Abstract
During sporulation in Saccharomyces cerevisiae, the dityrosine transporter Dtr1p, which is required for formation of the outermost layer of the spore wall, is specifically expressed and transported to the prospore membrane, a novel double-lipid-bilayer membrane. Dtr1p consists of 572 amino acids with predicted N- and C-terminal cytoplasmic extensions and 12 transmembrane domains. Dtr1p missing the largest internal cytoplasmic loop was trapped in the endoplasmic reticulum in both mitotically dividing cells and cells induced to sporulate. Deletion of the carboxyl 15 amino acids, but not the N-terminal extension of Dtr1p, resulted in a protein that failed to localize to the prospore membrane and was instead observed in cytoplasmic puncta. The puncta colocalized with a cis-Golgi marker, suggesting that Dtr1p missing the last 15 amino acids was trapped in an early Golgi compartment. Deletion of the C-terminal 10 amino acids resulted in a protein that localized to the prospore membrane with a delay and accumulated in cytoplasmic puncta that partially colocalized with a trans-Golgi marker. Both full-length Dtr1p and Dtr1p missing the last 10 amino acids expressed in vegetative cells localized to the plasma membrane and vacuoles, while Dtr1p deleted for the carboxyl-terminal 15 amino acids was observed only at vacuoles, suggesting that transport to the prospore membrane is mediated by distinct signals from those that specify plasma membrane localization. Transfer-of-function experiments revealed that both the carboxyl transmembrane domain and the C-terminal tail are important for Golgi complex-to-prospore membrane transport.
Saccharomyces cerevisiae sporulation is a complex sexual differentiation program initiated in MATa/MATα diploid cells by depletion of nitrogen in the presence of a nonfermentable carbon source (16). During sporulation, four haploid gametes, packaged as spores, are produced within the mother cell to form the ascus. Sporulation requires that meiosis, the process whereby the ploidy is reduced, and spore formation be exquisitely coordinated to ensure the production of viable gametes. During the second meiotic division, new proteinaceous structures termed the meiotic outer plaques (MOPs) are formed on the cytoplasmic face of the spindle pole bodies (SPBs), which are analogous to vertebrate centrosomes, and serve as platforms for vesicles to dock and fuse to form a novel double-lipid bilayer, the prospore membrane (PSM) (12, 14). PSMs develop as sacs by continuous fusion of vesicles to engulf each daughter nucleus, followed by PSM closure at the opposite side of each SPB (29). The inner lipid bilayer of the PSM becomes the plasma membrane (PM) of the spore, while the outer layer eventually lyses during the course of spore wall formation. The completion of PSM formation triggers the initiation of spore wall synthesis. The spore wall is composed of four distinct layers, namely, mannan and glucan, which form the inner layers, and chitosan and dityrosine, which form the spore-specific outer layers, formed outward one by one (6). The mature spore wall confers resistance to severe environmental conditions, such as heat, desiccation, ultraviolet light, and toxic chemicals.
Due in large part to yeast's facile genetics, sporulation has served as a paradigm for elucidating the underlying molecular mechanisms of developmental networks. Consequently, a number of genetic and genomic screens have led to the identification of many of the genes that mediate this complex process. For example, Mpc54p, Spo21p/Mpc70p, and Spo74p are essential for the initiation of PSM formation and are components of the MOP (1, 14, 31), while leading-edge protein coat components, i.e., Don1p, Ady1p, and Ssp1p, are required for the development and closure of the PSM (19, 21). Lipids and enzymes that modify lipids also play essential roles in sporulation. Spo14p, the major yeast phospholipase D, generates phosphatidic acid, which is essential for PSM precursor vesicle fusion (27, 38). Spo14p is positively regulated during sporulation by the Arf-GTPase activating protein, Gcs1p, which functions in vesicle transport in vegetative cells (8, 36), and by the lipid phosphatidylinositol 4,5-bisphosphate (38). Furthermore, a unique subset of the vesicle fusion machinery is required for PSM formation. Two soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), Spo20p and Sso1p, specifically mediate vesicle fusion at the SPB during PSM formation (27, 30). Interestingly, Spo20p is recruited to PSM precursor vesicles via phosphatidic acid binding (26). PSM localization of the v-SNARE Snc1p suggests that Snc1p also participates in vesicle fusion for PSM synthesis (23).
PSM formation requires redirection of trafficking pathways that move proteins, lipids, and other materials within the cell. Existing data suggest that Golgi complex-to-PSM transport is the major route for supplying vesicles for PSM formation. Inactivation of early SEC genes, such as SEC14, encoding a lipid transfer protein involved in the formation of secretory vesicles at the trans-Golgi apparatus, completely blocks PSM formation, strongly suggesting that PSM precursor vesicles are derived from the Golgi apparatus (27). However, endocytosis (22) and endosomal recycling pathways (26) are also important for transport during sporulation. Analysis of a mutant simultaneously deleted for the Golgi complex-associated retrograde protein (GARP) complex and the retromer complex, both of which are important for recycling through the Golgi complex (7, 42), indicated that Snc1p and Dtr1p are transported to the SPB via different pathways (26). These results suggest that different populations of vesicles, taking different routes, are trafficked to the SPB and contribute to PSM formation. However, the mechanisms underlying the dynamic rearrangements of membrane trafficking pathways during sporulation have not been elucidated.
To determine how proteins are redirected within the cell during sporulation, we analyzed the cis signals on the sporulation-specific transmembrane protein Dtr1p. Localization of truncated forms of Dtr1p-green fluorescent protein (Dtr1p-GFP) revealed that the C-terminal 15 amino acids, but not the N-terminal cytoplasmic extension, are essential for transport to the PSM. Deletion of 10 amino acids at the C-terminal end as well as a mutation that disrupts a dileucine motif in the C terminus showed partial defects in PSM transport. Analysis of chimeric proteins revealed that neither the C-terminal tail nor the last transmembrane domain of Dtr1p is sufficient to direct transport to the PSM. However, a chimeric protein containing both the last predicted transmembrane domain and the cytoplasmic C-terminal tail was observed at the PSM as well as in the cytoplasm, indicating that these regions of Dtr1p contain signals that direct the protein to the PSM during sporulation.
MATERIALS AND METHODS
Strains, media, and plasmids.
