used in this study are summarized in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| mnt1-Afu5g10760-5-fwd | TTGCGGCCGCTCTTCGGCTGGAGATTGGACG |
| mnt1-Afu5g10760-5-rev | TGGCCTGAGTGGCCCAAAGGATGATGTTGCCTCCG |
| mnt1-Afu5g10760-3-fwd | TGGCCATCTAGGCCCTATTGTCGGGTCGCTCTTCC |
| mnt1-Afu5g10760-3-rev | TTGCGGCCGCATGCCGTGTATCGCAGCGAG |
| mnt1-5′ | ATGTCTTCGGGACCGAAATAT |
| mnt1-3′ | TAGTCAACTCTGTTCTTCAGC |
| mnt1-Afu5g10760-5-cast | GGTAGGGTAGTATCAAAGCCG |
| hph-3-SmaI | TCCCGGGCTATTCCTTTGCCCTCGGACGAG |
| trpCt-fwd | CAGAATGCACAGGTACACTTG |
| mnt1-3-UTR-rev | ATGTAGGATACAGACCCTTACTG |
| mnt1-mRFP1-fu-rev | GCCGCCGCACTCTGTTCTTCAGCCCCCT |
| mRFP1-5-PmeI | AAACATGGCCTCCTCCGAGGAC |
| mRFP1-3-SmaI | TCCCGGGGGTACCTTAGGCGCCGGTG |
The PCR product was cloned, sequenced (Eurofin, Medigenomix, Munich, Germany), and compared to the predicted mRNA sequence derived from the A. fumigatus genome project using the BlastN algorithm. To construct a suitable replacement cassette, the protocol of Kämper (13) was followed. For this purpose, a 3.5-kb hygromycin resistance cassette was excised from pSK346 (14) using the SfiI restriction enzyme. The flanking regions of the afmnt1 gene (approximately 1.1 kb each) were amplified by PCR from chromosomal DNA using the oligonucleotide pairs mnt1-Afu5g10760-5-fwd/mnt1-Afu5g10760-5-rev and mnt1-Afu5g10760-3-fwd/mnt1-Afu5g10760-3-rev. These oligonucleotides harbored incompatible SfiI sites. After digestion with SfiI, a ligation of the three fragments (the resistance cassette and flanking regions) yielded a 5.6-kb deletion cassette that was purified using the PrepEase gel extraction kit (USB, Cleveland, OH). The fragment was cloned into the pBluescript KS vector (Stratagene, La Jolla, CA) using oligonucleotide-derived NotI sites. Purified NotI fragments from the resulting plasmid were used for transformation.
For complementation, the afmnt1 gene was amplified using oligonucleotides mnt1-5′ and mnt1-3′. The PCR product was purified and cloned into pSK379 to drive expression from the gpdA promoter. The resulting plasmid was isolated using the Pure Yield Plasmid Midiprep system (Promega, Mannheim, Germany). A. fumigatus protoplasts were generated, and transformation was performed essentially as described previously (26). The resulting protoplasts were transferred to AMM plates containing 1.2 M sorbitol and either 200 μg/ml hygromycin (Roche Applied Science, Mannheim, Germany) or 0.1 μg/ml pyrithiamine (Sigma, Deisenhofen, Germany).
Genomic DNA analysis.
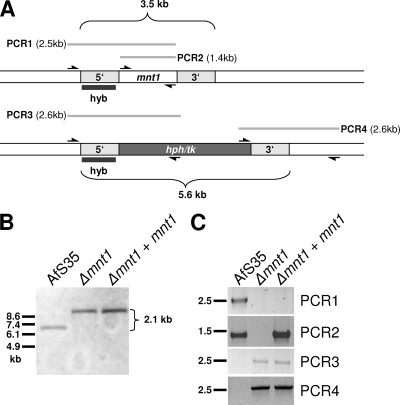
A. fumigatus clones that showed the expected resistance on selective plates were analyzed further by PCR. In the first PCR, one oligonucleotide (mnt1-Afu5g10760-5-cast) that hybridized upstream of the 1.1-bp 5′-flanking region of the afmnt1 gene was combined with a second primer (mnt1-3′) localized at the 3′ end of the afmnt1 gene (see Fig. 2). This reaction (designated PCR1) was used to detect the afmnt1 gene in its wild-type context. The core afmnt1 gene was amplified using oligonucleotides mnt1-5′ and mnt1-3′ (PCR2). The correct integration of the hygromycin resistance cassette was analyzed at the 5′ end using oligonucleotides mnt1-Afu5g10760-5-cast and hph-3-SmaI (PCR3) and at the 3′ end using oligonucleotides trpCt-fwd and mnt1-3-UTR-rev (PCR4) (see Fig. 2).
FIG. 2.
Construction of the Δafmnt1 mutant and the complemented strain (Δafmnt1 afmnt1). (A) Structure of the genomic A. fumigatus mnt1 gene and the deleted mnt1::hph/tk locus. A total of 1.1 kb of the 5′ and 3′ regions of afmnt1 was used for the construction of the deletion cassette. The probe used for the Southern blot (hyb), the positions of the primers used for PCR amplifications, and the resulting PCR products (PCR1 to PCR4) are indicated. (B) Genomic DNA of strain AfS35, the deletion mutant (Δafmnt1), and the complemented mutant (Δafmnt1 afmnt1) were digested with the restriction enzyme EheI and separated by agarose gel electrophoresis. After blotting and immobilization, membranes were hybridized with the digoxigenin-labeled 5′-specific probe indicated in panel A (hyb). The expected size shift is indicated (2.1 kb). (C) Equal amounts of genomic DNA of AfS35, Δafmnt1, and Δafmnt1 afmnt1 were used as the template for PCR amplification of the regions indicated in panel A (PCR1 to PCR4).
Genomic DNA was isolated from mycelia grown in liquid culture (AMM supplemented with 1% glucose) using the MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI) and digested using EheI. This enzyme cuts solely outside of the afmnt1 region (including the 1.1-kb flanking regions) and the hygromycin resistance cassette. Complete digestion was verified on agarose gels, and the DNA restriction fragments were transferred to nylon membranes (Hybond-N+; GE Healthcare, Freiburg, Germany). A 1.1-kb probe corresponding to the upstream flanking region of the afmnt1 gene was amplified by PCR using oligonucleotides Afu5g10760-5-fwd and mnt1-Afu5g10760-5-rev and digoxigenin-labeled nucleotides (Roche Applied Science, Mannheim, Germany). The separation of the samples and the hybridization and detection of the probe were performed according to standard procedures and the instructions of the vendor.
Construction of a strain expressing AfMnt1-mRFP1.
The sequence of monomeric red fluorescence protein 1 (mRFP1) was amplified using oligonucleotides mRFP1-5-PmeI and mRFP1-3-SmaI and the template vector pMT-mRFP1 (32). The resulting 695-bp fragment was cloned into pSK379 by retaining a PmeI restriction site at the 5′ end of the integrated fragment, resulting in pJW101. The genomic sequence of afmnt1 without the stop codon was amplified using oligonucleotides mnt1-5′ and mnt1-mRFP1-fu-rev. The resulting 1,378-bp fragment was cloned into pJW101 using the PmeI restriction site. The resulting plasmid pJW102 was transformed into A. fumigatus protoplasts as described above. To test whether the AfMnt1-mRFP-containing organelles are sensitive to brefeldin A, hyphae grown in minimal medium (AMM) at 37°C were further incubated for 6 h in the presence of 20 μg/ml brefeldin A (Sigma, Deisenhofen, Germany). For microscopic analysis, samples were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 5 min. Samples were embedded using Vectashield mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI; Vector, Burlingame, CA) and analyzed using an SP-5 confocal laser-scanning microscope (Leica Microsystems, Heidelberg, Germany).
