Abstract
The predominant cell wall melanin of Wangiella dermatitidis, a black fungal pathogen of humans, is synthesized from 1,8-dihydroxynaphthalene (D2HN). An early precursor, 1,3,6,8-tetrahydroxynaphthalene (T4HN), in the pathway leading to D2HN is reportedly produced directly as a pentaketide by an iterative type I polyketide synthase (PKS). In contrast, the bluish-green pigment in Aspergillus fumigatus is produced after the enzyme Ayg1p converts the PKS product, the heptaketide YWA1, to T4HN. Previously, we created a new melanin-deficient mutant of W. dermatitidis, WdBrm1, by random molecular insertion. From this strain, the altered gene WdYG1 was cloned by a marker rescue strategy and found to encode WdYg1p, an ortholog of Ayg1p. In the present study, two gene replacement mutants devoid of the complete WdYG1 gene were derived to eliminate the possibility that the phenotype of WdBrm1 was due to other mutations. Characterization of the new mutants showed that they were phenotypically identical to WdBrm1. Chemical analyses of mutant cultures demonstrated that melanin biosynthesis was blocked, resulting in the accumulation of 2-acetyl-1,3,6,8-tetrahydroxynaphthalene (AT4HN) and its oxidative product 3-acetylflaviolin in the culture media. When given to an albino W. dermatitidis strain with an inactivated WdPKS1 gene, AT4HN was mostly oxidized to 3-acetylflaviolin and deacetylated to flaviolin. Under reduced oxygen conditions, cell-free homogenates of the albino converted AT4HN to D2HN. This is the first report of evidence that the hexaketide AT4HN is a melanin precursor for T4HN in W. dermatitidis.
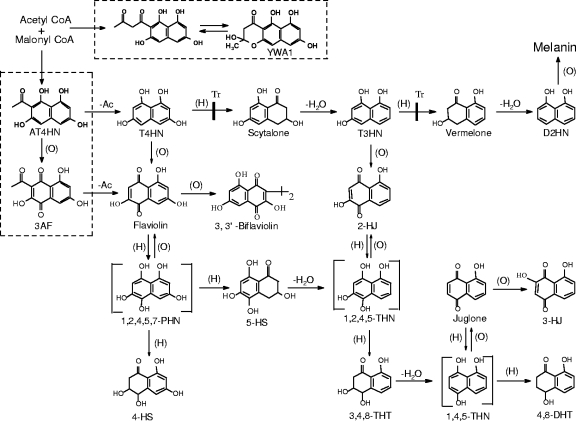
Melanins are dark pigments widely produced by fungi and other organisms. In fungi, they are frequently found in the cell wall. While not essential for growth and development, these complex polymers seem to enhance the survival and competitive abilities of fungi in certain environments. They are composed of various types of phenolic monomers and are often complexed with protein and, less often, carbohydrates (12, 22, 29). The melanins in fungi are named according to their composition and the way they are synthesized and include dihydroxyphenylalanine melanin, catechol melanin, γ-glutaminyl-4-hydroxybenzene melanin, and 1,8-dihydroxynaphthalene (D2HN) melanin (23, 26). The best characterized of these fungal melanins is probably D2HN melanin, which is synthesized by related polyketide pathways (Fig. 1 and 2A and B). The D2HN melanin pathways start with one acetyl-coenzyme A (acetyl-CoA) molecule and four malonyl-CoA molecules, or solely with malonyl-CoA molecules, which undergo a head-to-tail joining and cyclization catalyzed by an iterative type I polyketide synthase (PKS) to initially form 1,3,6,8-tetrahydroxynaphthalene (T4HN) (16). From T4HN, multiple sequential enzyme-catalyzed steps produce D2HN, which is then polymerized to form melanin by a poorly characterized oxidase/laccase reaction (5, 7, 22).
FIG. 1.
Structures and abbreviated names for metabolites in the D2HN melanin biosynthetic pathway and related shunt pathways in W. dermatitidis. Previously, T4HN was believed to be made directly from acetyl-CoA and malonyl-CoA; however, the present study shows that T4HN is a deacetylation product of AT4HN, which is made via a WdPks1p enzyme from malonyl-CoA and acetyl-CoA or possibly malonyl-CoA only (16). The metabolites shown in the two rectangles with dashed lines have not previously been reported for W. dermatitidis or other brown/black fungi; however, YWA1 is a known melanin precursor in A. nidulans and A. fumigatus. The reactions that metabolize T4HN to D2HN and then melanin have been reported for Verticillium dahliae, Magnaporthe grisea, and other fungi (5, 7), as well as for W. dermatitidis (5, 20, 44). Reaction types are indicated as follows: (−Ac), deacetylation; (O), oxidation; (H), reduction; and −H2O, dehydration. Tricyclazole (Tr) specifically inhibits the pathway at the sites indicated. The proposed intermediates in brackets are extremely unstable and have not been isolated from fungi. Other abbreviations: 3AF, 3-acetylflaviolin; T3HN, 1,3,8-trihydroxynaphthalene; 4-HS, 4-hydroxyscytalone; 5-HS, 5-hydroxyscytalone; 2-HJ, 2-hydroxyjuglone; 3-HJ, 3-hydroxyjuglone; 4,8-DHT, 4,8-dihydroxytetralone; 3,4,8-THT, 3,4,8-trihydroxytetralone; YWA1, 2,3-dihydro-2,5,6,8-tetrahydroxy-2-methyl-4H-naphtho[2,3-b]pyran-4-one.
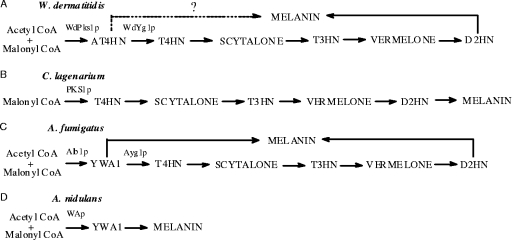
FIG. 2.
Related polyketide pigment pathways that synthesize T4HN, AT4HN, and/or YWA1 in W. dermatitidis (A), C. lagenarium (B), A. fumigatus (C), and A. nidulans (D). Early PKSp enzymes for these fungi are designated WdPks1p, PKS1p, Alb1p, and WAp, respectively. The enzymes that convert AT4HN and YWA1 to T4HN are named WdYg1p in W. dermatitidis and Ayg1p in A. fumigatus. Chemical structures for the metabolites depicted are shown in Fig. 1. Note that A. fumigatus reportedly makes melanin from YWA1 via two pathways, as shown in panel C, and that AT4HN may similarly contribute to melanin production in W. dermatitidis via two pathways, as shown in panel A.
In Colletotrichum lagenarium, T4HN is made directly from malonyl-CoA by PKS1p, as shown in Fig. 2B (16). In contrast, in the bluish-green fungus Aspergillus fumigatus, a pigment-producing polyketide synthase, Alb1p, uses acetyl-CoA and/or malonyl-CoA to produce a heptaketide naphthopyrone designated YWA1 (Fig. 1), which is then enzymatically converted to T4HN, as shown in Fig. 2C (17, 40). YWA1 is metabolized to T4HN by a post-PKS polyketide shortening mechanism (17) involving a protein called Ayglp. This reaction occurs upstream of the T4HN reductase step that is required for the synthesis of scytalone. Other compounds made in A. fumigatus include 1,3,8-trihydroxynaphthalene (T3HN), vermelone, and D2HN (6, 17). The reductase that converts T4HN to scytalone can be blocked by a specific pathway inhibitor known as tricyclazole (20). Using this blocking tool, the reductase step was shown to be present not only in the bluish-green fungus A. fumigatus but also in all the brown to black fungi that make D2HN melanins (5, 10, 36, 40). In A. fumigatus, a second type of polymeric subunit made upstream from YWA1 (Fig. 2C) is also believed to be incorporated into the final melanin polymer (17, 39). Details of this second pathway are unknown at this time. In contrast to the results with A. fumigatus, the fungus Aspergillus nidulans does not make T4HN or D2HN (Fig. 2D), but instead, it uses YWA1 directly in the synthesis of its green conidial pigment (42). Again, the polymerization reaction that forms the melanin is poorly characterized.