All strains used in this study, unless noted otherwise, are derived from the sporulation-proficient SK-1 strain background and are listed in Table 1. Solid and liquid yeast media were YPAD (1% yeast extract, 2% peptone, 2% dextrose, and 0.1 mM adenine), SC (synthetic complete medium) lacking the indicated amino acids (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 2% dextrose), and SPO (1% potassium acetate, 0.1% dextrose, 0.125% yeast extract, and 0.05% complete amino acid dropout powder) to induce sporulation.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| AN120a | MATa/MATα ura3/ura3 his3/his3 leu2/leu2 trp1/trp1 arg4/ARG4 rme1::LEU2/RME1 |
| Y893b | MATa/MATα leu2-27/leu2-1 ura3-50/ura3-50 spo13-1/spo13-1 can1-101/can1-101 lys2::ura3ΔNcoI/lys2::ura3ΔNcoI his3Δ/his3Δ his4/his4 |
| Y6582 | Y893 plus YEp352 (2μm URA3) |
| Y6583 | Y893 plus pME2441 (DTR1-GFP 2μm URA3) |
| Y5120 | MATα ura3 his3 leu2 trp1 arg4 rme::LEU2 sso1::HIS3 DTR1-GFP::TRP1 |
| Y7047b | MATa/MATα dtr1::KanMX/dtr1::KanMX leu2-x/leu2-y his4-x/his4-y trp1/trp1 ura3-1/ura3-1 thr1/THR1 lys2/lys2 CYH1/CYH1 ADE2/ade2-1 |
| Y7006 | AN120, but homozygous MPC54-RFP::URA3 |
| Y7730 | Y7006 plus pRS424-DTR1-GFP (2μm TRP1) |
| Y7419 | AN120 plus pME2599 (dtr1NΔ-GFP 2μm TRP1) |
| Y7747 | Y7006 plus pME2599 (dtr1NΔ-GFP 2μm TRP1) |
| Y7732 | Y7006 plus pME2611 (dtr1C10Δ-GFP 2μm TRP1) |
| Y7733 | Y7006 plus pME2630 (dtr1C15Δ-GFP 2μm TRP1) |
| Y7735 | Y7006 plus pME2585 (dtr1T568A-GFP 2μm TRP1) |
| Y7749 | Y7006 plus pME2635 (dtr1LA-GFP 2μm TRP1) |
| Y7784 | Y7006 plus pME2667 (dtr1LATA-GFP 2μm TRP1) |
| Y7884 | Y7006 plus pME2686 (dtr1LA-C10Δ-GFP 2μm TRP1) |
| Y7479 | AN120 plus pME2606 (CLB2p-DTR1-GFP 2μm TRP1) |
| Y7557 | AN120, but homozygous vps54::KanMX plus pME2606 (CLB2p-DTR1-GFP 2μm TRP1) |
| Y7592 | AN120 plus pME2619 (CLB2p-dtr1C10Δ-GFP 2μm TRP1) |
| Y7777 | AN120 plus pME2649 (CLB2p-dtr1C15Δ-GFP 2μm TRP1) |
| Y7807 | AN120, but homozygous end3::KanMX4 plus pME2649 (CLB2p-dtr1C15Δ-GFP 2μm TRP1) |
| Y7752 | AN120 plus pME2653 (CLB2p-dtr1LA-GFP 2μm TRP1) |
| Y7612 | AN120 plus pME2620 (CLB2p-dtr1T568A-GFP 2μm TRP1) |
| Y7803 | AN120 plus pME2669 (CLB2p-dtr1LAT568A-GFP 2μm TRP1) |
| Y7889 | AN120 plus pME2689 (CLB2p-dtr1LA-C10Δ-GFP 2μm TRP1) |
| Y7657 | AN120 plus pME2629 (dtr1Loop6Δ-RFP 2μm TRP1) and pCP217 (pRS316 SEC63-GFP CEN URA3) |
| Y7526 | AN120 plus pME2611 (dtr1C10Δ-GFP 2μm TRP1) |
| Y7637 | AN120 plus pME2630 (dtr1C15Δ-GFP 2μm TRP1) |
| Y7782 | AN120 plus pME2626 (DTR1-RFP 2μm TRP1) and pME2665 (dtr1C10Δ-GFP 2μ URA3) |
| Y8206 | AN120 plus pME2626 (DTR1-RFP 2μm TRP1) and pME2726 (GFP-DTR1 2μm URA3) |
| Y8207 | AN120 plus pME2626 (DTR1-RFP 2μm TRP1) and pME2730 (GFP-dtr1C10Δ 2μm URA3) |
| Y7820 | AN120, but heterozygous SEC7-dsRED::TRP1 plus pME2665 (dtr1C10Δ-GFP 2μm URA3) |
| Y7744 | AN120, but heterozygous SEC7-dsRED::TRP1 plus pME2650 (dtr1C15Δ-GFP 2μm URA3) |
| Y8018 | AN120 plus pME2650 (dtr1C15Δ-GFP 2μm URA3) and pME2697 (mRFP-GOS1 2μm TRP1) |
| Y8044 | AN120 plus pME2650 (dtr1C15Δ-GFP 2μm URA3) and pME2698 (mRFP-SED5 2μm TRP1) |
| Y7449 | AN120, but homozygous VPS10-GFP::KanMX4 |
| Y7451 | AN120, but homozygous VPS10-CΔ-GFP::KanMX4 |
| Y7750 | AN120, but homozygous VPS10-CΔ-DTR1C16-GFP::KanMX4 |
| Y8007 | AN120, but homozygous VPS10-TMΔCΔ-DTR1TMC10-GFP::KanMX4 |
| Y8190 | AN120, but homozygous VPS10-TMΔCΔ-DTR1TM-GFP::KanMX4 |
From reference 30.
Derived from S288C.
The sequences of oligonucleotide primers used in this study are available upon request. DNA-mediated transformation of yeast cells was performed using the lithium acetate procedure (13). The dtr1::KanMX4 deletion mutant, in which the entire Dtr1p open reading frame was replaced with that for KanMX4, was synthesized by PCR amplification using primers P527 and P535 from a knockout strain collection purchased from Research Genetics. Vps10p chimeric GFP fusions were constructed by PCR, using pKT127 (43) and primers P577, P578, P579, P597, and P580. The Vps10-TMΔCΔ-DTR1TM-C10-GFP fusion was synthesized by PCR with P613 and P615, using Y5120 genomic DNA as the template. The Vps10-TMΔCΔ-DTR1TM-GFP fusion was synthesized in two rounds of PCR. The first amplification used pME2611 (dtr1C10Δ-GFP) as a template with primers P525 and P613. The product was subsequently used in a second round with primer P615, with pKT127 (44) as the template. All integrants were confirmed by PCR using appropriate primers (P581, P582, P525, P614, and P615) and by sequence analysis.
Plasmid ME2441 (DTR1-GFP 2μm URA3) was constructed by digesting pRS424-DTR1-GFP (27) with BamHI and KpnI and inserting it into the corresponding sites of YEp352. pME2599 (dtr1N330Δ-GFP 2μm TRP1) was constructed in several steps. First, a 1.5-kb DTR1 fragment was amplified from pRS424-DTR1-GFP with P526 and P525 and digested with BglII and EcoRI. The resultant fragment was inserted into the BglII/EcoRI sites of pRS424-DTR1-GFP to generate pRS424-dtr1N92-280Δ-GFP. A 550-bp DTR1 fragment immediately upstream of the open reading frame was amplified from pRS424-DTR1-GFP with P539 and P541 and digested with BamHI and BglII. The resultant fragment was inserted into the BamHI/BglII sites of pRS424-DTR1-GFP to generate pRS424-dtr1N7-280Δ-GFP. A 1.5-kb DTR1 fragment was amplified from pRS424-DTR1-GFP with P566 and P525 and digested with BglII and EcoRI. The resultant fragment was inserted into the BglII/EcoRI sites of pRS424-dtr1N3-280Δ-GFP to generate pME2599.
To construct pME2611 (dtr1C10Δ-GFP 2μm TRP1), P527 and P589 were used to PCR amplify a 2-kb fragment, which was subsequently digested with BglII and EcoRI and inserted into the BglII and EcoRI sites of pRS424-DTR1-GFP. To construct pME2630 (dtr1C15Δ-GFP 2μm TRP1), a PCR-amplified 1.8-kb fragment using P593 and P599 was digested with BglII and EcoRI and inserted into the corresponding sites of pRS424-DTR1-GFP. To construct pME2589 (dtr1Loop6Δ-GFP 2μm TRP1) and pME2629 (dtr1Loop6Δ-RFP 2μm TRP1), two fragments, upstream and downstream of loop 6, were amplified by PCR with the primer pairs P541/P567 and P568/P525, respectively. The upstream and downstream fragments were digested with BamHI/XbaI and XbaI/EcoRI, respectively, and inserted into the BamHI/EcoRI sites of pRS424-DTR1-GFP. The BglII/EcoRI-digested fragment of dtr1Loop6Δ was cloned into the BglII/EcoRI sites of pME2626 (DTR1-RFP 2μm TRP1) to generate pME2629.