Transmission electron microscopy.
For transmission electron microscopy, samples grown at 37°C were fixed in a fixation solution containing 5% formaldehyde and 2% glutaraldehyde in cacodylate buffer (0.1 M cacodylate, 0.01 M CaCl2, 0.01 M MgCl2, 0.09 M sucrose, pH 6.9) and washed with cacodylate buffer. Pellets were embedded in 2% water agar and cut into small cubes. Dehydration was achieved with a graded series of acetone (10, 20, and 50%) for 30 min on ice, followed by contrasting with 2% uranyl acetate in 70% acetone overnight at 4°C and further dehydrated with 90 and 100% acetone. Samples in the 100% acetone step were allowed to reach room temperature and were infiltrated with the epoxy resin according to Spurr's formula for a medium resin (30): 1 part 100% acetone/1 part resin overnight, 1 part 100% acetone/2 parts resin for 8 h, pure resin overnight, and several changes the following 2 days. Samples then were transferred to resin-filled gelatin capsules and polymerized for 8 h at 75°C. Ultrathin sections were cut with a diamond knife, picked up with formvar-coated copper grids (300 mesh), and counterstained with 4% aqueous uranyl acetate. After being air dried, samples were examined in a Zeiss transmission electron microscope TEM910 at an acceleration voltage of 80 kV. Images were recorded digitally with a slow-scan charge-coupled display camera (1,024 by 1,024 pixels; ProScan, Scheuring, Germany) with ITEM software (Olympus Soft Imaging Solutions, Münster, Germany), and cell wall thickness was measured by applying the measurement function of the software at the calibrated ×20,000 magnification step.
Field emission scanning electron microscopy.
For scanning electron microscopy, A. fumigatus strains were grown on glass coverslips in AMM at 48°C. Samples were fixed in 5% formaldehyde-50 mM Tris, pH 7.0, for 10 min, and then incubated in 3% glutaraldehyde-50 mM Tris, pH 7.0, for 4 h. Samples were washed several times with cacodylate buffer and subsequently washed with TE buffer (20 mM Tris, 1 mM EDTA, pH 6.9) before being dehydrated in a graded series of acetone (10, 30, 50, 70, 90, and 100%) on ice for 15 min for each step. Samples in the 100% acetone step were allowed to reach room temperature before another change in 100% acetone. Samples then were subjected to critical-point drying with liquid CO2 (CPD 30; Balzers Union, Liechtenstein). Dried samples were mounted onto conductive carbon adhesive tabs on an aluminum stub and sputter coated with a thin gold film (SCD 40; Balzers Union, Liechtenstein). Samples then were examined in a field emission scanning electron microscope (Zeiss DSM 982 Gemini) using the Everhart Thornley SE detector and the in-lens SE detector at a 50/50 ratio at an acceleration voltage of 5 kV and at calibrated magnifications.
Immunofluorescence microscopy.
Conidia of the respective A. fumigatus strain were transferred to a well of a 24-well plate containing a glass coverslip and 1 ml of AMM. Plates were incubated in sealed chambers to prevent excessive evaporation. After incubation for 15 h at 37°C, the cultures were shifted to 48°C, cultured for another 6 h, and fixed using 3.7% formaldehyde-PBS for 10 min. Samples then were blocked using 2% normal goat serum in PBS at 37°C for 30 min. To detect the acidic ribosomal proteins, the coverslips were incubated in a moistening chamber. In the first step, the samples were covered with culture supernatant of the hybridoma B8-C4 (29) and incubated for 30 min at 37°C. After three washing steps using PBS, bound antibodies were visualized with a Cy3-conjugated secondary antibody (Dianova, Hamburg, Germany). Samples were examined using a Leica SP5 confocal laser-scanning microscope (Leica Microsystems, Mannheim, Germany).
Phenotypic characterization of the mutant.
Isolated conidia were counted using a Neubauer chamber. For plate assays, defined numbers of conidia were spotted onto agar plates. These plates were supplemented with the indicated agents and incubated at the indicated temperatures. For the quantification of the radial growth assays, plates were run in triplicate.
Adhesion to epithelial cells was examined using the human lung epithelial cell line A549. Infection was performed in 24-well plates. Semiconfluent monolayers of 2 × 105 cells grown on glass coverslips were infected with 2 × 106 conidia. After 2 and 4 h at 37°C, cells were fixed using 3.7% formaldehyde-PBS. After permeabilization (0.2% Triton X-100-PBS for 1 min), the F-actin cytoskeleton of the epithelial cells was visualized using fluorescein isothiocyanate-labeled phalloidin (Sigma, Deisenhofen, Germany). Epithelial cells and bound conidia were counted using a Leica DMLB microscope (Leica, Wetzlar, Germany) equipped with epifluorescence.
Susceptibility testing.
Etest strips of voriconazole, posoconazole, caspofungin, and amphotericin were obtained from Inverness Medical (Cologne, Germany). A total of 25,000 conidia were plated onto YG (pH 6.0) plates and incubated for 48 h at 37°C. The MIC was determined according to the instructions of the vendor.
Stimulation of TNF-α in murine macrophages.
Bone marrow cells were isolated from C57/Bl6 mice according to standard procedures. The cells were differentiated to macrophages in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% L929 cell-conditioned medium and 10% fetal calf serum for 8 days. Conidia of the respective strains were seeded in 96-well plates at 2.5 × 104 conidia per well. The plates were further incubated in DMEM without serum for 24 h at 37°C in a humidified CO2 incubator. The resulting hyphae were killed using UV light, and the samples were washed with PBS; 2.5 × 105 macrophages were added per well in 0.2 ml fresh DMEM (containing 10% fetal calf serum) per well, and the cultures were further incubated for 15 h at 37°C in a humidified CO2 incubator. Supernatants were harvested and analyzed using a murine tumor necrosis factor alpha (TNF-α) enzyme-linked immunosorbent assay kit (R&D Systems, Bad Nauheim, Germany).
Mouse infection experiments.
Conidia (1 × 106) of the Δafmnt1 mutant, the parental strain Afs35, or the complemented mutant in a final volume of 300 μl PBS containing 0.02% Tween 20 were injected retroorbitally into male CD-1 and B6.CD45.1 mice. The viability of the inoculum was controlled by incubating aliquots of conidia overnight in YG medium and subsequent microscopic inspection. The survival of infected animals was monitored once a day. Kaplan-Meier survival curves were compared using the log-rank test (SPSS 15.0 software). P values of <0.05 were considered statistically significant.
RESULTS
The genome of A. fumigatus harbors only three putative α-1,2-mannosyltransferase genes.
In an attempt to analyze the functional importance of mannosyltransferases in A. fumigatus, we used sequences of the known members of the Ktr family of α-1,2-mannosyltransferases from S. cerevisiae and the BlastP algorithm and analyzed the A. fumigatus genome database at the Central Aspergillus Data Repository for homologous sequences. We identified three predicted open reading frames with significant homology. All of them were already annotated as putative α-1,2-mannosyltransferases (AFUA_5G10760, AFUA_5G12160, and AFUA_5G02740). The size of the Ktr family differs significantly between different yeast and fungal species, being nine in S. cerevisiae (ScKre2/ScMnt1, ScKtr1, ScKtr2, ScKtr3, ScKtr4, ScKtr5, ScKtr6, ScKtr7, and ScYur1) (20), five in C. albicans (CaMnt1, CaMnt2, CaMnt3, CaKtr2, and CaKtr4), but only three in A. fumigatus.