The D2HN melanin biosynthetic pathway was initially discovered in Verticillium dahliae (4) and then identified in many other phytopathogens and saprophytes (45). The existence of the pathway in human-pathogenic fungi was demonstrated first in Wangiella dermatitidis, a dematiaceous (melanized) agent of phaeohyphomycosis (20), and shortly thereafter in a number of other related species (36). The pathway in W. dermatitidis from T4HN to D2HN and then to melanin (Fig. 1) was characterized (20, 44) via the chemical identification of key intermediates or shunt metabolites that accumulated in cultures of different melanin-deficient (Mel−) mutant strains, the inhibitor tricyclazole, metabolic cross-feeding by the different Mel− strains, and enzymatic studies. Subsequently, the D2HN melanin was found to be polymerized exclusively in the cell walls of W. dermatitidis, and the earliest enzymatic step identified in its synthesis was determined to involve a PKS encoded by the gene WdPKS1 (14, 44). Importantly, the pathway has been documented to be a virulence factor in W. dermatitidis by virtue of the finding that all Mel− mutants tested thus far are less virulent than the wild type in mouse models of acute infection (12, 13, 14), less resistant to the phagolysosomal activities of human neutrophils (14, 33), and more susceptible to antifungal compounds (30). The results of these studies of W. dermatitidis suggest that the D2HN melanins, and possibly also melanins synthesized from l-3,4-dihydroxyphenyalanine, of the more than 100 other dematiaceous pathogens of humans known or suspected to be melanized by D2HN melanin are important to virulence (30, 34, 35).
The objective of the present study was to delineate further the biosynthesis of D2HN melanin in W. dermatitidis. Of interest were early reactions in the pathway that occur prior to synthesis of T4HN and that might provide evidence to confirm our previous preliminary result (47) that suggested that the early part of the pathway to D2HN melanin in W. dermatitidis involves WdYg1p, an ortholog of Ayglp in A. fumigatus. As such, metabolites of the disruption strain WdBrm1, generated by a random genetic insertion in the preliminary study, and two related melanin-deficient gene deletion strains, generated for this study, were identified. In addition, the relationships among those compounds and the other known D2HN melanin metabolites were determined.
MATERIALS AND METHODS
Strains.
The eight strains of W. dermatitidis used in this study are listed in Table 1. They include the well-described laboratory wild-type strain 8656 (ATCC 34100; Exophiala dermatitidis CBS 525.76), which is usually black and sometimes dark brown, and an albino mutant, WdPks1 (Wdpks1Δ-1 strain), derived previously from the wild type (14, 24). Also listed are WdBrm1 (Wdyg1Δ-1), which is an integrative disruption mutant strain described previously (47); two additional yellow-to-brown, pigment-producing deletion strains, designated WdYel2 (Wdyg1Δ-2) and WdYel3 (Wdyg1Δ-3); and the same three Wdyg1Δ mutants complemented with the wild-type WdYG1 gene, designated the Wdyg1Δ-1WdYG1, Wdyg1Δ-2WdYG1, and Wdyg1Δ-3WdYG1-reconstituted strains. An Escherichia coli strain, DH5, was used for all W. dermatitidis DNA cloning and manipulation procedures. The Aspergillus oryzae strain M-2-3 (21), which is unable to make T4HN or other D2HN metabolites, was used as the heterologous host for the expression of the melanin WdPKS1 gene from W. dermatitidis.
TABLE 1.
Wangiella dermatitidis strains used in this research
| W. dermatitidis straina | Parent strain | Genotype | Reference or source |
|---|---|---|---|
| 8656 | Wild type | ATCC 34100 | |
| WdPks1 (Wdpks1Δ-1) | W. dermatitidis 8656 | Wdpks1::hph | 14 |
| WdBrm1 (Wdyg1Δ-1) | W. dermatitidis 8656 | Wdyg1::hph | 47 |
| WdYel2 (Wdyg1Δ-2) | W. dermatitidis 8656 | Wdyg1::hph | This study |
| WdYel3 (Wdyg1Δ-3) | W. dermatitidis 8656 | Wdyg1::hph | This study |
| Wdyg1Δ-1WdYG1 | WdBrm1 | Wdyg1::hph WdYG1::sur | This study |
| Wdyg1Δ-2WdYG1 | WdYel2 | Wd::hph1yg WdYG1::sur | This study |
| Wdyg1Δ-3WdYG1 | WdYel3 | Wdyg1::hph WdYG1::sur | This study |
Alternate designations are shown in parentheses. Δ, disruption/deletion.
Media and growth conditions.
Routine culture of W. dermatitidis for most of the molecular studies was done at 25°C with yeast-peptone-dextrose agar and broth media (2) or with synthetically defined broth or agar (SDA) medium containing 0.17% (wt/vol) yeast nitrogen base without amino acids and ammonium sulfate, 0.2% ammonium nitrate, 0.1% asparagine, and 1% glucose (49). When selection of transformants was required, media were supplemented with 50 μg/ml hygromycin B (Sigma, St. Louis, MO) or 50 μg/ml chlorimuron ethyl (VWR, West Chester, PA), thus allowing the determination of resistance in strains having acquired the hygromycin phosphotransferase (hph) gene or the sulfonylurea receptor (sur) gene, respectively. Cultures of E. coli were grown at 37°C in Luria-Bertani (LB) medium (2), which was supplemented as necessary with 100 μg/ml ampicillin or 25 μg/ml chloramphenicol for selection of transformants. Experimental cultures of W. dermatitidis and A. oryzae for physiological and biochemical studies were initiated from stock cultures maintained in the dark for 7 to 14 days at 25°C on 25 ml Difco Czapek Dox agar (Becton Dickinson, Sparks, MD) with 0.1% (wt/vol) Difco yeast extract (CDYA) and on Difco potato dextrose agar (PDA), respectively, for 14 to 30 days at 25°C. Cultures prepared to investigate the accumulation of melanin metabolites produced by W. dermatitidis were grown in Difco Czapek Dox broth with 0.1% (wt/vol) Difco yeast extract (CDYB) or on CDYA without or with tricyclazole, which was added to the autoclaved medium in ethanol (EtOH) at a final concentration of 30 μg/ml tricyclazole (Dow Elanco, Indianapolis, IN), unless otherwise stated. Control cultures without tricyclazole contained the same amount of EtOH (1% [vol/vol]). AOIC medium was used to grow the A. oryzae strain expressing the WdPKS1 gene and the Wdyg1Δ strains in order to produce 2-acetyl-1,3,6,8-tetrahydroxynaphthalene (AT4HN) and 3-acetylflaviolin (3-AF), which was identified in this study. The AOIC medium consisted of 3 g NaNO3, 2 g KCl, 1 g KH2PO4, 0.5 g MgSO4·7H2O, 0.01 g FeSO4·7H2O, 10 g peptone P (United States Biological, Swampscott, MA), and 20 g soluble starch (Sigma-Aldrich, St. Louis, MO) brought up to 1,000 ml in deionized H2O and then adjusted to pH 5.5 with H3PO4. For the Wdyg1Δ strains to make AT4HN, the soluble starch was omitted and EtOH was added (1% [vol/vol]). For the accumulation of 3-AF, AOIC that lacked both starch and exogenous EtOH was used. The resulting CDYB and AOIC cultures were generally grown in 250-ml Erlenmeyer flasks after inoculation of 50 ml of media with 0.3 ml H2O containing 1 × 107 W. dermatitidis propagules obtained from 24-h-old CDYB cultures via a previously described serial transfer technique (20). Unless stated otherwise, CDYB and AOIC cultures without added EtOH were grown in the dark in a reciprocating shaker at 200 rpm and 28°C for 5 days, whereas AOIC cultures amended with EtOH were grown for 84 h. CDYA was inoculated with 0.2 ml of evenly spread propagules (2 × 106) of W. dermatitidis from CDYB cultures, after which the resulting plates were incubated in the dark for 10 days at 25°C. To grow A. oryzae for metabolite analysis, conidia (1 × 106) obtained from PDA cultures were added to 25 ml of growth medium in a 125-ml Erlenmeyer flask and incubated for 3 days in the dark at 200 rpm and 28°C. The growth medium contained dextrin from 10 g corn (Sigma, St. Louis, MO), 10 g peptone P, 5 g Difco yeast extract, 5 g KH2PO4, and 0.5 g MgSO4·7H2O brought up to 1,000 ml in deionized H2O. Fungal hyphae and other cells from cultures were then collected on Whatman no. 3 filter paper (Whatman International Ltd., Maidstone, England) and washed with H2O. Approximately half of the fungal material was transferred to 50 ml AOIC in a 250-ml flask and then incubated at 28°C and 200 rpm for 2 days.