The T568A mutation was introduced during PCR with mutagenic primers (P527 and P565). The resulting mutant fragment was digested with BglII and EcoRI and inserted into the BglII and EcoRI sites of pRS424-DTR1-GFP to generate pME2585 (dtr1T568A-GFP 2μm TRP1). The dileucine (L560 and L561)-to-alanine mutations (LA; CTT to GCT and CTC to GCC) were generated using mutagenic primers P594 and P599. The resulting amplified sequences were digested with BglII and EcoRI and inserted into the BglII and EcoRI sites of pRS424-DTR1-GFP to generate pME2635 (dtr1LA-GFP 2μm TRP1). To generate pME2667 (dtr1LATA-GFP 2μm TRP1), primer P607 or P608 and P599 were used in PCR. The resulting fragments were digested with BglII and EcoRI and inserted into the BglII and EcoRI sites of pRS424-DTR1-GFP.
The CLB2 promoter-driven versions of the DTR1 mutants were constructed in several steps. First, DTR1 sequences were amplified with P541 and P536 and digested with BamHI and BglII, and the 97-bp fragment was inserted into the BamHI and BglII sites of pRS424-DTR1-GFP to generate a DTR1 version lacking the promoter and containing a BamHI site immediately upstream of the initiation codon. The CLB2 promoter was inserted into the BamHI site, whose ends had been filled in with the Klenow fragment of DNA polymerase, by digesting pFA6a-pCLB2-3HA-KanMX6 (17) with PacI, whose resultant overhanging ends were removed with T4 DNA polymerase, followed by digestion with BglII, whose resultant ends were filled in with the Klenow fragment of DNA polymerase, to generate pME2606 (CLB2p-DTR1-GFP 2μm TRP1). This was subsequently used for PCR amplification with primers P590 and P536. The PCR product was digested with BamHI and BglII and inserted into the corresponding sites of pRS424-DTR1-GFP mutant plasmids.
N-terminal fusions between GFP and Dtr1p were generated by amplifying the GFP sequence with in-frame BglII sites, using primers P626 and P627, and digesting the resulting product with BglII. The fragment was inserted into the N-terminal BglII sites of full-length DTR1 (pME2724) and dtr1C10Δ (pME2725) plasmids, which lack the C-terminal GFP sequence, to generate pME2726 and pME27310, respectively. All constructs were verified by sequence analysis.
Analysis of sporulation.
Sporulation on solid and liquid media was carried out as described previously (22). For analysis of GFP fusion proteins, live cells were examined by fluorescein-5-isothiocyanate optics at various times after induction of sporulation. Rhodamine optics was used to visualize monomeric red fluorescent protein (mRFP)-Sed5p, mRFP-Gos1p, Sec7p-dsRED, Dtr1p-RFP, and Dtr1L6Δp-RFP.
FM4-64 staining.
Cells were induced to sporulate in liquid SPO and subjected to FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide; Molecular Probes, Eugene, OR] staining as described by Morishita et al. (22).
Protein purification, SAP treatment, and immunoblot analysis.
Approximate 1 × 109 cells induced in sporulation medium or grown in appropriate SC dropout medium were collected by centrifugation, resuspended in 200 μl breaking buffer (10 mM Tris, pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol) containing 1% Triton X-100, protein inhibitor cocktail (1 tablet/10 ml; Roche Diagnostics Corp., Indianapolis, IN), 0.5 mM phenylmethylsulfonyl fluoride, and phosphatase inhibitors (50 mM NaF, 10 mM sodium pyrophosphate, and 40 mM β-glycerophosphate), and disrupted by vigorous vortexing with glass beads for 5 min at 4°C. Cell lysates were collected by centrifugation and subjected to further centrifugation at 14,000 rpm for 20 min at 4°C, following the addition of 100 μl cell breaking buffer. The lysates (∼600 μg) were normalized with extraction buffer to a final volume of 500 μl and incubated with Aequorea victoria anti-GFP polyclonal antibody (Clontech, Mountain View, CA) at a 1:200 dilution with rocking at 4°C for 2 h. Fifty microliters of 50% protein A immunoglobulin G beads (Pierce, Rockford, IL) was added to each sample and incubated overnight with rocking at 4°C. Following incubation, the beads were washed four times with 1 ml breaking buffer without phosphatase inhibitors and once with shrimp alkaline phosphatase (SAP) buffer. Protein A beads resuspended in 1 ml SAP buffer were split into three tubes, and the buffer was removed. SAP treatment was performed at 30°C for 30 min in the absence or presence of SAP (U.S. Biological, Swampscott, MA), with or without phosphatase inhibitors (1 mM sodium pyrophosphate, 0.5 mM EDTA, and 0.5 mM EGTA). The reaction was terminated by the addition of 2× sample buffer. Ten microliters of each sample was loaded and separated in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred to a nitrocellulose membrane. Dtr1p-GFP was detected with N86/38 (NeuroMab, Davis, CA) followed by horseradish peroxidase-conjugated goat anti-rabbit secondary antibody at a 1:1,000 dilution (Antibody Incorporated, Davis, CA).
RESULTS
The large internal loop 6 of Dtr1p is required for ER exit.
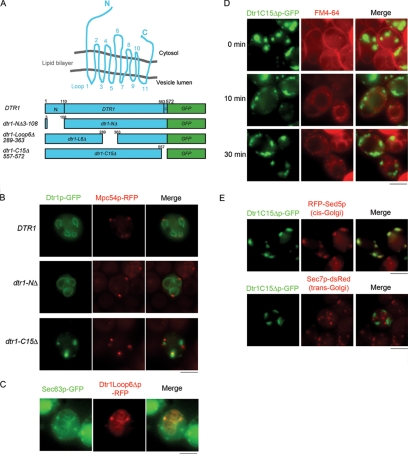
During sporulation, the integral transmembrane dityrosine transporter Dtr1p is synthesized in the endoplasmic reticulum (ER) and then transits the Golgi complex before reaching its final destination at the PSM (27). Dtr1p contains 572 amino acids, with 12 predicted transmembrane domains, cytoplasmic N and C termini, and a large cytoplasmic loop in the middle; the predicted topology of Dtr1p in the lipid bilayer is shown in Fig. 1A. To determine which regions of Dtr1p are important for transport to the PSM, we constructed several truncations/in-frame deletions of Dtr1p fused with GFP. These included versions lacking the amino-terminal region (NΔ), the large cytoplasmic loop in the middle (L6Δ), and the carboxyl 15 amino acids (C15Δ) (Fig. 1A). While the wild-type fusion is functional, none of the mutant Dtr1p-GFP fusions are functional, based on complementation of the dtr1Δ mutant phenotype (data not shown). We examined the fusions by fluorescence microscopy in living wild-type cells, induced to sporulate, expressing the MOP component Mpc54p-RFP, which is expressed only once cells have progressed to the second meiotic division (14). Full-length Dtr1p-GFP and Dtr1NΔp-GFP initially colocalized with Mpc54p-RFP and then outlined the developing PSM (Fig. 1B), indicating that the N-terminal extension does not play a role in the localization of the protein to the PSM. In contrast, Dtr1L6Δp-GFP failed to localize to the PSM and instead showed a fluorescence pattern similar to that of ER proteins (data not shown). To confirm ER localization of Dtr1L6Δp, we expressed Dtr1L6Δp-RFP in cells harboring the resident ER protein Sec63p-GFP, which is essential for signal recognition particle translocation (32). Dtr1L6Δp-RFP signals largely overlapped with Sec63p-GFP at the peripheries of nuclei and of spores, indicating that Dtr1L6Δp was trapped in the ER (Fig. 1C).
FIG. 1.