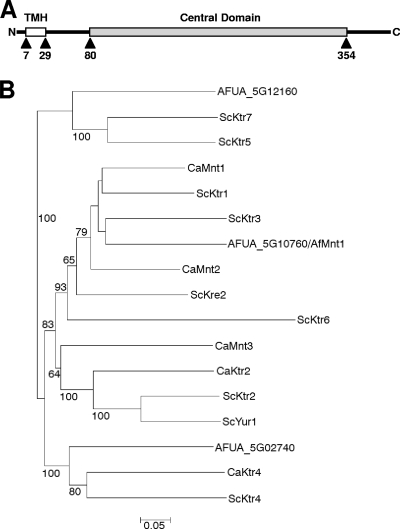
Using the AFUA_5G10760 sequence and the BlastP algorithm, we detected high homologies to the Ktr1 mannosyltransferase domain (COG5020; E value, 2e-150) and the α-1,2-mannosyltransferase domain (Pfam 01793; E value, 2e-141). Interestingly, no protein with significant homology was found in the human genome database. Members of the Ktr family are type II membrane proteins consisting of a highly variable N-terminal part that comprises a short cytoplasmic tail and a transmembrane domain, a conserved central domain characteristic for glycolipid 2-α-mannosyltransferases (Pfam 01793), and a variable C-terminal tail (19). Since both the N- and the C-terminal sequence stretches are highly variable, we generated a phylogenetic tree based on the sequences of the conserved central domains. The partial sequences used for the phylogenetic analysis were defined according to a previous study in which a phylogenetic tree of the Ktr proteins of S. cerevisiae was generated (19) (e.g., the central domain of AFUA_5G10760 comprises residues 80 to 354) (Fig. 1A).
FIG. 1.
(A) Schematic representation of the AfMnt1 (AFUA_5G10760) protein. The putative transmembrane helix (TMH) and the conserved central domain of AfMnt1 (residues 80 to 354) are indicated. (B) Phylogenetic tree of the conserved central domains of AfMnt1 (AFUA_5G10760) and the homologous domains of A. fumigatus (AFUA_5G12160 and AFUA_5G02740), S. cerevisiae (ScKtr1, ScKtr2, ScKtr3, ScKtr4, ScKtr5, ScKtr6, ScKtr7, ScKre2, and ScYur1), and C. albicans (CaMnt1, CaMnt2, CaMnt3, CaKtr2, and CaKtr4). Percentages are bootstrap values (relative to 2,000). Only bootstrap values of at least 50% are shown. A branch length standard is indicated.
The phylogenetic tree comprising the conserved domains of the three putative α-1,2-mannosyltransferase genes of A. fumigatus and the homologous sequences of S. cerevisiae and C. albicans shows that AFUA_5G10760 is closely related to S. cerevisiae ScKtr1 and ScKtr3 and C. albicans CaMnt1 (Fig. 1B). Candida albicans CaMnt2 and S. cerevisiae ScKre2 appear to be closely related to this cluster, but the bootstrap values already indicate a significant evolutionary distance. ScKtr1, ScKtr3, ScKre2, CaMnt1, and CaMnt2 are proven α-1,2-mannosyltransferases with overlapping functions. Experimental evidence has been published that demonstrates that these enzymes collectively add the second and third mannosyl residue to O-linked carbohydrates and also are jointly involved in N-linked glycosylation (10, 11, 17, 20, 22, 27).
The two other putative A. fumigatus mannosyltransferases, AFUA_5G12160 and AFUA_5G02740, were localized in two separate gene clusters. AFUA_5G12160 turned out to be closely related to the putative mannosyltransferases ScKtr5 and ScKtr7, whereas AFUA_5G02740 shared the highest homologies with the putative mannosyltransferases CaKtr4 and ScKtr4 (Fig. 1).
AFUA_5G10760 tentatively has been named Kre2 in the A. fumigatus genome database. The KRE2 gene of S. cerevisiae originally was identified in a screen of mutants resistant to the K1 killer toxin (20). It has not been shown so far that A. fumigatus is sensitive to the K1 killer toxin, and differences in the cell walls of yeasts and filamentous fungi argue against such sensitivity. We therefore propose to use the alternative designation AfMnt1, for A. fumigatus mannosyltransferase 1. This nomenclature is used for the homologous C. albicans proteins (3), and MNT1 is an alternative name for the KRE2 gene in S. cerevisiae (10).
Disruption of the afmnt1 gene by homologous recombination.
We amplified the afmnt1 mRNA from cDNA using oligonucleotides mnt1-5′ and mnt1-3′ (Table 1). A sequence analysis of the cloned PCR product confirmed the predicted structure of the afmnt1 gene with four exons and three introns. The obtained nucleotide sequence was identical to the predicted mRNA sequence of the A. fumigatus reference strain 293 (24).
A deletion construct was generated containing 1.1-kb regions upstream and downstream of the afmnt1 gene and a hygromycin resistance cassette (Fig. 2A). This linear deletion construct was used for the transformation of protoplasts of strain AfS35. AfS35 is a derivative of the wild-type strain D141 that lacks akuA, an essential gene of the nonhomologous end-joining apparatus, and therefore enables high frequencies of homologous integrations (14). Several hygromycin-resistant clones were obtained and tested for correct integration by PCR (Fig. 2C). To complement the deletion mutant, the afmnt1 gene was cloned into the plasmid pSK379, which harbors a pyrithiamine resistance gene (ptrA), and the plasmid was introduced into the afmnt1 mutant. Several resistant clones were obtained and analyzed by PCR (Fig. 2C). The Δafmnt1 mutant and the complemented strain then were verified by Southern blot analysis (Fig. 2B). The data revealed the expected difference in the length of the hybridizing fragments of the parental strain and the Δafmnt1 mutant and thereby confirmed a correct and unique integration of the deletion construct.
Deletion of the afmnt1 gene results in thinner hyphal cell walls.
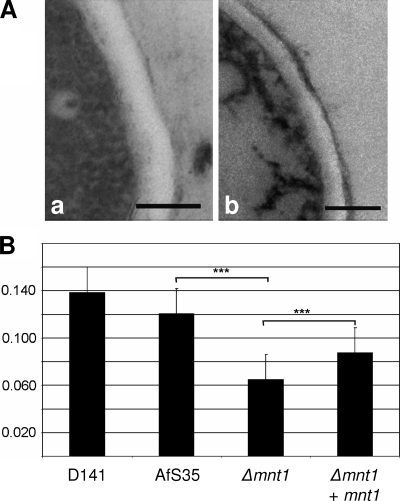
Under standard growth conditions, the Δafmnt1 mutant and the complemented mutant showed a comparable radial growth on AMM plates. We observed neither a difference in the sporulation of the tested strains at 37°C nor a delayed conidial germination (data not shown), as has been reported very recently for an A. fumigatus mutant in the protein O-mannosyltransferase 1 (36). Immunofluorescence studies revealed no differences between the Δafmnt1 mutant and the corresponding control strains with respect to the surface exposure of β1-3 glucan or galactomannan, or in the staining pattern obtained with calcofluor white (data not shown). To further analyze a potential influence of the afmnt1 gene on the A. fumigatus cell wall, we used transmission electron microscopy. Images of cross-sections obtained from hyphae of the different strains grown at 37°C were used to determine the thickness of the cell walls. Representative images of the parental strain AfS35 and the Δafmnt1 mutant are shown in Fig. 3A. The data obtained for the thickness of the cell walls are summarized in Fig. 3B. They strongly suggest that the cell wall of the Δafmnt1 mutant is significantly thinner than that of the parental and the complemented strains. This defect was largely restored in the complemented strain. The observed difference between the parental and the complemented strain might be due to a position effect.