Nucleic acid manipulations and computational analyses.
The E. coli transformations with W. dermatitidis DNA were done by the TSS method (9). The methods for transformation of W. dermatitidis by electroporation of intact, competent yeast cells were described previously (41, 49). The isolation of genomic DNA from W. dermatitidis was done by a glass bead method adapted from the work of Ausubel et al. (2) as described previously (27). Total RNA from W. dermatitidis was isolated by Tri reagent (Sigma, St. Louis, MO) extraction of cells according to the manufacturer's instructions and prior to spheroplasting with Zymolyase 20T (ICB Biomedicals, Inc., Aurora, OH). DNA contamination was removed from the RNA by incubation with RQ1 DNase (Fisher Scientific, Suwanee, GA) at 37°C for 1 h, followed by acid phenol-chloroform extraction, ethanol precipitation, and washing. Southern blotting experiments were carried out using standard methods (2). Prior to transfer to nylon Nytran N membranes (Schleicher & Schuell, Keene, NH), genomic DNA was digested with appropriate restriction enzymes and separated in 1% agarose gels. DNA dot blots were obtained by using a Bio-Dot SF microfiltration apparatus (Bio-Rad, Hercules, CA) and its manufacturer's protocol. After genomic DNA (0.5 to 1 μg) was transferred to the Nytran N membranes, hybridization was performed according to standard procedures (2). Blots were then hybridized and washed under low-stringency conditions at 65°C. Assembly and editing of DNA sequences were performed using Lasergene software (DNAStar, Madison, WI). Sequence similarity searches were carried out using the BLAST algorithm (1) and the GenBank nucleotide and protein databases (http://www.ncbi.nlm.nih.gov). Protein sequences were aligned using the CLUSTAL W program (37). The phylogram was constructed using MegAlign (DNAStar, Madison, WI).
PCR and RT-PCRs.
PCRs were carried out with a model 2720 thermal cycler (Applied Biosystems, Foster City, CA) and a Perkin-Elmer PCR system (Norwalk, CT). PCR mixtures usually contained 50 pmol of each primer, 100 ng of sample DNA, deoxynucleoside triphosphates at a final concentration of 200 μM (each), 2 mM MgCl2, and 1 U of TaKaRa Taq DNA polymerase in the manufacturer's buffer (Takara Bio, Inc., Shiga, Japan). The thermocycling conditions used for the PCR were as follows: initial denaturation at 94°C for 2 min; 36 cycles of denaturation (94°C for 40 s), annealing (55°C for 1 min), and extension (72°C for 2 min); and a final extension at 72°C for 8 min. Specific gene probes for the Southern and dot blot analyses were prepared by PCR amplifications using the following primers: for the 287-bp YG1 probe, forward primer FYG1 (5′-CAAAGTCCTCCTGGTCAATGGG-3′) and reverse primer RYG1 (5′-CCTCTAGTATTTAGTCCGGCTGG-3′); and for the 203-bp hph probe (HG), FHYG (5′-ATAAGAATGCGGCCGCACGGTCGACGTTAACTGGGTTCC-3′) and RHYG (5′-GCTCTAGAATTTCGATGATGCAGCTGGCGC-3′), carrying the restriction sites (underlined) for NotI and XbaI, respectively. The latter also facilitated the cloning of HG into the complementation plasmid pDACYG (Fig. 3E). All probes were labeled with [α-32P]dATP, using a Deca Prime II DNA labeling kit (Ambion, Austin, TX). Reverse transcription-PCRs (RT-PCRs) were carried out using a one-step RT-PCR kit (Qiagen, Valencia, CA) and appropriate primers enclosing the WdYG1 gene. To generate the WdYG1 cDNA and to aid in cloning, primer RTF (5′-GCTCTAGAACTATGGGAAACAAAATGGAAGAAGCC-3′) was designed to add an XbaI restriction site (underlined) at the 5′ end, and primer RTR (5′-GACTAGTAATGCATGCTCTGTACCTTATCTGTACTAC-3′) was designed to add an SpeI restriction site (underlined) at the 3′ end. Each RT-PCR involved RT at 50°C for 30 min, after which the PCR was begun as described above. To ensure that the samples were not contaminated with genomic DNA, PCR was also carried out with total RNA and Taq polymerase. The PCR and RT-PCR products were eluted from gels, ligated with the pGEM-T Easy (Promega, Madison, WI) or pBluescript SK(−) (New England Biolabs, Ipswich, MA) vector, cloned, and then analyzed by DNA sequencing. Sequencing of the PCR and RT-PCR gene products was carried out by the core facility of the Institute for Cellular and Molecular Biology, the University of Texas at Austin (Austin, TX).
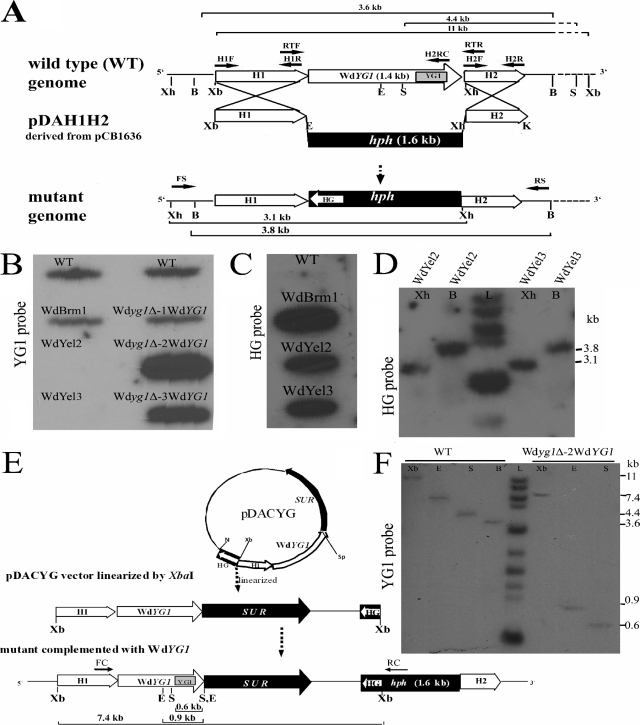
FIG. 3.