Internal loop 6 and the C-terminal region of Dtr1p are essential for PSM localization. (A) Cartoon of predicted topology of Dtr1p in the lipid bilayer and the truncation/deletion mutant proteins used in this study. Predicted amino acid positions of transmembrane domains are as follows: TM1, 111 to 133; TM2, 148 to 160; TM3, 177 to 200; TM4, 203 to 226; TM5, 237 to 260; TM6, 265 to 287; TM7, 367 to 382; TM8, 404 to 426; TM9, 447 to 469; TM10, 473 to 495; TM11, 508 to 531; and TM12, 540 to 562 (http://db.yeastgenome.org/cgi-bin/protein). (B) Localization of full-length Dtr1p-GFP, Dtr1NΔp-GFP, and Dtr1C15Δp-GFP in sporulating cells (Y7730, Y7747, and Y7733, respectively) expressing Mpc54p-RFP, a MOP marker. Dtr1C15Δp-GFP failed to be transported to the PSM and showed cytoplasmic puncta in the vicinity of Mpc54p-RFP signals. (C) Dtr1Loop6Δp-RFP localized to the ER, as demonstrated by colocalization with the ER marker Sec63p-GFP, in sporulating cells (Y7657). Sec63p-GFP showed dispersed staining in the cytoplasm in addition to ER staining. (D) FM4-64 staining of sporulating cells expressing Dtr1C15Δp-GFP (Y7637) 5 h after the induction of sporulation. FM4-64 stained PM (0 min), was internalized into the cytoplasm (10 min), and was transported to vacuoles (30 min). Dtr1C15Δp-GFP puncta did not overlap with FM4-64 signals at any time point. (E) Localization of Dtr1C15Δp-GFP in sporulating cells expressing mRFP-Sed5p (Y8044) or Sec7p-dsRed (Y7744). Dtr1C15Δp-GFP did not colocalize with Sec7p-dsRed, but signals overlapped with mRFP-Sed5p. Bars, 5 μm.
Previously, it was reported that Dtr1p expressed in vegetative cells localizes to the PM (11, 28). To examine whether deletion of Dtr1p loop 6 also impaired transport during vegetative growth, we expressed Dtr1L6Δp-GFP under the control of the CLB2 promoter (17). Similar to what was observed in cells induced to sporulate, Dtr1L6Δp-GFP was found predominantly in the ER in mitotically dividing cells (data not shown). Thus, loop 6 is important for exit from the ER in both vegetative and sporulating cells.
The Dtr1p carboxyl-terminal 15 amino acids are essential for transport through the Golgi complex.
Deletion of the carboxyl-terminal 15 amino acids resulted in a protein that failed to reach the PSM but was instead detected in cytoplasmic puncta in the vicinity of Mpc54p-RFP fluorescence (Fig. 1B). To determine the nature of the Dtr1C15Δp-GFP punctate structures, we stained cells expressing Dtr1C15Δp-GFP with the lipophilic endocytosis dye FM4-64 (46). FM4-64 is bound to the PM (Fig. 1D, 0 min), internalized by endocytosis (Fig. 1D, 10 min), and transported to the vacuolar membrane (Fig. 1D, 30 min) (22). Dtr1C15Δp-GFP puncta did not overlap with FM4-64-labeled compartments at any time point (Fig. 1D), indicating that Dtr1C15Δp-GFP is not trapped in endosomal or vacuolar compartments.
We also coexpressed Dtr1C15Δp-GFP and the cis-Golgi marker mRFP-Sed5p (20), the medial-Golgi marker mRFP-Gos1p (20), or the trans-Golgi marker Sec7p-dsRed (18) in wild-type cells induced in sporulation medium. We observed extensive colocalization between Dtr1C15Δp-GFP and mRFP-Sed5p; Dtr1C15Δp-GFP puncta did not overlap with Sec7p-dsRed (Fig. 1E). While mRFP-Gos1p showed the expected punctate pattern in vegetative cells, during sporulation mRFP-Gos1p was observed primarily in the vacuole and thus also did not colocalize with Dtr1C15Δp-GFP (data not shown). Nonetheless, the colocalization experiments suggest that Dtr1C15Δp-GFP is trapped in an early Golgi compartment.
Dtr1C15Δp-GFP is transported directly to vacuoles in mitotically dividing cells.
To examine whether deletion of the Dtr1p C terminus also affects the localization of the protein during vegetative growth, we expressed Dtr1p-GFP and Dtr1C15Δp-GFP under the control of the CLB2 promoter (17). As expected, CLB2 promoter-driven Dtr1p-GFP localized to the PM and was also observed in vacuoles in vegetative cells (Fig. 2A). In contrast to the wild type, CLB2 promoter-driven Dtr1C15Δp-GFP was not detected at the PM but was found only in vacuoles, with a low intensity of GFP fluorescence (Fig. 2B).
FIG. 2.
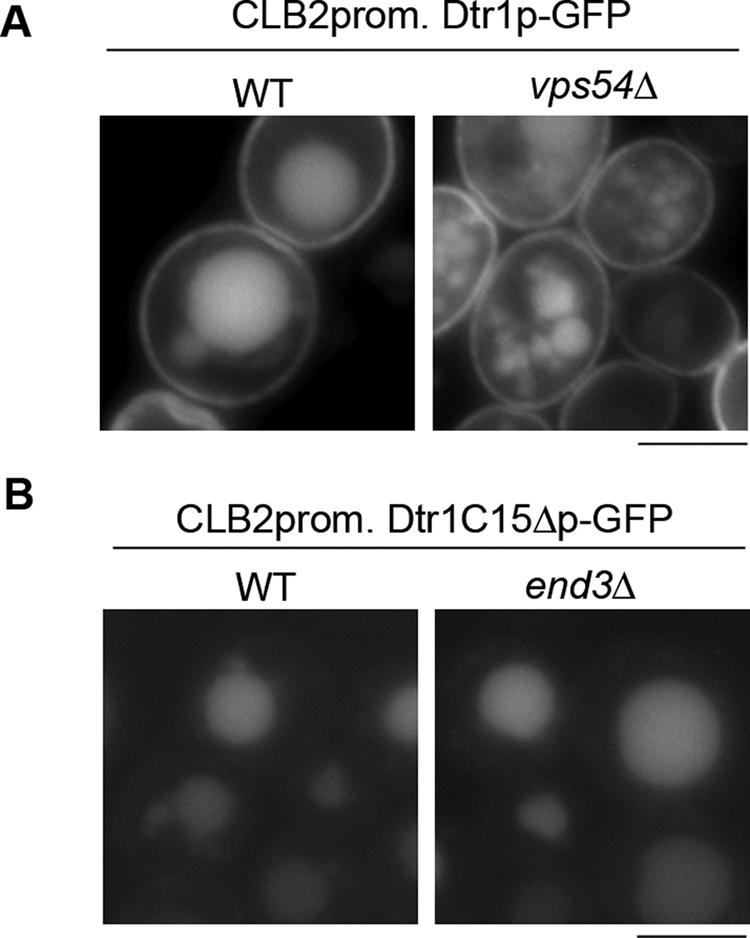
Dtr1C15Δp-GFP is transported directly to vacuoles in vegetative cells. (A) Dtr1p-GFP was expressed under the control of the CLB2 promoter in wild-type (WT) (Y7479) and vps54Δ (Y7557) cells. Dtr1p-GFP in vps54Δ cells showed an identical GFP pattern, localized in the PM and vacuoles, to that in wild-type cells, suggesting that Dtr1p-GFP is not recycled through the endosome. (B) Dtr1C15Δp-GFP expressed in vegetative cells was detected at the vacuoles in wild-type cells (Y7777). An identical staining pattern was seen in the endocytosis-defective end3Δ mutant (Y7807), suggesting Dtr1C15Δp-GFP was not transported to the PM. Bars, 5 μm.