FIG. 3.
Analysis of the effect of the afmnt1 deletion on the hyphal cell wall using transmission electron microscopy. (A) Representative micrographs of the cell walls of the parental strain AfS35 and the Δafmnt1 mutant grown at 37°C are shown in images a and b, respectively. Bars represent 0.2 μm. (B) Graphic representation of the mean values and the corresponding standard deviations of the thickness of the cell walls (in micrometers) determined for the wild-type strain D141, the parental strain AfS35, the Δafmnt1 mutant, and the complemented mutant Δafmnt1 afmnt1. The cell wall was measured 20 times for each strain. ***, P < 0.0001 by Student's t test.
A C. albicans mutant in the CaMnt1 mannosyltransferase showed a reduced binding to epithelial cells (22). We therefore analyzed the binding of conidia to the human lung epithelial cell line A549 but observed no significant difference in the binding of the Δafmnt1 mutant, its parental strain, or the complemented mutant, which suggests that the AfMnt1 protein is not required for the adhesion of conidia to epithelial cells (data not shown).
The Δafmnt1 mutant shows increased sensitivity to cell wall stress.
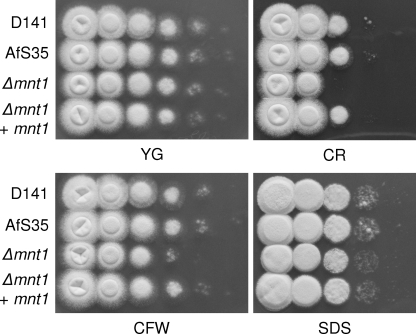
As we observed a difference in the thickness of the cell walls of the Δafmnt1 mutant and the parental AfS35, we speculated that this mutation also affected the robustness of the cell wall. To address this question, we tested the different strains for sensitivity to well-known cell wall stressors in plate assays at 37°C. We indeed observed that the Δafmnt1 mutant showed an increased sensitivity to calcofluor white (60 μg/ml) and Congo red (30 μg/ml), whereas no difference was found between the Δafmnt1 mutant and the parental AfS35 strain in the presence of 0.01% sodium dodecyl sulfate (SDS) (Fig. 4) or H2O2 (data not shown). The observed sensitivity of the Δafmnt1 mutant to cell wall stress could be completely restored by complementation, demonstrating that this phenotype is associated specifically with the lack of the afmnt1 gene (Fig. 4).
FIG. 4.
Effect of SDS and the cell wall stressors calcofluor white (CFW) and Congo red (CR) on fungal growth. From series of tenfold dilutions derived from starting suspensions of 4 × 107 conidia/ml, aliquots of 5 μl were spotted onto a YG agar plates (pH 6.0). Plates were supplemented with 60 μg/ml calcofluor white, 30 μg/ml Congo red, or 0.01% SDS and incubated at 37°C for 24 h. The depicted plates are representative of a set of three independent experiments.
The cell wall of the Δafmnt1 mutant is unstable at elevated temperatures.
Aspergillus fumigatus is thermotolerant and grows at temperatures of up to 50°C. To analyze whether an increased temperature affects the growth of the mutant, we measured growth at elevated temperatures. As mentioned above, we observed no obvious difference at 37°C. In contrast, plating series of tenfold dilutions on plates that subsequently were incubated at 48°C revealed a striking growth defect of the Δafmnt1 strain that was abolished in the complemented mutant (Fig. 5A). Osmotic stabilization due to the presence of 1.2 M sorbitol in the plates restored the growth of the Δafmnt1 mutant, but the resulting colonies remained white, suggesting a dramatically reduced sporulation (Fig. 5B). A closer inspection of these colonies by scanning electron microscopy revealed that the mutant still was able to form vesicles and phialides, whereas only very few conidia were found (data not shown).
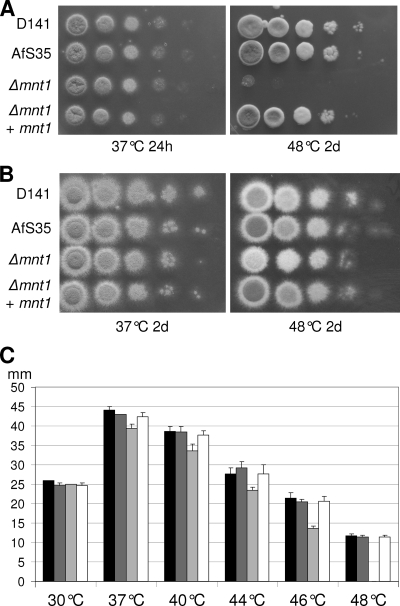
FIG. 5.
Impairment of growth of the Δafmnt1 mutant is restricted to elevated temperatures. From series of tenfold dilutions derived from starting suspensions of 4 × 107 conidia/ml (A) or 4 × 106 conidia/ml (B), aliquots of 5 μl were spotted onto AMM agar plates with (B) or without (A) 1.2 M sorbitol. Images were taken after growth at 37 or 48°C for the indicated time. 2d, 2 days. (C) Determination of the radial growth of colonies at the indicated temperatures. Equal numbers of conidia were spotted onto AMM plates and grown at the indicated temperatures. The graph shows the average colony diameters after 72 h determined in three independent experiments. D141 (black bars), AfS35 (dark gray bars), the Δafmnt1 mutant (light gray bars), and the complemented mutant Δafmnt1 afmnt1 (white bars) are shown.
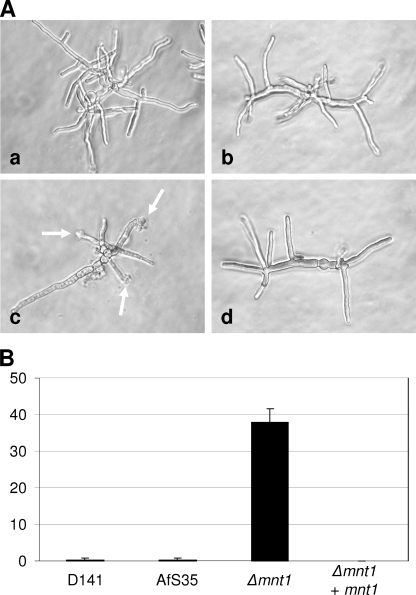
The quantification of the radial growth of the different strains at temperatures between 30 and 48°C on plates revealed a marginal delay in the growth of the Δafmnt1 mutant between 37 and 44°C, whereas a dramatic growth defect was apparent at 48°C (Fig. 5C). Although we observed only a slight reduction in the growth of the Δafmnt1 mutant at 44°C, the resulting colonies were white, indicating a defect in sporulation already at this temperature. Defects in growth and sporulation were completely restored in the complemented mutant (Fig. 5). A microscopic inspection of hyphae grown for 12 h at 37°C and then for an additional 6 h at 48°C revealed bulky structures at the tips of the Δafmnt1 mutant that were hardly observed for the parental or complemented strains (Fig. 6A). Quantification analysis revealed that after 6 h at 48°C, approximately 38% of the hyphal tips of the Δafmnt1 mutant had an abnormal morphology (Fig. 6B).
FIG. 6.
Detection of amorphous structures at the terminal tips of Δafmnt1 mutant hyphae. Conidia of the different strains were grown in AMM in 24-well plates. After 12 h at 37°C, the cultures were shifted to 48°C and incubated for another 6 h. The fixed samples were analyzed by light microscopy. (A) Micrographs demonstrating the appearance of amorphous structures at the hyphal tips of the Δafmnt1 (arrows). Image a, D141; image b, parental strain AfS35; image c, Δafmnt1 mutant; and image d, the complemented mutant Δafmnt1 afmnt1. (B) Percentage of hyphal tips showing an abnormal morphology.