Strategies used for the deletion of WdYG1 and for mutant complementation in W. dermatitidis and Southern blot analyses. (A) Deletion of WdYG1 in the wild-type strain was done by gene replacement with a hygromycin phosphotransphorase (hph) selection marker contained in pDAH1H2. Construction of pDAH1H2 was carried out by inserting targeting sequences H1 and H2 into pCB1636 at positions flanking the hph selection marker and into engineered restriction sites for XbaI/EcoRI and XhoI/KpnI, respectively. (B) Southern dot blot analysis of genomic DNA with the 32P-labeled YG1 probe. (C) Southern dot blot analysis of genomic DNA hybridized with the 32P-labeled HG probe to detect the hph cassette. (D) Genomic DNAs isolated from mutants WdYel2 and WdYel3 were digested with BamHI (B) and XhoI (Xh) and hybridized with the 32P-labeled HG probe. (E) Complementation with WdYG1 was done by transformation of the null WdYG1 mutant strains and WdBrm1 with linearized pDACYG, which was derived from vector pCB1532 containing the full-length WdYG1 gene, the 2.8-kb sur selection marker, and portions of the targeting WdYG1 (H1) and hph (HG) sequences adequate to bring about complementation by site-directed integration into the Wdyg1Δ genomic locus. (F) Southern blot analysis of genomic DNA with a specific YG1 32P-labeled probe. Genomic DNAs from the wild-type (WT) and WdYG1-complemented strains (Wdyg1Δ-2WdYG1) were digested separately with XbaI (Xb), EcoRI (E), SalI (S), and BamHI (B). Lane L, 2-log DNA ladder (New England Biolabs). In panels A and E, the shaded YG1 boxes and HG nonshaded arrow show the locations of corresponding probes used for Southern blotting. The arrows FS and RS and the arrows FC and RC indicate the primers used for the PCR amplifications and for screening and verification of the knockout transformants and the complemented strains, respectively. The bold arrows H1F, H1R, H2F, H2R, and H2RC show the locations of the primers used for PCR amplification of DNA fragments used for construction of disruption and reconstitution vectors. Arrows RTF and RTR show the primers used for cDNA amplification by RT-PCR. The thin horizontal lines show the sizes of DNA expected by Southern analysis after restriction enzyme digestion of DNAs from the wild-type, WdYel2, WdYel3, and Wdyg1Δ-2WdYG1 complemented strains; here it should be noted that the complementation of WdBrm1, unlike the complementation of the null WdYG1 mutants, was apparently ectopic, as indicated by Southern analyses (data not shown). Restriction enzymes are abbreviated as follows: Xb, XbaI; E, EcoRI; K, KpnI; Xh, XhoI; B, BamHI; S, SalI; N, NotI; Sp, SpeI.
Identification of the gene disrupted in WdBrm1 and the production of related strains.
The WdYel2 and WdYel3 strains were produced by gene manipulations based in part on preliminary results about the disrupted gene in WdBrm1, which was isolated by a marker rescue approach and then sequenced (47). The WdBrm1 strain itself was produced by the unintended insertion into the WdYG1 gene of W. dermatitidis of an XbaI-linearized disruption vector, pLZ70 (48, 49), which was constructed by incorporating a 500-bp PCR fragment of the WdURA5 gene into the multiple cloning site of pCB1004, which contains genes for resistance to chloramphenicol and hygromycin B. Among the hygromycin B-resistant mutants produced, WdBrm1 was retained for further study because it secreted a yellow-brown pigment. After the genomic DNA was extracted from WdBrm1, the WdYG1 gene was cloned by a marker rescue strategy. Briefly, this strategy involved the digestion of the WdBrm1 genomic DNA with SacI and KpnI, followed by self-ligation of the resulting two fragments with DNA ligase and, finally, the rescue of the two cyclized plasmids, pCQBrmK1 and pCQBrmS16, in E. coli by selection with chloramphenicol. Sequencing then showed that pCQBrmK1 contained considerable lengths of both the 5′-end and 3′-end portions of the entire WdYG1 gene, whereas pCQBrmS16 contained only a single large portion of WdYG1 starting from its 5′ end (data not shown). For construction of the WdYG1 replacement knockout vector (Fig. 3A), a homologous upstream 700-bp region (H1) and a homologous downstream 290-bp region (H2) for targeting the WdYG1 gene were generated by PCR amplifications, using primers H1F (5′-GCTCTAGACCAGTGTCAGTGCTTGAACTCCG-3′) and H1R (5′-GGAATTCGAGGTCTTTTGAGCCAAGGGG-3′), which introduced sites (underlined) for XbaI and EcoRI, respectively, for the HI fragment amplification. Also used were primers H2F (5′-CCGCTCGAGGCATCCAACACAGCGTCTCAGTAGG-3′) and H2R (5′-GGGGTACCGTCTCACGATCGTGACGGG-3′), which introduced sites (underlined) for XhoI and KpnI, respectively, for H2 fragment amplification. After amplification, the H1 and H2 targeting fragments were cloned into the corresponding sites of vector pCB1636 (kindly provided by J. Sweigard, Dupont Co., Wilmington, DL), which has the hph gene marker for selection, to produce the knockout replacement vector pDAH1H2. After being linearized with XbaI and KpnI, pDAHIH2 was used to transform competent W. dermatitidis cells by electroporation. Identification of potential Wdyg1Δ disruption strains was achieved by observation of resistance to hygromycin on SDA selection medium and by their production of yellow-brown secretion products. The specific gene deletions were verified by PCR, using specific primers for WdYG1 gene amplifications, and by Southern blotting analyses using probes for both WdYG1 and hph. The complementation vector pDACYG (Fig. 3E) was constructed by ligating a 0.2-kb hph gene fragment, generated by digestion with NotI and XbaI, and a 2.1-kb DNA fragment, generated by PCR with primers H1F and H2RC (5′-GACTAGTCTAATGATGATGATGATGATGGTATTTAGTCCGGCTGGG-3′), into the multiple cloning site of pCB1532 (Fungal Genetics Stock Center, Kansas City, KS). Thus, a 0.7-kb homologous targeting region (HI) and the 1.4-kb full-length WdYG1 gene were inserted. After the resulting pDACYG complementation vector was linearized with XbaI, it was transformed into the WdBrm1 and the Wdyg1Δ mutant strains by electroporation. Putative reconstituted Wdyg1Δ strains were selected by observation of resistance on SDA to both hygromycin B and chlorimuron ethyl and by their wild-type phenotype. Transformants were subsequently confirmed to contain the intact WdYG1 gene by PCR using the FC and RC primers and also by Southern blotting with a WdYG1 probe.
Generation of an A. oryzae strain expressing the WdPKS1 gene.
The WdPKS1 gene amplified from the genomic DNA of the W. dermatitidis wild-type strain by PCR was cloned into the fungal expression vector pTAex3 (19), using Gateway cloning technology (Invitrogen, Carlsbad, CA). After sequence confirmation, the constructed expression plasmid, pTA-WdPKS, was introduced into A. oryzae M-2-3 to generate the strain expressing the WdPKS1 gene by the method reported previously (18).
Analysis and isolation of melanin metabolites from W. dermatitidis and A. oryzae.
To determine the accumulated known D2HN melanin precursors or related metabolites, the W. dermatitidis or A. oryzae cultures were extracted with ethyl acetate (EtOAc), and the extracts were dried in vacuo. The resulting residues were dissolved in methanol (MeOH) and then analyzed by high-performance liquid chromatography (HPLC) and, occasionally, thin-layer chromatography (TLC) (20, 46). Authentic D2HN melanin metabolite standards obtained previously, including YWA1 and AT4HN, were used to confirm HPLC peak and TLC spot identities. Known standards of 3-AF and the shunt metabolites 1,2,4,5,7-pentahydroxynaphthalene (1,2,4,5,7-PHN), 1,2,4,5-tetrahydroxynaphthalene (1,2,4,5-THN), and 1,4,5-trihydroxynaphthalene (1,4,5-THN), however, were not available for the study. HPLC was also performed directly on supernatants from liquid fungal cultures. For these analyses, 1.0 ml of culture was centrifuged in a Napco 2002 microcentrifuge (Napco, Winchester, VA) for 10 min at 13,000 × g and room temperature, and then a portion of the clear supernatant was injected into the HPLC instrument. To isolate 3-AF for structure identification, AOIC cultures of WdYel2 were processed as for metabolite analysis. The resulting metabolites in MeOH were applied to TLC plates of silica gel G/HR (Mallinckrodt, Baker, Inc., Phillipsburg, NJ) with 2.5% (wt/wt) zinc silicate phosphor (Sigma, St. Louis, MO). The plates then were developed in ethyl ether-hexane-formic acid (78:21:1). After scraping of the orange compound, later identified as 3-AF, as a band from the plates, it was eluted in MeOH, separated from the silica gel by filtration, and dried in vacuo. Further purification of the compound was carried out by HPLC, using the same separation method as that for metabolite analysis (46). Mass and UV-visible spectra of the purified compound were obtained by electron impact (70 eV), using a Trace DSQ mass spectrometer via direct insertion probe (Thermo Electron Corp., Austin, TX), and in MeOH with a model G1103A UV-visible spectrometer (Hewlett Packard), respectively.