The localization of Dtr1C15Δp-GFP to vacuoles could be due to a defect in forward transport from the Golgi complex, enhanced endocytosis and transport to the vacuole, or a failure in retrieval of the protein after endocytosis from the PM. To distinguish between these possibilities, we first determined if Dtr1p expressed in vegetative cells is recycled via the endosome. To that end, we examined the localization of CLB2 promoter-driven Dtr1p-GFP in the vps54Δ mutant; Vps54p is a component of the GARP tethering complex that is involved in recycling membrane proteins such as the v-SNARE Snc1p (33). Dtr1p-GFP localized to the PM and vacuoles in vps54Δ cells, similar to the case for wild-type cells, suggesting that Dtr1p-GFP is not recycled through the endosome in a GARP-dependent manner (Fig. 2A). To determine if Dtr1C15Δp is transported to the PM before accumulating in vacuoles, we examined Dtr1C15Δp-GFP in the endocytosis-defective end3Δ mutant (34). As shown in Fig. 2B, Dtr1C15Δp-GFP was observed only in vacuoles in the end3Δ mutant, indicating that Dtr1C15Δp is transported directly to vacuoles from the Golgi complex. These results suggest that deletion of the C-terminal 15 amino acids affects the localization of Dtr1p during both sporulation and vegetative growth; however, distinct localization patterns are observed under these two conditions.
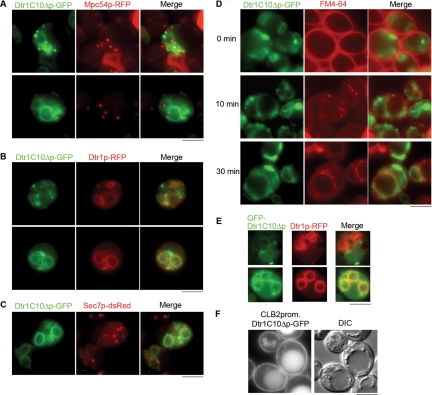
Deletion of the C-terminal 10 amino acids in Dtr1p causes a delay in transport to the PSM.
To determine the specific transport signals within the last 15 amino acids, we constructed a Dtr1p fusion lacking only the C-terminal 10 amino acids (Dtr1C10Δp-GFP) and examined the localization of the protein during sporulation and in vegetative cells. In cells induced to sporulate, GFP signals were detected as punctate structures as well as some diffuse staining in the cytoplasm (Fig. 3A). However, as sporulation proceeded, Dtr1C10Δp-GFP was found at the PSM. The Dtr1C10Δp-GFP punctate structures partially colocalized with the trans-Golgi marker Sec7p-dsRed (Fig. 3C) and with vacuole compartments marked by FM4-64 (Fig. 3D), but not with RFP-Sed5p (cis-Golgi) (data not shown), suggesting that some portion of Dtr1C10Δp-GFP was in the trans-Golgi apparatus as well as in vacuoles. To confirm that Dtr1C10Δp-GFP is delayed for transport to the PSM, we coexpressed Dtr1C10Δp-GFP with full-length Dtr1p-RFP and observed clear PSM localization of Dtr1p-RFP in cells where Dtr1C10Δp-GFP was seen in puncta. Colocalization of Dtr1C10Δp-GFP and Dtr1p-RFP at PSMs was seen only in cells in which PSMs were fully developed at a late stage of sporulation (Fig. 3B). To determine if GFP appended to the C terminus of Dtr1C10Δp affected localization, we also examined N-terminally tagged GFP-Dtr1p and GFP-Dtr1C10Δp fusions. GFP-Dtr1p behaved similarly to Dtr1p-GFP (data not shown), while GFP-Dtr1C10Δp showed punctate structures, vacuole staining, and delayed transport to the PSM, as observed with the C-terminal fusion (Fig. 3E). During vegetative growth, CLB2 promoter-driven Dtr1C10Δp-GFP showed an identical localization pattern to that of full-length Dtr1p-GFP (Fig. 3F), suggesting that the C-terminal 10 amino acids specifically promote PSM localization independent of PM localization.
FIG. 3.
Deletion of the Dtr1p C-terminal 10 amino acids causes a delay in transport to the PSM. (A) Localization of Dtr1C10Δp-GFP in sporulating cells expressing Mpc54p-RFP (Y7732). In addition to PSM signals, cytoplasmic punctate signals were detected near PSMs. (B) Sporulating cells coexpressing Dtr1C10Δp-GFP and Dtr1p-RFP (Y7782) showed a delay in transport of Dtr1C10Δp-GFP to PSMs (top panels; early in sporulation) and colocalization with Dtr1p-RFP at late stages of spore formation (bottom panels; late in sporulation). (C) Localization of Dtr1C10Δp-GFP in cells expressing Sec7p-dsRed (Y7820). Dtr1C10Δp-GFP puncta were partially overlapping with Sec7p-dsRed signals. (D) FM4-64 staining in sporulating cells expressing Dtr1C10Δp-GFP (Y7526). Cells were induced to sporulate and stained with FM4-64 as described in the legend to Fig. 1. Some overlap of Dtr1C10Δp-GFP and FM4-64 signals was seen at the vacuole. (E) Sporulating cells coexpressing GFP-Dtr1C10Δp and Dtr1p-RFP (Y8207) show GFP-Dtr1C10Δp in puncta and Dtr1p-RFP at PSMs (top panels; early in sporulation), with colocalization at PSMs at late stages of spore formation (bottom panels; late in sporulation). (F) CLB2 promoter-driven Dtr1C10Δp-GFP localized to the PM and vacuoles in vegetative cells (Y7592). A differential interference contrast (DIC) image of the same cells is shown. Bars, 5 μm.
Dtr1p is a phosphoprotein, but the predicted phosphorylation site in the C terminus does not influence PSM localization.
Protein phosphorylation is a common mechanism used to regulate protein function in sporulation (15, 19, 39, 41), and of particular importance, it has been shown to influence protein localization (37). Dtr1p contains 22 serine/threonine residues that are predicted to be phosphorylated (http://www.cbs.dtu.dk/services/NetPhos/). Eleven of these residues reside in the N-terminal extension, and one residue is located at position 568 (T568), within the last C-terminal 10 amino acids (see Fig. 5A). To determine if Dtr1p phosphorylation is a signal for transport of the protein to the PSM, we first examined whether Dtr1p is phosphorylated in vivo. To that end, CLB2-driven Dtr1p-GFP isolated from vegetative cells and Dtr1p-GFP isolated from cells induced in sporulation medium were immunoprecipitated and treated with SAP in the absence or presence of phosphatase inhibitors. The reaction mixtures were subjected to SDS-PAGE and immunoblot analysis. The mobility of untreated Dtr1p-GFP was very heterogeneous in nature, suggesting extensive posttranslational modifications. Dtr1p-GFP mobility increased after SAP treatment (Fig. 4, lanes 2 and 6) compared to that for both untreated cells and SAP treatment in the presence of phosphatase inhibitors (Fig. 4, lanes 1, 3, 5, and 7) when the protein was isolated from either vegetative or sporulating cells. Even following SAP treatment, Dtr1p-GFP migrated as a heterogeneous population, suggesting that Dtr1p is subject to other posttranslational modifications in addition to phosphorylation. It is unlikely that the GFP sequences are modified posttranslationally, as the changes in mobility are also seen with TAP (tandem affinity purification) versions of Dtr1p (data not shown). Since 11 of the predicted phosphorylation sites reside in the N-terminal extension, we also examined the N-terminal deletion and found that there was little change in mobility in the presence and absence of phosphatase (Fig. 4, lanes 8 to 10). Taken together, these results indicate that Dtr1p is a phosphoprotein and that the change in mobility observed on SDS-PAGE is due to phosphorylation of the N-terminal domain.
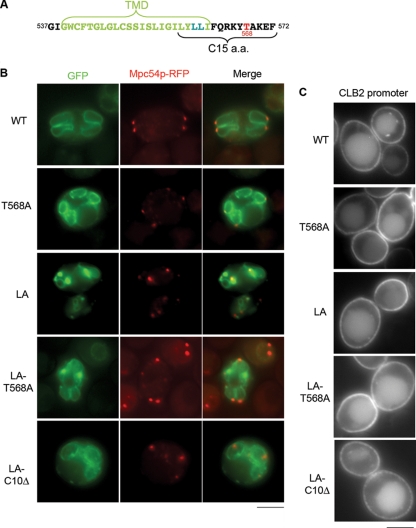
FIG. 5.