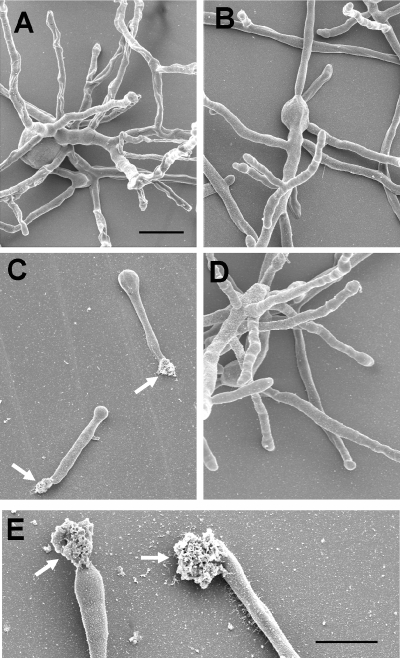
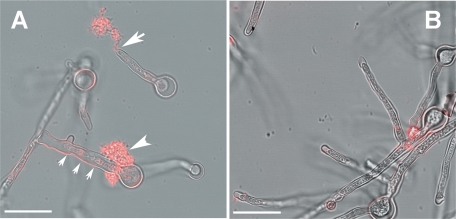
The scanning electron microscopy of fungal cells grown at 48°C revealed that the Δafmnt1 mutant is able to germinate at this temperature, but the resulting germ tubes were short and characterized by amorphous structures at their terminal tips (Fig. 7C and E). In contrast, the parental strain and the complemented mutant showed normal growth and intact hyphal tips (Fig. 7A, B, and D). To determine whether the amorphous material at the hyphal tips is released cytoplasm, we stained Δafmnt1 samples using the monoclonal antibody B8-C4, which specifically recognizes an epitope in the conserved domain of the acid ribosomal proteins P0, P1, and P2 (29). Immunofluorescence analysis revealed a strong labeling of the amorphous material at the hyphal tips of the Δafmnt1 mutant (Fig. 8A) but no staining with intact hyphae and hyphal tips of strains D141, AfS35, and the complemented strain (Fig. 8B and data not shown) and in a control experiment with mutant hyphae and the secondary antibody alone (data not shown). These data strongly suggest that the cell wall at the growing hyphal tip of the Δafmnt1 mutant is unstable at 48°C, resulting in a localized disruption of the cell wall and a subsequent cellular leakage.
FIG. 7.
Scanning electron microscopy of hyphae grown at 48°C. Conidia of the different strains were seeded on glass coverslips and grown in AMM for 16 h at 48°C. (A) D141; (B) parental strain AfS35; (C) Δafmnt1 mutant; (D) complemented mutant Δafmnt1 afmnt1; and (E) higher magnification of the Δafmnt1 mutant. Arrows in panels C and E indicate amorphous material released at the hyphal tips. The bars in panels A and E represent 20 μm. The bar in panel A is also valid for panels B to D.
FIG. 8.
Detection of cytosolic proteins that are released from the hyphal tips of the Δafmnt1 mutant. Conidia of different strains were seeded in 24-well plates containing glass coverslips. The fungi were grown at 37°C for 8 h and then shifted to 48°C. After 4 h, the samples were fixed and the A. fumigatus acid ribosomal proteins P0, P1, and P2 were stained using the monoclonal antibody B8-C4 and an appropriate Cy3-labeled secondary antibody (red). (A) Δafmnt1 mutant; (B) complemented strain Δafmnt1 afmnt1. The typical breakage of a hyphal tip is indicated by a large arrow. Unusual leakage at the base of a germ tube is indicated by an arrowhead. Note the shrunk cytoplasm in the damaged hyphae indicated by three small arrows. Bars represent 10 μm.
Intracellular localization of the AfMnt1 protein.
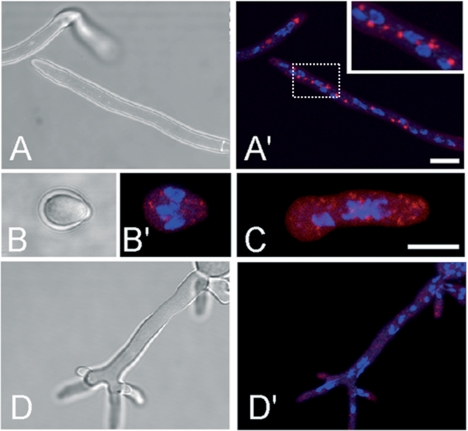
The high homologies of AfMnt1 to the conserved domains of the members of the Ktr family strongly suggest that AfMnt1 is an α-1,2-mannosyltransferase. These enzymes are engaged in protein glycosylation and are supposed to reside within the Golgi equivalents of fungi. For the S. cerevisiae Kre2/Mnt1 protein, this localization has been proven by immunofluorescence (18). To analyze the localization of AfMnt1, we generated a pSK379 derivative in which the afmnt1 gene was fused to the 5′ end of the mrfp1 reporter gene. After transformation into the Δafmnt1 mutant, we obtained pyrithiamine-resistant clones that showed a striking localization of the RFP fusion protein within distinct intracellular structures in hyphae grown at 37°C (Fig. 9A ′) and 48°C (data not shown), whereas a control construct containing the mrfp1 gene alone led to a diffuse cytoplasmic distribution of the red fluorescence (data not shown). Interestingly, the RFP fusion protein was able to fully complement the temperature-sensitive phenotype of the Δafmnt1 mutant, thereby providing evidence for a correct targeting of the fusion protein. An analysis of swollen conidia and germlings revealed a localization of the fusion protein in small spot-like structures (Fig. 9B′ and C, respectively), suggesting that Golgi equivalents already exist in swollen conidia.
FIG. 9.
Intracellular localization of an AfMnt1-RFP fusion protein. Hyphae of the Δafmnt1 mutant strain harboring plasmid pJW102 encoding an AfMnt1-RFP fusion protein were grown in minimal medium at 37°C. The AfMnt1-RFP fusion protein is shown in red, and DAPI-stained nuclei are shown in blue. The localization of AfMnt1-RFP in hyphae is shown in panel A′. The corresponding bright-field image is depicted in panel A. An enlargement of the region indicated by the dotted box is shown as an inset in the upper right part of panel A′. Micrographs of a swollen conidium (B and B′) and a germling (C) are shown. The localization of AfMnt1-RFP in hyphae treated with 20 μg/ml brefeldin A for 6 h is shown in panel D′. The corresponding bright-field image is depicted in panel D. Bars represent 5 μm. The bar in panel A′ is also valid for panels A, D, and D′.
Brefeldin A is known to disrupt the Golgi apparatus in mammals and fungi (4). Low doses of brefeldin A (20 μg/ml) led to a striking redistribution of the AfMnt1-RFP fusion protein, indicating the disruption of the corresponding organelles (Fig. 9D′).
The Δafmnt1 mutant shows an increased sensitivity to azole antifungals.