Cross-feeding between the albino Wdpks1Δ strain and the Wdyg1Δ and reconstituted Wdyg1Δ strains.
Typically, nontouching, alternating parallel streaks of the albino Wdpks1Δ strain were grown in close proximity to streaks of the complemented and noncomplemented Wdyg1Δ strains on CDYA with or without tricyclazole in 100- by 15-mm petri dishes. After growth for 10 days at 25°C, metabolites were extracted from the cultures and identified by HPLC (46). Studies were also carried out with a combined cellular suspension of the albino and Wdyg1Δ strains in the presence of tricyclazole to determine if the albino was able to make D2HN melanin metabolites in CDYB or AOIC. These cultures were prepared by inoculating 50 ml of medium in a 250-ml Erlenmeyer flask with 0.3 ml of a 2-to-1 (vol/vol) ratio of the albino and Wdyg1Δ strain propagules obtained from 24-h-old CDYB cultures of each strain prepared serially. After 6 days of growth in the dark at 25°C on a rotary shaker at 250 rpm, extracts of the mixed cultures were analyzed by HPLC.
Treatment of albino WdpksΔ mutant cultures with AT4HN and 3-AF.
AT4HN and 3-AF obtained from AOIC cultures of W. dermatitidis strain WdYel2 were fed to CDYB cultures of the albino Wdpks1 strain. In these assays, additions of 1 mg of AT4HN or 3-AF dissolved in 0.05 ml MeOH were added to 10 ml of the cultures (some containing tricyclazole) in 50-ml wide-based Erlenmeyer (Kimax 26502) flasks at 96 h and again at 108 h. These cultures were incubated on a shaker at 25°C and 250 rpm in the dark for 36 h after the second addition of substrate. They were then extracted and analyzed by HPLC for metabolites.
Treatment of homogenates of the albino Wdpks1Δ mutant with AT4HN, 3-AF, T4HN, and flaviolin.
Cell-free buffered homogenates of the albino Wdpks1 strain were provided AT4HN, 3-AF, T4HN, and flaviolin under reduced oxygen conditions, as described previously (44), to determine their enzymatic fates. Specifically, 5 ml of cell-free albino homogenate in a 20-ml culture tube was diluted with 4.9 ml of 20 mM potassium phosphate buffer at pH 6.8. Next, 5 μmol NADPH was added, and the tube was sealed and backfilled with N2 gas. Enzymatic reactions with the desired substrates were initiated by adding 2 mg of AT4HN or 3-AF or 1 mg of flaviolin or T4HN in 0.1 ml MeOH through a syringe needle without opening the tube seal. The enzymatic solution was then incubated at 25°C in the N2 environment. At the end of 16 h, a sample was taken from the enzymatic solution with a syringe through the tube seal, sealed into an HPLC vial under a stream of N2 gas, and then immediately analyzed by HPLC for melanin-related metabolites. This procedure allowed for the minimization of any oxidation processes in the homogenates during feeding and during preparation for analysis. At the end of the experiment, the full enzymatic solution in the tube was extracted for HPLC analysis.
Nucleotide sequence accession numbers.
The sequence of WdYG1 was submitted to the GenBank database (accession number AY667610). The revised sequence of WdPKS1 was also submitted to the GenBank database (accession number AF130309.3).
RESULTS
Genetic characterization of Wdyg1Δ mutant strains.
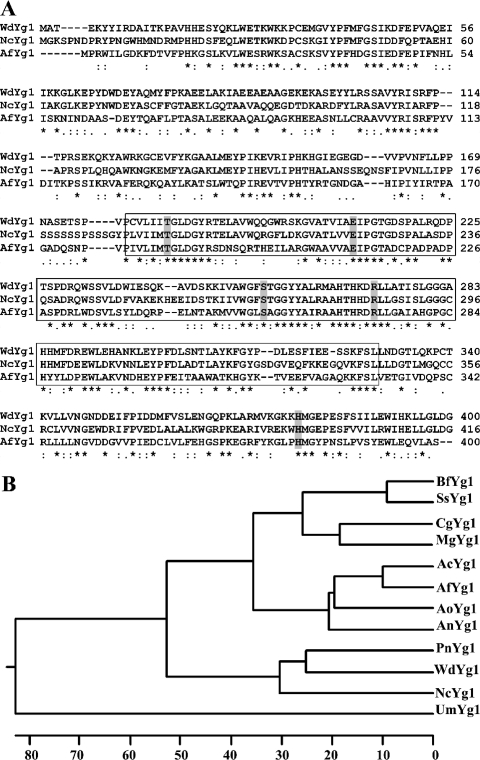
The first yellow-to-brown pigment-secreting strain, WdBrm1, was generated by an unintended insertion into the W. dermatitidis genome of pLZ70, which was designed with the intention of disrupting its WdURA5 gene (47, 48, 49). The WdBrm1 mutant was maintained for further study because it was obviously different from the wild-type strain and any previously derived Mel− and Wdpks1Δ mutant strains with defects in the D2HN melanin biosynthetic pathway (10, 14, 20). After the gene putatively disrupted in WdBrm1 was isolated by a marker rescue strategy, sequencing showed that it was a homolog of the Ayg1 gene of A. fumigatus. This gene in A. fumigatus encodes an enzyme responsible for the chain shortening of the heptaketide generated in a pathway leading to the bluish-green pigment of its conidial cell walls (38, 39). Therefore, the gene identified in W. dermatitidis was named WdYG1, and its encoded protein was named WdYg1p. Sequencing of a cDNA clone corresponding to the entire predicted coding region showed that it consisted of a single open reading frame of 1,248 bp and contained three introns of 49, 54, and 63 bp. Initial BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST) indicated that the derived protein sequence of WdYg1p had its closest relationship (56% identity) with the deduced sequence of a hypothetical protein from Neurospora crassa (GenBank accession no. XP_960081) and significant sequence similarity (38.7% identity) with Ayg1p of A. fumigatus (Fig. 4A). Phylogenetic and sequence analyses revealed additional homologies with other functionally characterized and uncharacterized orthologs in the database (Fig. 4B). The analyses also showed the presence of hydrolase (COG0412), esterase/lipase (Aes) (COG0657.1), and dipeptidylaminopeptidase/acylaminoacyl-peptidase (COG1506.1) domains, and amino acid alignments of WdYg1p with Aes family proteins revealed the presence of conserved amino acid residues T184, E211, S256, R272, and H378 (Fig. 4A), which are all reported to be important for their enzyme activity (17).
FIG. 4.