The dileucine motif in the C terminus of Dtr1p plays a role in PSM localization. (A) Sequence of the C-terminal tail of Dtr1p. The dileucine motif (blue) and the threonine (red) predicted to be phosphorylated are indicated. (B) Localization of mutant Dtr1 proteins during sporulation. Dtr1LAp-GFP, Dtr1T568Ap-GFP, Dtr1LA-T568Ap-GFP, and Dtr1LA-C10Δ-GFP were expressed in wild-type (WT) cells harboring MPC54-RFP (Y7749, Y7735, Y7784, and Y7884, respectively). The threonine-to-alanine (T568A) mutation did not affect PSM localization, while the dileucine mutation to alanine (LA) showed partial defects in PSM localization. The combination of the T568A and LA mutations showed an identical localization pattern to that of the LA mutant, while the LA-C10Δ mutation showed defects similar to those with the C10Δ mutation (Fig. 3). (C) Vegetative localization of mutant Dtr1p-GFPs at PM. Strains: LA, Y7752; T568A, Y7612; LA-T568A, Y7803; LA-C10D, Y7889. Bars, 5 μm.
FIG. 4.
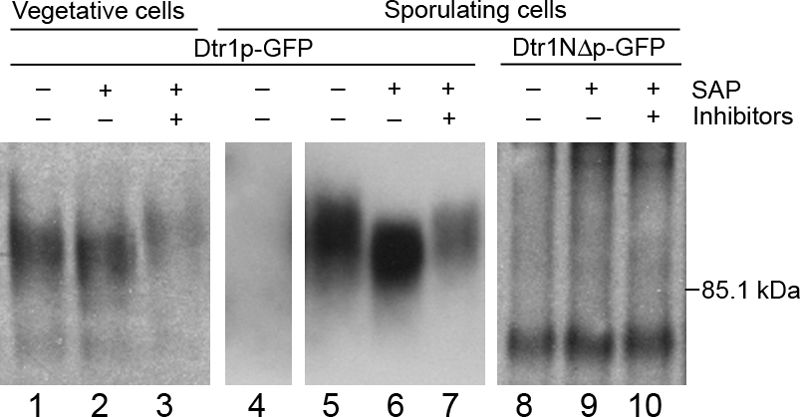
Dtr1p-GFP is a phosphoprotein. CLB2-driven Dtr1p-GFP extracted from vegetative cells (Y7479) (lanes 1 to 3), untagged Dtr1p extracted from sporulating cells (Y6582) (lane 4), and Dtr1p-GFP extracted from sporulating cells (Y6583) (lanes 5 to 7) were treated with (+) or without (−) SAP in the presence (+) or absence (−) of phosphatase inhibitors and subjected to SDS-PAGE followed by immunoblot analysis. Dtr1p-GFP showed a change in mobility following treatment with SAP that was blocked in the presence of inhibitors, indicating that Dtr1p is a phosphoprotein. Analysis of Dtr1NΔp-GFP (Y7419) (lanes 8 to 10) suggests that the change in mobility observed on SDS-PAGE is due to phosphorylation of the N-terminal cytoplasmic domain.
To test whether phosphorylation of T568 in the C terminus contributes to PSM localization, we mutated the threonine to alanine (T568A), which mimics the nonphosphorylated state (Fig. 5A), and analyzed localization of the mutant protein. Dtr1T568Ap-GFP localized to the PSM similarly to the wild type (Fig. 5B), suggesting that phosphorylation of T568 is not necessary for Dtr1p transport to the PSM.
A dileucine in the C terminus plays a role in PSM localization.
Dileucine motifs serve as signals for the translocation of membrane proteins (3). The C-terminal 15 amino acids of Dtr1p-GFP contain two leucines, at positions 560 and 561 (Fig. 5A). To determine if this dileucine pair is important for PSM transport of Dtr1p, both leucines were mutated to alanines (LA) and the resulting fusion protein was examined in sporulating cells. Dtr1LAp-GFP was slightly impaired for localization, as cytoplasmic puncta were observed in addition to PSM staining (Fig. 5B), suggesting that this motif contributes to efficient PSM localization. Dtr1LAp-GFP also mutated for T568A (LA-T568A) behaved similarly to Dtr1LAp-GFP, suggesting that there is no additive effect of the dileucine and T568A mutations. Both Dtr1LAp-GFP and Dtr1LA-T568Ap-GFP localized to the PM and vacuoles in vegetative cells, similar to the wild type (Fig. 5C). When the LA mutation was combined with the C10Δ mutation (Dtr1LA-C10Δp-GFP), a similar localization pattern to that for Dtr1C10Δp-GFP was observed during both sporulation and vegetative growth (Fig. 5B and C). These results suggest that the dileucine motif in the Dtr1p C terminus contributes to but is not essential for PSM transport.
The C-terminal region is partially sufficient for Dtr1p localization to the PSM.
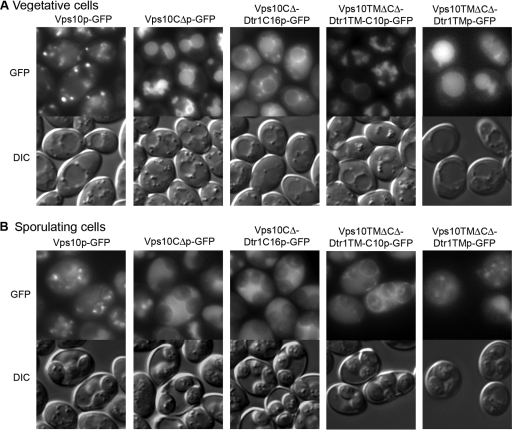
To examine whether the Dtr1p C-terminal tail is sufficient to recruit a transmembrane protein to the PSM, we constructed chimeric proteins between Vps10p and Dtr1p. Vps10p, a type I transmembrane receptor protein involved in sorting the soluble vacuolar hydrolase carboxypeptidase Y, cycles between endosomes and the Golgi complex (9). The cytoplasmic tail of Vps10p is important for stability, function, and localization (4); removal of the Vps10p C-terminal domain (Vps10CΔp) results in its vacuolar delivery and degradation (44). We constructed a GFP-tagged chimera of Vps10CΔp fused to the C-terminal 16 amino acids of Dtr1p (Vps10CΔ-Dtr1C16p-GFP). As previously reported, Vps10p-GFP was observed in punctate structures, which presumably represent endosome/Golgi compartments, while Vps10CΔp-GFP was detected at vacuoles in vegetative cells (Fig. 6A). During sporulation, Vps10p-GFP and Vps10CΔp-GFP did not alter their localization patterns in spores and cytoplasm of the asci (Fig. 6B). The Vps10CΔ-Dtr1C16p-GFP chimera was observed dispersed in the cytoplasm with some bright puncta, in addition to vacuolar staining in both vegetative and sporulating cells (Fig. 6B), indicating that there was only a subtle effect of the Dtr1p C terminus on the transport of Vps10CΔp.
FIG. 6.
The C-terminal region is partially sufficient for Dtr1p localization to the PSM. Strains harboring the indicated chimeras were incubated on YPAD solid medium (vegetative cells) (A) and SPO solid medium (sporulating cells) (B). During both vegetative growth and sporulation, Vps10p-GFP showed the typical pattern for endosome/Golgi proteins and Vps10CΔp-GFP was found at vacuoles. Vps10CΔDtr1C16p-GFP showed vacuolar signals with some cytoplasmic puncta and diffuse staining in the cytoplasm of vegetative cells, with a relatively lower intensity of vacuolar signals in sporulating cells. Vps10TMΔCΔ-Dtr1TM-C10p-GFP was observed in vacuoles in vegetative cells and at PSMs as well as in vacuoles in sporulating cells. Vps10TMΔCΔ-Dtr1TM-GFP was observed in vacuoles in vegetative and sporulating cells. Strains: Y7449, VPS10-GFP; Y7451, VPS10CΔ-GFP; Y7750, VPS10CΔ-DTR1C16-GFP; Y8007, VPS10TMΔCΔ-DTR1TM-C10-GFP; and Y8190, VPS10TMΔCΔ-DTR1TM-GFP. Bars, 5 μm.