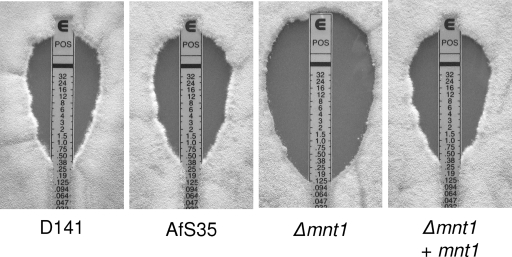
We next tested the potential impact of the loss of the afmnt1 gene for the susceptibility of A. fumigatus to antifungals, which represent current therapeutic options in cases of invasive aspergillosis. Using commercially available Etest strips, we evaluated the susceptibility of the D141 wild-type strain, the parental strain, the afmnt1 mutant, and the complemented strain. The inhibition ellipses obtained with amphotericin B were small and indistinguishable, whereas striking differences were observed with the azoles posoconazole and voriconazole (Fig. 10 and data not shown). The MICs of posoconazole and voriconazole for the Δafmnt1 mutant were 0.125 and 0.19 μg/ml, respectively. Identical MICs of 0.38 μg/ml for posoconazole and voriconazole were obtained for the parental and complemented strain. Thus, the deletion of the afmnt1 gene led to an increase in the sensitivity to posoconazole and voriconazole. For the echinocandine caspofungin we observed similar zones of inhibition, but the characteristic trailing growth within these zones (6) clearly was reduced for the Δafmnt1 mutant (data not shown), suggesting a slightly increased sensitivity.
FIG. 10.
Susceptibility to antifungal agents. YG agar plates were inoculated with 25,000 conidia of the indicated strains, Etest strips were applied, and plates were incubated at 37°C for 48 h. Representative pictures showing the inhibition ellipses obtained with posoconazole Etest strips are shown. Experiments with all antifungal drugs were performed in triplicate.
Cytokine response of murine macrophages to the Δafmnt1 hyphae.
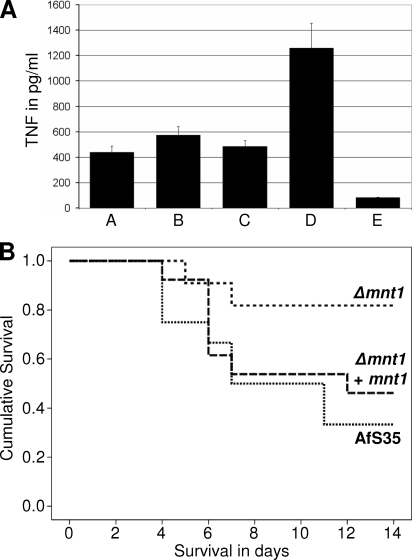
For C. albicans it has recently been shown that a camnt1 camnt2 double mutant is partially impaired in its ability to trigger a proinflammatory cytokine response (23). Due to the relatively small number of putative α-1,2-mannosyltransferases in A. fumigatus, AfMnt1 is likely to be the functional equivalent of CaMnt1 and CaMnt2 (Fig. 1). We therefore analyzed the cytokine response of murine bone marrow-derived macrophages to hyphae of the Δafmnt1 mutant and the corresponding parental and complemented strains. As shown in Fig. 11A, we observed no significant difference in the production of the proinflammatory cytokine TNF-α.
FIG. 11.
TNF-α responses of murine bone marrow-derived macrophages to hyphae of the wild type (bar A), the Δafmnt1 mutant (bar B), and the complemented mutant (bar C) are shown in panel A. Results obtained after stimulation with 100 ng lipopolysaccharide/well and with an unstimulated control are shown in bars D and E, respectively. (B) Results obtained in a mouse model of infection. CD-1 mice were infected retroorbitally with 1 × 106 conidia of the Δafmnt1 mutant (n = 11), the parental strain Afs35 (n = 12), and the complemented mutant Δafmnt1 afmnt1 (n = 13). The survival of mice is depicted over time.
The Δafmnt1 mutant is attenuated in a murine infection model.
Several lines of evidence indicate that even at 37°C the cell wall of the Δafmnt1 mutant is less robust than that of the wild type. Since stress resistance is a characteristic feature of many successful pathogens, we infected mice intravenously with conidia of the Δafmnt1 mutant, the corresponding parental strain, and the complemented strain. We observed that mice were significantly more resistant to infection with the Δafmnt1 mutant than to infection with the parental strain (P = 0.025) or the complemented strain (P = 0.091) (Fig. 11B). Very similar results were observed in a parallel experiment using B6.CD45.1 mice (P = 0.008 for the wild type and P = 0.056 for the complemented strain). Although the route of infection used in this study does not allow drawing conclusions on the initial phase of infection in the lung, the data nevertheless clearly demonstrate an impairment of the Δafmnt1 mutant to establish a successful systemic infection in immunocompetent mice.
DISCUSSION
Systemic infections caused by A. fumigatus are a major risk for severely immunocompromised patients, e.g., after stem cell transplantation, and due to the progress in modern medicine, the incidence of invasive aspergillosis steadily increased in the 1990s (21, 33). Although several new antifungals have been approved in recent years, there is still an urgent need to improve and expand the limited set of therapeutic options, and due to its uniqueness, the fungal cell wall has received much interest as a potential source for new therapeutic targets.
Mannosyltransferases add mannosyl residues to N- or O-linked glycans of proteins, but they also are involved in the synthesis of other glycoconjugates of the fungal cell wall. Using the conserved central domains of the members of the Ktr family from S. cerevisiae and C. albicans, we identified three A. fumigatus proteins in the current database that shared high homologies. Obviously, there is a striking difference in the number of these mannosyltransferases, with nine in S. cerevisiae, five in C. albicans, and only three in A. fumigatus. One of the latter, the AfMnt1 (AFUA_5G10760) protein, is the subject of this study.
A phylogenetic analysis of the conserved domains of AfMnt1 and the other members of the Ktr family in S. cerevisiae, C. albicans, and A. fumigatus revealed the presence of at least four clusters of closely related proteins. One cluster, comprising ScKtr2, ScYur1, CaMnt3, and CaKtr2, has no A. fumigatus member. AFUA_5G12160 forms a cluster together with ScKtr5 and ScKtr7. AFUA_5G02740 turned out to be especially related to CaKtr4 and ScKTR4, whereas AfMnt1 (AFUA_5G10760) shows the highest homologies to ScKtr3, ScKtr1, ScKre2, CaMnt1, and CaMnt2. Interestingly, experimental evidence for an α-1,2-mannosyltransferase activity has been published for all proteins of the AfMnt1 cluster (3, 10, 11, 17, 22, 27). This and the high homology of the core domain of AFUA_5G10760 to the α-1,2-mannosyltransferase domain Pfam 01793 strongly suggest that this protein is a bona fide α-1,2-mannosyltransferase. The AFUA_5G10760 gene tentatively was named kre2 according to the homologous ScKRE2/ScMNT1 gene. However, a cytotoxic effect of yeast killer toxin K1 has not yet been demonstrated for Aspergillus, and therefore AfMnt1 seems to be a more appropriate designation.
To examine the biological function of the AfMnt1 protein, we generated a knockout mutant of the ΔakuA strain AfS35, a derivative of the wild-type strain D141. Strains that lack either akuA or akuB show a dramatic reduction in the frequency of nonhomologous recombination events compared to that of the respective wild-type strains but no differences with respect to growth properties or virulence (5, 14).