Amino acid alignment and phylogenetic analysis of WdYg1p with other yellowish-green fungus-like orthologs. (A) Alignment of WdYg1p of W. dermatitidis with Ayg1p of Aspergillus fumigatus (GenBank accession no. AAF03353) and NcYg1p of Neurospora crassa (GenBank accession no. XP_960081). In the consensus line, identical amino acids are marked by asterisks, conserved substitutions are marked by colons, and semiconserved substitutions are marked by dots. The numbers at the right represent the number of amino acids depicted from the first methionine for each derived protein. The boxed areas identify the Aes enzymatic domain, in which the conserved amino acids for Aes family members are indicated by the dark shading. (B) The phylogenetic tree was inferred by applying a Clustal W algorithm by the neighbor-joining method (32). The units at the bottom of the tree indicate the approximate numbers of substitution events. The fungal source and accession number of each sequence are as follows: BfYg1, Botryotinia fuckeliana, XP_001552894.1; SsYg1, Sclerotinia sclerotiorum, XP_001597188.1; CgYg1, Chaetomium globosum, XP_001222736.1; MgYg1, Magnaporthe grisea, XP_001522572.1; AcYg1, Aspergillus clavatus, XP_001276032.1; AfYg1, A. fumigatus, AF116902_1; AoYg1, A. oryzae, BAE55407.1; AnYg1, A. niger, XP_001401158.1; PnYg1, Phaeosphaeria nodorum, EAT80224.2; WdYg1, Wangiella (Exophiala) dermatitidis, AAT81166.2; NcYg1, Neurospora crassa, XP_965534.2; UmYg1, Ustilago maydis, XP_758182.1.
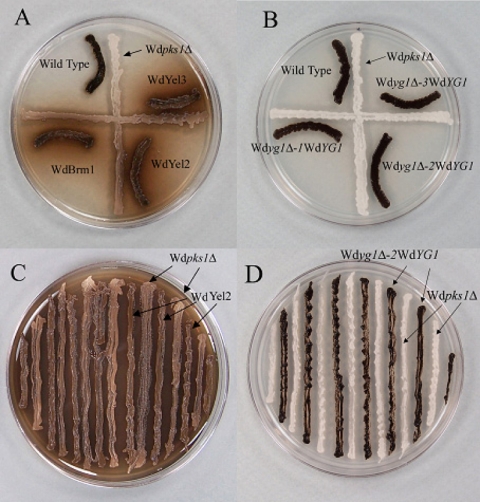
Because the WdYG1 gene in WdBrm1 was disrupted by the unintended integration of a vector in the W. dermatitidis genome, a complete deletion of the gene was carried out by an allelic one-step insertion-replacement strategy to confirm that the phenotype of WdBrm1 was not due to other mutations (Fig. 3A). Among the transformants produced, two (WdYel2 and WdYel3) had the same phenotypic characteristics as WdBrm1 (Fig. 5A). Comparisons of the microscopic characteristics of the transformants and those of the wild type and the Wdpks1Δ mutant showed that except for their pigment differences, all were identical (data not shown). The deletion of WdYG1 was then confirmed by PCR amplifications using primers FS and RS, which flanked the upstream (H1) and downstream (H2) homology regions, respectively, and by DNA sequencing (data not shown). Southern blotting analyses showed that the WdYG1 signals were absent from the WdYel2 and WdYel3 strains, but not the WdBrm1 strain, when the blots were probed with the YG1 probe (Fig. 3B). Additionally, the hph replacement fragments were present in all three strains when the blots were probed with the hph probe (Fig. 3C and D). Southern blotting also confirmed that WdYG1 existed as a single gene in the W. dermatitidis wild-type genome (Fig. 3F). Evidence for the importance of WdYg1p to melanin biosynthesis in W. dermatitidis was provided by showing that both WdBrm1 and the WdYg1Δ null mutants (WdYel2 and WdYel3) were restored to the wild-type colony phenotype (Fig. 5B) after transformation with a linearized complementation plasmid, pDACYG, containing the full WdYG1 open reading frame and the sur selection marker (Fig. 3E) and selection on yeast-peptone-dextrose agar that contained both hygromycin B and chlorimuron ethyl. PCR using primers FC and RC, DNA sequencing (data not shown), and Southern blotting with the YG1 probe (Fig. 3F) confirmed that all three complemented WdYg1Δ strains contained the WdYG1 gene.
FIG. 5.
CDYA cultures of eight strains of W. dermatitidis grown in the dark at 25°C for 10 days and used in paired cross-feeding studies. (A) Albino Wdpks1Δ mutant grown adjacent to the wild-type and Wdyg1Δ strains. (B) Albino mutant adjacent to the wild-type and reconstituted Wdyg1Δ-WdYG1 strains. (C) Albino mutant adjacent to the WdYel2 mutant, which produces a brown pigment. (D) Albino mutant adjacent to the WdYel2 strain reconstituted with WdYG1. Note that the wild-type and reconstituted Wdyg1Δ-WdYG1 strains are identical in appearance, as are all the Wdyg1Δ strains which produce brown pigments.
AT4HN and 3-AF are identified in pigment secretions of Wdyg1Δ mutants.
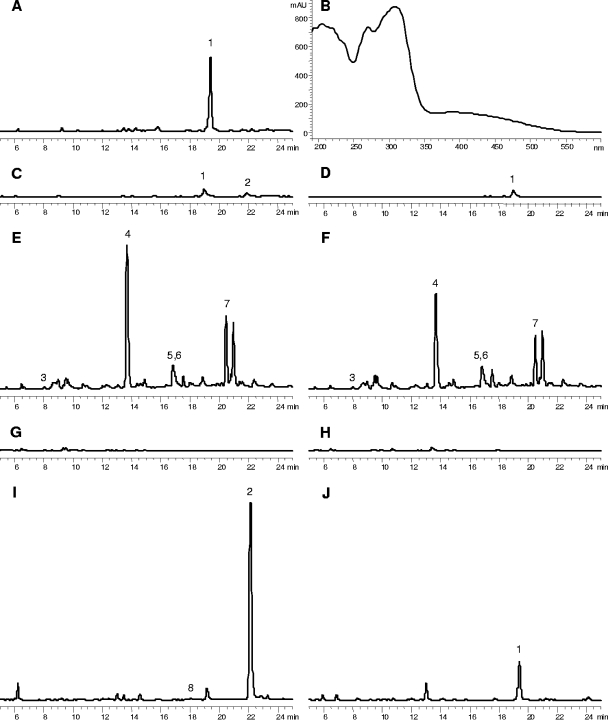
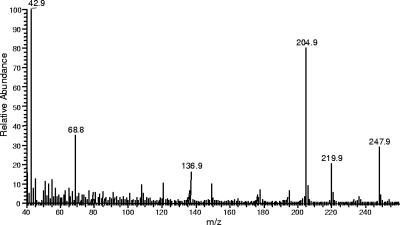
All three Wdyg1Δ strains (WdBrm1, WdYel2, and WdYel3) had cells that were light brown and secreted soluble yellow pigments during the first 2 to 4 days of growth at 25°C in CDYB. By 5 to 6 days, the cells were a darker brown and the soluble pigments were brown instead of yellow. In contrast, the wild type and the reconstituted Wdyg1Δ strains at the end of 5 to 6 days had cells that were dark gray to black and did not produce appreciable amounts of soluble pigment. Cells and pigments obtained from 10-day-old CDYA cultures were similar in appearance to those obtained from 6-day-old CDYB cultures. Analysis of the extracts from CDYB cultures showed that the Wdyg1Δ strains had not produced any known melanin metabolites, except for occasional trace amounts of flaviolin. They did, however, secrete compounds not previously reported in studies with W. dermatitidis. Specifically, extracts of the Wdyg1Δ cultures usually contained (Fig. 6A) a large amount of an unknown compound (HPLC tR = 19.4 min) with a UV-visible spectrum (Fig. 6B) distinctly different from those of AT4HN, YWA1, and any of the other relevant melanin metabolites. From WdYel2 cultures grown in AOIC, this compound was extracted into EtOAc, but upon concentration of the extract, a dark, almost black, solid precipitated. While the concentrated extract dried to an orange solid that was soluble in organic solvents (e.g., EtOAc, MeOH, EtOH, acetonitrile, etc.), the black precipitate was found to be soluble only in water, where it produced an orange color. Upon acidification of the water solution to pH 3 to 4, the orange material could be partitioned into EtOAc and the resulting extract dried to an orange solid. The unknown compound, analyzed via probe insertion mass spectrometry, had a parent ion peak at m/z 248 and the fragmentation pattern shown in Fig. 7. Experiments revealed that the unknown compound appeared to be formed from AT4HN (molecular weight, 234) by nonenzymatic oxidation. Interestingly, the conversion of AT4HN to the unknown compound was strongly influenced by pH. That is, only a small amount of AT4HN was converted to the unknown in 0.2 M potassium phosphate at pH 5.0 over 60 min, whereas AT4HN in pH 7.0 buffer was converted completely to the unknown in 10 min. These results led to identification of the unknown compound as 3-AF.