To determine if additional signals are important for transport to the PSM, we constructed a chimera that replaced the transmembrane domain of Vps10p with the 12th Dtr1p transmembrane domain followed by the C-terminal tail (Vps10TMΔCΔ-Dtr1TMCp-GFP). Interestingly, Vps10TMΔCΔ-Dtr1TMCp-GFP was observed in the cytoplasm during vegetative growth and in the cytoplasm and at PSMs late in sporulation (Fig. 6B). To determine if the transmembrane domain alone could confer PSM localization, we also generated a chimera that replaced the Vps10p transmembrane domain with the Dtr1p transmembrane domain (Vps10TMΔCΔ-Dtr1TMp-GFP). This chimera behaved similarly to Vps10CΔp in that it was transported to vacuoles (Fig. 6). Taken together, these results suggest that both the last transmembrane domain and the C-terminal tail play a role in transport to the PSM.
DISCUSSION
Intracellular transport pathways not only are responsible for maintaining the normal compartmentalization of all eukaryotic cells but also are altered during development for the formation of cell-type-specific organelles. The formation of the PSM during yeast sporulation is an excellent model for elucidating how membrane trafficking pathways are modified to transport lipids, proteins, and other material for the de novo synthesis of cell-type-specific organelles. While a number of proteins required for formation of the PSM have been identified, the cis-acting signals that direct proteins to this novel membrane compartment have not been investigated. In this study, we dissected the signals on the sporulation-specific integral membrane dityrosine transporter, Dtr1p, that promote its transport to the PSM. Based on the predicted topology of Dtr1p in the membrane, Dtr1p contains three significant cytoplasmic regions, including an N-terminal extension, a large loop in the middle of the protein, and the C-terminal tail (Fig. 1A). Individual deletion of these predicted cytoplasmic domains revealed that both the C terminus and the large internal loop are required for transport of the protein to the PSM (Fig. 1). Although the N-terminal domain does not appear to influence the ability of Dtr1p-GFP to localize to the PSM, it is important for the dityrosine transport function of the protein.
ER exit.
Following synthesis in the ER, transmembrane proteins are incorporated into transport vesicles that move through the endomembrane system to their final destination. Exit from the ER is accomplished by the incorporation of cargo proteins into COPII-coated vesicles, either by binding COPII directly or through specific export receptors (2). Cytoplasmic ER export signals are diverse and include short hydrophobic motifs, longer sequences, folded determinants, or combinations of these signals (2). Deletion of the large cytoplasmic loop 6 of Dtr1p resulted in a protein that was trapped in the ER, suggesting that this region contains a signal(s) for exit from the ER. Two copies of the hydrophobic motif, FF (F = phenylalanine), are present in loop 6; this motif has been shown to be a signal for ER export in a number of proteins (2). Thus, these hydrophobic amino acid pairs may represent the signal(s) that mediates Dtr1p incorporation into COPII-coated vesicles and exit from the ER. Alternatively, removal of the large internal loop may cause misfolding of the protein, resulting in its retention in the ER.
Surprisingly, it was recently shown that exit of proteins from the ER has an altered genetic requirement during sporulation, as simultaneous deletion of the ER export receptors, Erv14p and Erv15p, blocks the exit of a number of proteins, including Dtr1p, in cells induced to sporulate but not in mitotically dividing cells (28). However, since deletion of the Dtr1p internal loop blocks exit from the ER in both vegetative and sporulating cells, either this must cause general misfolding or this domain binds COPII and/or receptors under both conditions. If the latter is true, this would suggest either that the same signal(s) is used or that distinct sorting signals for ER exit during sporulation and in dividing cells reside within this cytoplasmic loop.
Transit through Golgi cisternae.
After exit from the ER, transmembrane proteins are trafficked through the Golgi complex, which serves as the main sorting organelle of the cell. Removal of the last 15 amino acids of Dtr1p resulted in a protein that accumulated in an early Golgi compartment during sporulation, colocalizing with the cis-Golgi SNARE mRFP-Sed5p (20). In contrast to a deletion mutant lacking the internal loop, which was trapped in the ER in both vegetative and sporulating cells, Dtr1p lacking the last 15 amino acids did not accumulate in the cis-Golgi in vegetative cells but was instead found at vacuoles. In addition, while the medial-Golgi SNARE Gos1p localized as expected in mitotically dividing cells, it accumulated in vacuoles in cells induced to sporulate (data not shown). These observations suggest that there is an overall change in the organization of the Golgi complex during sporulation and/or that some proteins are altered with respect to their steady-state levels within a particular Golgi compartment. Thus, as recently reported for organelle inheritance (45), there appear to be distinct changes in the Golgi complex during this developmental program.
Deletion of the last 15 amino acids of Dtr1p is predicted to remove not only the C-terminal cytoplasmic tail but also a few amino acids of the last transmembrane domain. Transmembrane domains form alpha helices that span the lipid bilayer and are approximately 20 amino acids in length. Based on prediction programs, the last Dtr1p transmembrane domain comprises 23 amino acids. However, the exact boundaries of transmembrane domains need to be verified experimentally, as current programs do not always correctly predict which amino acids are contained within the bilayer (5). Thus, it remains possible that deletion of the last 15 amino acids removes only the cytoplasmic tail.
Contained within the last 15 amino acids is a dileucine pair, which has been shown to be a motif for sorting proteins to the vacuole. This dileucine does not fit consensus motifs, defined as [DE]XXXL[LI] or DXXLL (X = any amino acid), seen in vacuolar proteins such as Vam3p and ALP, whose sorting is dependent on AP-3 (10). The leucine-to-alanine mutations showed only subtle defects in PSM localization, suggesting that this dileucine pair plays a minor role in PSM transport. This may be due to disruption of the transmembrane domain or alteration of specific sequences.
Previous work has demonstrated a strong correlation between transmembrane domain lengths of single-transmembrane-domain proteins and localization in specific compartments, such that longer domains favor localization at the PM, while shorter domains favor localization in internal membranes (24, 25, 35). If transmembrane domains also influenced sorting of multiple-transmembrane-domain proteins, then deletion of the last 15 amino acids of the multipass Dtr1p protein might shorten the transmembrane domain and may explain why it is retained in the Golgi complex. The finding that this deletion differentially influences sorting in vegetative cells and during sporulation raises the possibility that transmembrane domain length may influence the sorting between the PM and the PSM (see below).
Golgi complex-to-PSM transport.
Dtr1p lacking only the last 10 amino acids exhibited a delay in transport to the PSM and accumulated in a trans-Golgi compartment during sporulation. In contrast, the same protein expressed in vegetative cells was found at the PM, similar to the wild-type protein. These results suggest that the C-terminal tail plays a specific role in transport from the trans-Golgi to the PSM and provides further evidence that protein sorting at the Golgi complex is altered during sporulation.