The deletion of the afmnt1 gene and subsequent complementation were confirmed by PCR and Southern blotting. For complementation studies, a single copy of the afmnt1 gene (under the control of the gpdA promoter) was introduced into the mutant by targeted insertion. The resulting four strains, the wild-type D141, the parental strain AfS35, the Δafmnt1 mutant, and the complemented mutant (Δafmnt1 afmnt1) were analyzed. At 37°C, all strains grew and sporulated normally, indicating that the Δafmnt1 mutant is not obviously impaired under standard conditions. However, the transmission microscopy of hyphae hinted toward a reduced thickness of the cell wall of the Δafmnt1 mutant at 37°C, a phenotype that resembles that recently reported for a C. albicans camnt1 camnt2 double mutant (22). This already suggested a higher sensitivity to cell wall stress, and we indeed observed a reduced growth of the mutant in the presence of calcofluor white and Congo red. This phenotype resembles that of a C. albicans camnt1 mutant (22) and a very recently described A. fumigatus mutant that lacks the afpmt1 gene. AfPmt1 encodes an O-mannosyltransferase that initiates the O-glycosylation of proteins (36), a step that precedes the addition of mannosyl residues by an α-1,2-mannosyltransferase. Apart from phenotypes observed with calcofluor white and Congo red, this mutant shares another characteristic with the Δafmnt1 mutant: both are severely impaired in growth at higher temperatures (48 to 50°C), a phenotype that resembles that of S. cerevisiae mutants with multiple deletions in PMT genes (7). Interestingly, the growth inhibition at 42°C was dramatic for the Δafpmt1 mutant (36), whereas the growth of the Δafmnt1 mutant was only slightly impaired at 42 and even 44°C. The apparent growth defect of the Δafpmt1 mutant at elevated temperatures could be rescued by osmotic stabilization (36), and the same applies to the Δafmnt1 mutant at 48°C. In both cases, the resulting colonies appeared white, indicating a dramatically reduced sporulation (36) (Fig. 6). Using electron microscopy, we analyzed Δafmnt1 colonies grown at 48°C in the presence of sorbitol in more detail. We observed that the mutant is able to form rudimentary conidiophores that contain vesicles and phialides, but only very few conidia.
As the reasons for the temperature sensitivity of the Δafpmt1 mutant have not been further elucidated, we tried to determine them for the Δafmnt1 mutant. With this mutant, we frequently observed bulky structures at the ends of hyphae grown at 48°C. Closer inspection by scanning electron microscopy revealed an amorphous material that appeared to be released from the fungus. Immunofluorescence analysis revealed the presence of ribosomal proteins in these structures, demonstrating that they consist of released cytoplasm. The striking colocalization of these leakages with the terminal ends of hyphae indicates that the growing tip is the most unstable part of the Δafmnt1 cell wall. Hyphal growth requires a localized reorganization of the cell wall at the tip to allow the incorporation of new material. During growth, the fungus has to find the delicate balance between the inevitable stability of the cell wall and the need for a dynamic reorganization. A dysfunction of the underlying fine-tuned network of enzymatic activities can result in instability and finally the leakage of the cell. Cell wall-associated proteins commonly are glycosylated, and defects in protein glycosylation may result in protein misfolding, instability, and/or a reduced enzymatic activity.
One characteristic feature of fungal α-1,2-mannosyltransferases is their localization in the so-called Golgi equivalents. We have analyzed the localization of an AfMnt1-RFP fusion protein and detected it in compact hyphal organelles both at 37 and 48°C. The expression of the fusion protein completely rescued the temperature-sensitive phenotype of the Δafmnt1 mutant, strongly suggesting that it is correctly targeted. The AfMnt1-containing organelles were sensitive to brefeldin A, an agent known to disrupt the ER and the Golgi apparatus. The ER of filamentous fungi is organized as a tubular network (4), a structure that is clearly different from the structures targeted by the AfMnt1-RFP fusion protein. The localization of the AfMnt1-RFP resembles that of a CopA-green fluorescent protein fusion protein that recently was shown to localize in the Golgi equivalents of A. nidulans (2). Taken together, our data provide evidence for a localization of AfMnt1 in the Golgi apparatus, which is a characteristic feature of α-1,2-mannosyltransferases.
The reduced thickness of the Δafmnt1 cell wall and the increased sensitivity of this mutant to cell wall stressors like calcofluor white suggest a latent instability of the mutant cell wall even at 37°C. An inhibition of the enzymatic activity of AfMnt1 therefore might be an interesting therapeutic option, especially as homologous α-1,2-mannosyltransferases do not exist in humans. AfPmt1 could be an alternative target, and potent inhibitors already have been identified for Pmt1 of C. albicans (25). However, in this case, homologous human protein O-mannosyltransferases do exist and might be a serious obstacle for the development of fungus-specific inhibitors.
We speculated that the latent instability of its cell wall renders the Δafmnt1 mutant more susceptible to stress exerted by therapeutic agents or the host immune response. In fact, we observed that the Δafmnt1 mutant showed a significantly enhanced sensitivity to posoconazole and voriconazole, two first-line antifungals used for the treatment of invasive aspergillosis. Azoles inhibit the cytochrome P450 enzyme lanosterol demethylase (34), a key enzyme in the biosynthesis of ergosterol, which in turn is a characteristic and essential component of the fungal membrane. Stress applied to the plasma membrane has been shown to activate the fungal cell wall integrity pathway (12, 35), demonstrating that stress targeted at the plasma membrane can be compensated for on the cell wall level. Thus, any stress applied to either the cell wall or plasma membrane is likely to affect the other. This functional interconnection of the structures seems to be pivotal for the maintenance of the fungal cell envelope and may explain the higher sensitivity of the Δafmnt1 mutant to azoles.
It has been shown recently that hyphae of a C. albicans double mutant lacking both MNT1 and MNT2 triggered a reduced production of proinflammatory cytokines, e.g., TNF-α. (23). Due to the limited number of members of the Ktr family, AfMnt1 seems to be the functional equivalent of both CaMnt1 and CaMnt2. We therefore challenged murine macrophages with hyphae of the Δafmnt1 mutant and the corresponding control strains, but we observed no differences. Mannoproteins are abundant in C. albicans and form a distinct layer on the surface of the cell wall, whereas these proteins seem to be less abundant and spatially differently organized in A. fumigatus (1), which might explain the obtained results.
To test whether the reduced robustness of the cell wall of the Δafmnt1 mutant has consequences for the resistance to host defense mechanisms, we infected mice intravenously with conidia. We observed an attenuated virulence for the Δafmnt1 mutant but not for the complemented strain, indicating that AfMnt1 activity is required for full virulence in this model of a systemic infection. Interestingly, similar results have been reported for the camnt1 camnt2 double mutant in a model of a systemic C. albicans infection (22). We cannot conclude from our data that the Δafmnt1 mutant is generally attenuated in virulence, since we have bypassed the initial phase of infection, but our data nevertheless demonstrate that the Δafmnt1 mutant is impaired in its ability to spread and colonize in infected mice, most likely due to an increased sensitivity to host defense mechanisms.
In conclusion, we found that the AfMnt1 protein is localized in Golgi equivalents and is required for the maintenance of the fungal cell wall. A lack of this protein leads to a thinner hyphal cell wall, an increased sensitivity to cell wall stressors, a growth defect at elevated temperature, and, as a consequence, a dramatically reduced sporulation and a disruption of the cell wall at its most dynamic part, the hyphal tip. The increased sensitivity of the Δafmnt1 mutant to certain antifungals and its attenuation in a systemic mouse model of infection identifies this bona fide α-1,2-mannosyltransferase as a potential drug target. Specific inhibitors could synergize with approved antifungal agents, and if used in combination therapy, they might establish an improved option for the treatment of invasive aspergillosis.
Acknowledgments
We thank Monika Schwienbacher for supplying the cDNA of Aspergillus fumigatus, Reinhard Fischer for generously providing plasmid pMT-mRFP1, and Kirsten Niebuhr for the critical reading of the manuscript. We thank Marzena Broniszewska, Ina Schleicher, and Verena Groβe for excellent technical assistance.
This work was supported by grants of the Deutsche Forschungsgemeinschaft to J.H. (SFB 576, TP B8) and to B.E. (KFO 146).