FIG. 6.
Representative HPLC traces (A and C to J) showing 3-AF, AT4HN, and other compounds from cultures of W. dermatitidis. (B) UV-visible spectrum of 3-AF. Only the 5- to 25-min regions of the full 39-min chromatograms acquired are shown, and all have the same response scale. The samples include the following: extract from a CDYB culture of WdYel2 grown for 6 days, showing the 3-AF peak (1) (A); supernatant from a 4-day-old culture of WdYel2 revealing the presence of AT4HN (2) in addition to 3-AF (1) (C); supernatant from the analysis in panel C after 16 h at room temperature, showing the loss of AT4HN during storage (D); extracts from tricyclazole-treated 6-day-old CDYB cultures of the wild-type and Wdyg1Δ-2WdYG1 strains, respectively, showing the presence of scytalone (3), flaviolin (4), tricyclazole (5), 2-HJ (6), and 3,3′-biflaviolin (7) (E and F); extracts from 6-day-old CDYB (no tricyclazole) cultures of the wild-type and Wdyg1Δ-2WdYG1 strains, respectively, with no melanin metabolites detectable (G and H); and extracts from 3-day-old AOIC cultures of the WdYel2 strain that were pretreated with EtOH (1% [vol/vol]) or not, respectively, illustrating how culture conditions can be used to selectively produce either AT4HN or 3-AF (I and J). Panel I also shows the presence of trace amounts of YWA1 at approximately 18 min (8).
FIG. 7.
Mass spectrum of 3-AF. The parent ion at m/z 248 fits that for 3-AF. As described for similar compounds by Becher et al. (3), the fragments at m/z 220 and 205 may arise from the sequential loss of a carbonyl and methyl group or the loss of a carbonyl group and an acetyl group (evidenced by the m/z 43 peak), respectively.
Importantly, although AT4HN was usually not detected in extracts of CDYB and CDYA cultures, it was found in supernatants of 4-day-old CDYB cultures of all three Wdyg1Δ strains when the supernatants were immediately and directly analyzed by HPLC (Fig. 6C). In contrast, when there was a delay between sampling and analysis, the quantity of AT4HN decreased or disappeared, while 3-AF appeared or increased (Fig. 6D). The presence of tricyclazole did not appreciably affect production of AT4HN or 3-AF in CDYA cultures of the Wdyg1Δ strains (without the albino) and did not cause flaviolin or other known DHN melanin metabolites to appear in the cultures.
Cultures of the wild-type and reconstituted Wdyg1Δ strains did not secrete appreciable amounts of soluble melanin metabolites, except during growth in the presence of tricyclazole, when they accumulated flaviolin, 3,3′-biflaviolin, and 2-hydroxyjuglone (2-HJ) (Fig. 6E to H). The accumulation of the melanin metabolites with the addition of tricyclazole to the CDYB or CDYA medium coincided with the appearance of a reddish brown coloration, not the gray-black appearance of the wild-type and reconstituted Wdyg1Δ cells (Fig. 6E and F; Table 2).
TABLE 2.
Melanin-related products from extracts of CDYB cultures containing single and combined strains of Wangiella dermatitidisa
| Strain(s)b | Productse |
|---|---|
| Wild type | ND |
| Wild type* | Flaviolin,c biflaviolin,c 4-HS, 5-HS, 2-HJ, scytalone |
| WdPks1 | ND |
| WdPks1* | ND |
| WdYel2 | 3-AF,c flaviolind |
| WdYel2* | 3-AF,c flaviolind |
| Wdyg1Δ-2WdYG1 | ND |
| Wdyg1Δ-2WdYG1* | Flaviolin,c biflaviolin,c 4-HS, 5-HS, 2-HJ, scytalone |
| WdYel2 + WdPks1 | 4-HS, 5-HS, 2-HJ, scytalone, 4,8-DHT |
| WdYel2 + WdPks1* | Flaviolin,c biflaviolin,c scytalone, 5-HS, 2-HJ, 3-HJ |
| Wild type + WdPks1 | ND |
| Wdyg1Δ-2WdYG1 + WdPks1 | ND |
The cultures were grown for 6 days on a rotary shaker at 250 rpm in the dark at 25°C.
*, Cultures contained 30 μg/ml tricyclazole.
3-AF, flaviolin, and 3,3′-biflaviolin were the main metabolites present.
Flaviolin was found only occasionally and only in trace amounts.
ND, not detected.
AT4HN is produced by WdPKS1 expressed in A. oryzae and in W. dermatitidis Wdyg1Δ strains cultured in AOIC.
An A. oryzae strain expressing WdPKS1 was used to identify any WdPks1p reaction product(s) that could be produced upon the expression of the cDNA of WdPKS1. This strain was used because it does not contain enzymes of the D2HN melanin pathway and thus is unable to make T4HN or other downstream precursors. When grown in AOIC at 28°C for 2 to 3 days, the WdPKS1-expressing strain of A. oryzae produced moderate to large amounts of AT4HN, small amounts of 3-AF, and occasionally, trace levels of YWA1. Surprisingly, when the Wdyg1Δ strains were grown similarly in AOIC, a medium not previously used for the culture of W. dermatitidis, they produced enhanced amounts of AT4HN and 3-AF and trace amounts of YWA1. The relative quantities of AT4HN, 3-AF, and YWA1 obtained from the Wdyg1Δ cultures depended on the presence or absence of EtOH in the AOIC medium. Extracts of EtOH-treated cultures generally contained large amounts of AT4HN, small or undetectable amounts of 3-AF, and trace amounts of YWA1 (Fig. 6I), whereas cultures without EtOH produced small or undetectable amounts of AT4HN, moderate to large amounts of 3-AF, and no detectable YWA1 (Fig. 6J). Coincident with the differences in the metabolites produced, cultures treated with EtOH were usually yellowish brown and had a pH of 4.9 to 6.6 at 3 days of growth, while those without EtOH were orange and had a pH of 6.5 to 7.8. With or without EtOH, the Wdyg1Δ strains grew well in AOIC and existed mostly as light brown, budding yeast-type cells.
Metabolites secreted by the Wdyg1Δ strains are metabolized by the albino Wdpks1Δ strain.
Cross-feeding experiments on CDYA between alternating parallel streaks of the albino and Wdyg1Δ strains showed that the Wdyg1Δ strains in the presence of 15 μg/ml tricyclazole secreted yellow-to-brown metabolites that diffused through the medium at the same time that the albino darkened (Fig. 5C). Under these conditions, the albino apparently metabolized one or more compounds from among the diffusible pigments, became brown in color, and made flaviolin, 3,3′-biflaviolin, 2-HJ, and trace amounts of 5-hydroxyscytalone (5-HS), as shown by HPLC analysis of culture extracts (Table 3). These metabolites were not found in streaked control cultures of the Wdyg1Δ strains where the albino strain was absent or in cultures of the albino by itself. We suggest that these results demonstrate that the Wdyg1Δ strains produced one or more upstream substrates that were converted by WdYg1p in the albino to T4HN or flaviolin and other related melanin metabolites. However, when pairings were made in the absence of tricyclazole, flaviolin, 4,8-dihydroxytetralone (4,8-DHT), and/or 4-hydroxyscytalone (4-HS) were detected and flaviolin was present in smaller amounts (Table 3). In contrast, the reconstituted Wdyg1Δ strains and the wild type did not secrete visible yellow-brown compounds in CDYA when they were streaked next to the albino (Fig. 5D). As a consequence, the albino failed to darken in these studies and did not produce downstream metabolites (Table 3). The results obtained on CDYA were similar to those obtained when propagules of the Wdyg1Δ strains and the albino were cocultured in CDYB medium and the albino made known DHN melanin metabolites from secretions of the Wdyg1Δ strains (compare Tables 2 and 3).