Examination of the last 10 amino acids does not reveal any striking motif; BLAST searches with this region of the protein did not identify other proteins that are transported to the PSM during sporulation. Consistent with this, while the C terminus is necessary, when it was appended onto Vps10p lacking its signal for retrieval from endosomes, the sequence was not sufficient to promote transport to the PSM. This suggests that additional sequences are required for directing proteins to the PSM. Previously, it was shown that the destinations of membrane proteins leaving the yeast Golgi complex are affected by the length and composition of the transmembrane domain (35). When both the last transmembrane domain and the C terminal tail are appended to Vps10p, a fraction of this chimera is found at the PSM (Fig. 6). However, when the last Dtr1p transmembrane domain is fused to Vps10p, the protein is still transported to the vacuole. Thus, it appears that the most C-terminal transmembrane domain and the cytoplasmic domain of Dtr1p are necessary and partially sufficient to promote Golgi complex-to-PSM transport. Since multiple transport signals are used even for targeting to the same organelle, it is not surprising that a “consensus” sorting signal for PSM localization was not revealed by these studies. The generality of the requirement for transmembrane and C-terminal cytoplasmic domains in PSM transport awaits future analyses.
Posttranslational modifications and sorting to the PSM.
Previously, it was shown that phosphorylation correlates with changes in protein localization during sporulation (37, 39). Dtr1p contains several predicted phosphorylation sites throughout the protein, including one in the C-terminal tail. The change in mobility of Dtr1p-GFP on SDS-PAGE in the presence and absence of phosphatase indicates that Dtr1p is a phosphoprotein. Analysis of the N-terminal deletion revealed that the mobility shift is primarily due to phosphorylation of the N-terminal cytoplasmic domain, which our studies have shown does not play a role in Dtr1p transport. Furthermore, changing the single predicted threonine phosphoacceptor site in the C-terminal cytosolic domain (T568) to an alanine, thereby preventing phosphorylation of this residue, had no effect on PSM transport, indicating that phosphorylation at T568 is dispensable for Dtr1p transport to the PSM. Thus, phosphorylation of the C-terminal sorting domain is unlikely to play a major role in Dtr1p transport to the PSM.
Even after phosphatase treatment, Dtr1p was very heterogeneous, suggesting that Dtr1p undergoes additional posttranslational modifications. Membrane proteins are glycosylated as they transit the endomembrane system; thus, it is likely that at least some of the Dtr1p heterogeneity is due to glycosylation. Interestingly, these carbohydrates can serve as sorting signals (40). Whether proteins are differentially modified by glycosylation or other posttranslational modifications for transport to the PSM during sporulation is a question for future investigation.
Acknowledgments
We thank B. Glick (University of Chicago), A. Nakano (RIKEN), and J. Nunnari (University of California, Davis) for the generous gifts of plasmids. We also thank A. Jaramillo-Lambert, R. Mendonsa, and J. Trimmer for useful advice, helpful discussions, and support.
This work was supported in part by National Institutes of Health research grant GM66124 to J.E.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Bajgier, B. K., M. Malzone, M. Nickas, and A. M. Neiman. 2001. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell 121611-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino, J. S., and B. S. Glick. 2004. The mechanisms of vesicle budding and fusion. Cell 116153-166. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72395-447. [DOI] [PubMed] [Google Scholar]
- 4.Cereghino, J. L., E. G. Marcusson, and S. D. Emr. 1995. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell 61089-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, Y., and J. B. Konopka. 2006. Accessibility of cysteine residues substituted into the cytoplasmic regions of the alpha-factor receptor identifies the intracellular residues that are available for G protein interaction. Biochemistry 4515310-15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coluccio, A., E. Bogengruber, M. N. Conrad, M. E. Dresser, P. Briza, and A. M. Neiman. 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 31464-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conibear, E., and T. H. Stevens. 2000. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11305-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly, J. E., and J. Engebrecht. 2006. The Arf-GTPase-activating protein Gcs1p is essential for sporulation and regulates the phospholipase D Spo14p. Eukaryot. Cell 5112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, A. A., and T. H. Stevens. 1996. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 133529-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darsow, T., C. G. Burd, and S. D. Emr. 1998. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol. 142913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sa-Correia, and P. Briza. 2002. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata, A., and C. Shimoda. 1994. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast 10173-183. [DOI] [PubMed] [Google Scholar]
- 13.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 193657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamoorthy, T., X. Chen, J. Govin, W. L. Cheung, J. Dorsey, K. Schindler, E. Winter, C. D. Allis, V. Guacci, S. Khochbin, M. T. Fuller, and S. L. Berger. 2006. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 202580-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces cell cycle, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 17.Lee, B. H., and A. Amon. 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300482-486. [DOI] [PubMed] [Google Scholar]
- 18.Losev, E., C. A. Reinke, J. Jellen, D. E. Strongin, B. J. Bevis, and B. S. Glick. 2006. Golgi maturation visualized in living yeast. Nature 4411002-1006. [DOI] [PubMed] [Google Scholar]
- 19.Maier, P., N. Rathfelder, M. G. Finkbeiner, C. Taxis, M. Mazza, S. Le Panse, R. Haguenauer-Tsapis, and M. Knop. 2007. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 261843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuura-Tokita, K., M. Takeuchi, A. Ichihara, K. Mikuriya, and A. Nakano. 2006. Live imaging of yeast Golgi cisternal maturation. Nature 4411007-1010. [DOI] [PubMed] [Google Scholar]
- 21.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 206946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 1701561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morishita, M., R. Mendonsa, J. Wright, and J. Engebrecht. 2007. Snc1p v-SNARE transport to the prospore membrane during yeast sporulation is dependent on endosomal retrieval pathways. Traffic 81231-1245. [DOI] [PubMed] [Google Scholar]
- 24.Munro, S. 1995. A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem. Soc. Trans. 23527-530. [DOI] [PubMed] [Google Scholar]
- 25.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 144695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi, H., P. de los Santos, and A. M. Neiman. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 151802-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakanishi, H., M. Morishita, C. L. Schwartz, A. Coluccio, J. Engebrecht, and A. M. Neiman. 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 1191406-1415. [DOI] [PubMed] [Google Scholar]
- 28.Nakanishi, H., Y. Suda, and A. M. Neiman. 2007. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 120908-916. [DOI] [PubMed] [Google Scholar]
- 29.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 14029-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neiman, A. M., L. Katz, and P. J. Brennwald. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 1551643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickas, M. E., C. Schwartz, and A. M. Neiman. 2003. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell 2431-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prinz, W. A., L. Grzyb, M. Veenhuis, J. A. Kahana, P. A. Silver, and T. A. Rapoport. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quenneville, N. R., T. Y. Chao, J. M. McCaffery, and E. Conibear. 2006. Domains within the GARP subunit Vps54 confer separate functions in complex assembly and early endosome recognition. Mol. Biol. Cell 171859-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 12055-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayner, J. C., and H. R. Pelham. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 161832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, M., P. P. Poon, C. Schindler, L. E. Murray, R. Kama, G. Gabriely, R. A. Singer, A. Spang, G. C. Johnston, and J. E. Gerst. 2006. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell 171845-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 14081-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudge, S. A., V. A. Sciorra, M. Iwamoto, C. Zhou, T. Strahl, A. J. Morris, J. Thorner, and J. Engebrecht. 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudge, S. A., C. Zhou, and J. Engebrecht. 2002. Differential regulation of Saccharomyces cerevisiae phospholipase D in sporulation and Sec14-independent secretion. Genetics 1601353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheiffele, P., and J. Fullekrug. 2000. Glycosylation and protein transport. Essays Biochem. 3627-35. [DOI] [PubMed] [Google Scholar]
- 41.Schindler, K., and E. Winter. 2006. Phosphorylation of Ime2 regulates meiotic progression in Saccharomyces cerevisiae. J. Biol. Chem. 28118307-18316. [DOI] [PubMed] [Google Scholar]
- 42.Seaman, M. N., J. M. McCaffery, and S. D. Emr. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21661-670. [DOI] [PubMed] [Google Scholar]
- 44.Strochlic, T. I., T. G. Setty, A. Sitaram, and C. G. Burd. 2007. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suda, Y., H. Nakanishi, E. M. Mathieson, and A. M. Neiman. 2007. Alternative modes of organellar segregation during sporulation in Saccharomyces cerevisiae. Eukaryot. Cell 62009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]