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Bowman, S. M., and S. J. Free. 2006. The structure and synthesis of the fungal cell wall. BioEssays 28799-808. [DOI] [PubMed] [Google Scholar]
- 2.Breakspear, A., K. J. Langford, M. Momany, and S. J. Assinder. 2007. CopA:GFP localizes to putative Golgi equivalents in Aspergillus nidulans. FEMS Microbiol. Lett. 27790-97. [DOI] [PubMed] [Google Scholar]
- 3.Buurman, E. T., C. Westwater, B. Hube, A. J. Brown, F. C. Odds, and N. A. Gow. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 957670-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, L., D. Davies, G. J. Hyde, and A. E. Ashford. 2000. Brefeldin A affects growth, endoplasmic reticulum, Golgi bodies, tubular vacuole system, and secretory pathway in Pisolithus tinctorius. Fungal Genet. Biol. 2995-106. [DOI] [PubMed] [Google Scholar]
- 5.da Silva Ferreira, M. E., M. R. Kress, M. Savoldi, M. H. Goldman, A. Härtl, T. Heinekamp, A. A. Brakhage, and G. H. Goldman. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinel-Ingroff, A. 2003. Evaluation of broth microdilution testing parameters and agar diffusion E-test procedure for testing susceptibilities of Aspergillus spp. to caspofungin acetate (MK-0991). J. Clin. Microbiol. 41403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentzsch, M., and W. Tanner. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 55752-5759. [PMC free article] [PubMed] [Google Scholar]
- 8.Gow, N. A., S. Bates, A. J. Brown, E. T. Buurman, L. M. Thomson, and C. Westwater. 1999. Candida cell wall mannosylation: importance in host-fungus interaction and potential as a target for the development of antifungal drugs. Biochem. Soc. Trans. 27512-516. [DOI] [PubMed] [Google Scholar]
- 9.Gozalbo, D., P. Roig, E. Villamón, and M. L. Gil. 2004. Candida and candidiasis: the cell wall as a potential molecular target for antifungal therapy. Curr. Drug Targets Infect. Disord. 4117-135. [DOI] [PubMed] [Google Scholar]
- 10.Häusler, A., L. Ballou, C. E. Ballou, and P. W. Robbins. 1992. Yeast glycoprotein biosynthesis: MNT1 encodes an alpha-1,2-mannosyltransferase involved in O-glycosylation. Proc. Natl. Acad. Sci. USA 896846-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill, K., C. Boone, M. Goebl, R. Puccia, A. M. Sdicu, and H. Bussey. 1992. Yeast KRE2 defines a new gene family encoding probable secretory proteins, and is required for the correct N-glycosylation of proteins. Genetics 130273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 91559-1571. [DOI] [PubMed] [Google Scholar]
- 13.Kämper, J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271103-110. [DOI] [PubMed] [Google Scholar]
- 14.Krappmann, S., C. Sasse, and G. H. Braus. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latgé, J. P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66279-290. [DOI] [PubMed] [Google Scholar]
- 17.Lussier, M., A. M. Sdicu, F. Bussereau, M. Jacquet, and H. Bussey. 1997. The Ktr1p, Ktr3p, and Kre2p/Mnt1p mannosyltransferases participate in the elaboration of yeast O- and N-linked carbohydrate chains. J. Biol. Chem. 27215527-15531. [DOI] [PubMed] [Google Scholar]
- 18.Lussier, M., A. M. Sdicu, T. Ketela, and H. Bussey. 1995. Localization and targeting of the Saccharomyces cerevisiae Kre2p/Mnt1p alpha 1,2-mannosyltransferase to a medial-Golgi compartment. J. Cell Biol. 131913-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lussier, M., A. M. Sdicu, E. Winnett, D. H. Vo, J. Sheraton, A. Düsterhöft, R. K. Storms, and H. Bussey. 1997. Completion of the Saccharomyces cerevisiae genome sequence allows identification of KTR5, KTR6 and KTR7 and definition of the nine-membered KRE2/MNT1 mannosyltransferase gene family in this organism. Yeast 13267-274. [DOI] [PubMed] [Google Scholar]
- 20.Lussier, M., A. M. Sdicu, and H. Bussey. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426323-334. [DOI] [PubMed] [Google Scholar]
- 21.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33641-647. [DOI] [PubMed] [Google Scholar]
- 22.Munro, C. A., S. Bates, E. T. Buurman, H. B. Hughes, D. M. Maccallum, G. Bertram, A. Atrih, et al. 2005. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2801051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netea, M. G., N. A. Gow, C. A. Munro, S. Bates, C. Collins, G. Ferwerda, R. P. Hobson, et al. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 1161642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyon, M. Brrima, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 4381151-1156. [DOI] [PubMed] [Google Scholar]
- 25.Orchard, M. G., J. C. Neuss, C. M. Galley, A. Carr, D. W. Porter, P. Smith, D. I. Scopes, et al. 2004. Rhodanine-3-acetic acid derivatives as inhibitors of fungal protein mannosyl transferase 1 (PMT1). Bioorg. Med. Chem. Lett. 143975-3978. [DOI] [PubMed] [Google Scholar]
- 26.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216447-457. [DOI] [PubMed] [Google Scholar]
- 27.Romero, P. A., M. Lussier, A. M. Sdicu, H. Bussey, and A. Herscovics. 1997. Ktr1p is an alpha-1,2-mannosyltransferase of Saccharomyces cerevisiae. Biochem. J. 321289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwienbacher, M., M. Weig, S. Thies, J. T. Regula, J. Heesemann, and F. Ebel. 2005. Analysis of the major proteins secreted by the human opportunistic pathogen Aspergillus fumigatus under in vitro conditions. Med. Mycol. 43623-630. [DOI] [PubMed] [Google Scholar]
- 29.Schwienbacher, M., L. Israel, J. Heesemann, and F. Ebel. 2005. Asp f6, an Aspergillus allergen specifically recognized by IgE from patients with allergic bronchopulmonary aspergillosis, is differentially expressed during germination. Allergy 601430-1435. [DOI] [PubMed] [Google Scholar]
- 30.Spurr, A. R. 1969. A low viscosity epoxy resin embedding medium for electron microsocopy. J. Ultrastruct. Res. 2631-43. [DOI] [PubMed] [Google Scholar]
- 31.Staib, F., S. K. Mishra, C. Rajendran, R. Voigt, J. Steffen, K. H. Neumann, C. A. Hartmann, and G. Heins. 1980. A notable Aspergillus from a mortal aspergilloma of the lung. New aspects of the epidemiology, serodiagnosis and taxonomy of Aspergillus fumigatus. Zentralbl. Bakteriol. A. 247530-536. [PubMed] [Google Scholar]
- 32.Toews, M. W., J. Warmbold, S. Konzack, P. Rischitor, D. Veith, K. Vienken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45383-389. [DOI] [PubMed] [Google Scholar]
- 33.Upton, A., and K. A. Marr. 2006. Emergence of opportunistic mould infections in the hematopoietic stem cell transplant patient. Curr. Infect. Dis. Rep. 8434-441. [DOI] [PubMed] [Google Scholar]
- 34.van den Bossche, H., G. Willemsens, W. Cools, W. F. Lauwers, and L. Le Jeune. 1978. Biochemical effects of miconazole on fungi. II. Inhibition of ergosterol biosynthesis in Candida albicans. Chem. Biol. Interact. 2159-78. [DOI] [PubMed] [Google Scholar]
- 35.Zakrzewska, A., A. Boorsma, D. Delneri, S. Brul, S. G. Oliver, and F. M. Klis. 2007. Cellular processes and pathways that protect Saccharomyces cerevisiae cells against the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 6600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, H., H. Hu, L. Zhang, R. Li, H. Ouyang, J. Ming, and C. Jin. 2007. O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 62260-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]