TABLE 3.
Melanin-related products from extracts of CDYA streaked cultures with single and paired strains of Wangiella dermatitidisa
| Strain(s)b | Product(s)c |
|---|---|
| WdYel2 | 3-AF |
| WdYel2* | 3-AF |
| WdPks1 | ND |
| WdPks1* | ND |
| WdYel2 + WdPks1 | Flaviolin, 4,8-DHT, 4-HS |
| WdYel2 + WdPks1* | Flaviolin, biflaviolin, 5-HS, 2-HJ |
| Wild type + WdPks1 | ND |
| Wdyg1Δ-2WdYG1 + WdPks1 | ND |
The cultures were grown for 10 days in the dark at 25°C. Cultures of the paired strains had 15 alternating parallel streaks.
*, Cultures contained 15 μg/ml tricyclazole.
5-HS was present only in trace quantities. ND, not detected.
AT4HN and 3-AF are metabolized by albino Wdpks1Δ cultures and homogenates.
When CDYB cultures of the albino were provided with AT4HN in the absence of tricyclazole, the AT4HN disappeared within 4 h, and in its place appeared 3-AF, flaviolin, 4-HS, and scytalone. In cultures amended with 15 μg/ml tricyclazole, scytalone and 4-HS were not present; however, 3-AF and flaviolin were still major products. Apparently, under aerobic culture conditions, the AT4HN was readily oxidized to 3-AF, which was then deacetylated to flaviolin (Fig. 1). Cultures of the albino also made flaviolin and 4-HS when they were treated directly with 3-AF in the absence of tricyclazole; however, 4-HS did not appear in cultures treated with tricyclazole. Under reduced oxygen conditions, cell-free homogenates of the albino Wdpks1Δ mutant metabolized AT4HN to D2HN but not to 3-AF (Table 4). The metabolism of AT4HN and T4HN to D2HN was prevented by tricyclazole. The homogenates metabolized 3-AF and flaviolin substrates to 4-HS, 5-HS, 2-HJ, 3,4,8-trihydroxytetralone, 4,8-DHT, and juglone unless tricyclazole was present to inhibit the production of most of the downstream metabolites, in which case only flaviolin, 5-HS, and/or 3-AF was found in the homogenates. When they were boiled for 10 min, the homogenates did not metabolize AT4HN, T4HN, 3-AF, or flaviolin, confirming that the reactions occurred enzymatically.
TABLE 4.
Known melanin metabolites obtained after T4HN and 3-AF were added to albino homogenates of Wangiella dermatitidis under anaerobic conditions in the absence and presence of tricyclazolea
| Substrate | Product(s)b |
|---|---|
| AT4HN | D2HN* |
| T4HN | D2HN* |
| 3-AF | 4-HS,* 5-HS, 2-HJ,* 3,4,8-THT,* 4,8-DHT,* juglone* |
| Flaviolin | 5-HS, 2-HJ,* 3,4,8-THT,* 4,8-DHT,* juglone* |
Initial concentration of AT4HN and 3-AF was 1.0 mM; initial concentration of T4HN and flaviolin was 0.5 mM. Reactions were carried out in 10-ml assay systems.
*, product production was blocked in assays containing 30 μg/ml tricyclazole.
DISCUSSION
Polyketide melanins in brown, black, green, and bluish-green fungi are the best-understood cell wall-bound fungal pigments, and knowledge of their biosynthetic pathways is extensive (7, 22, 26). Of these polyketide pigments, those made from D2HN exist in a large number of brown and black fungi. They have been characterized in most detail by genetic and biochemical evidence obtained from Verticillium dahliae (5), W. dermatitidis (12, 14, 20, 44), and Magnaporthe grisea or its anamorph, Pyricularia oryzae (5, 7, 22). Most of the enzymes, intermediate products, and shunt compounds in the melanin pathway of W. dermatitidis and V. dahliae are identical (Fig. 1). These compounds are also known to exist in numerous other brown/black fungi (5, 7, 26, 36).
In the present study and a preliminary one (47), we produced three Wdyg1Δ strains of W. dermatitidis that are different from all other melanin-deficient strains of W. dermatitidis studied previously (10). These new Wdyg1Δ strains have a common defective gene, WdYG1, and accumulate AT4HN or its newly identified oxidation product, 3-AF, in culture because they are unable to deacetylate AT4HN and 3-AF to T4HN and flaviolin, respectively. As a result of the absence of T4HN, the biosynthesis of other downstream melanin precursors or D2HN melanin does not occur. These findings are unique because AT4HN had not previously been reported as a melanin precursor in any fungus and because W. dermatitidis was assumed to make T4HN directly from either a combination of acetyl-CoA and malonyl-CoA or malonyl-CoA by itself (14, 20). Previously, AT4HN had only been identified in studies with a molecularly derived strain of A. oryzae, where it was described as a novel hexaketide produced from a chimeric PKS composed of WA from A. nidulans and PKS1 of C. lagenarium (43). While the A. oryzae strain used in that study produced three other known polyketides, it did not make YWA1 and T4HN, which are the usual products of WA and PKS1, respectively (Fig. 2B and C). Also of interest is the fact that 3-AF has not previously been reported as a metabolite in melanin biosynthesis. In our study, its identification was based on its mass spectrometry fragmentation pattern, together with its metabolic relationships with AT4HN and flaviolin (Fig. 1). Its mass spectrometric fragments at m/z 220 and 205 may have occurred because of the sequential loss of a carbonyl and methyl group and the loss of a carbonyl group and an acetyl group (evidenced by the m/z 43 peak), respectively, as described by Becher et al. (3) for other closely related compounds. In addition, the ions at m/z 69 and 137 are probably identical to those reported previously in the mass spectrum of flaviolin (4). The possibility that the unknown may be 2,5,7-trihydroxy-6-acetyl-1,4-naphthoquinone (6-acetylflaviolin), another oxidation product of AT4HN, was eliminated based on the distinctly different UV-visible and mass spectra of 6-acetylflaviolin (3, 28).
The eight strains of W. dermatitidis and the single strain of A. oryzae grown on a variety of media in this study showed significant differences in the accumulation of AT4HN and 3-AF. For example, in AOIC containing EtOH, the Wdyg1Δ strains usually accumulated AT4HN instead of 3-AF, whereas in AOIC without EtOH or in CDYB or CDYA, they accumulated 3-AF instead of AT4HN. We suspect that this difference was because the AOIC cultures containing EtOH grew under more acidic conditions than the other cultures, thus slowing the oxidation of AT4HN to 3-AF. The apparent oxidation process of AT4HN to 3-AF is supported by the HPLC results, wherein AT4HN was found in culture supernatants but not in extracts of the same cultures, which instead had 3-AF. It is noteworthy that this is probably the first reported instance of HPLC analyses being performed directly on melanin metabolite-containing culture supernatants. The analyses were easily carried out, and the only disadvantage noted was that concentrations of metabolites were reduced compared to those in extracts, to the extent that trace amounts of the metabolites might not be detectable.
The oxidation of AT4HN to 3-AF explains why the albino Wdpks1Δ strain made smaller amounts of melanin than expected in the cross-feeding experiments with the Wdyg1Δ strains on CDYA or in CDYB. That is, whereas some AT4HN was made in the cultures and metabolized to melanin, most was oxidized to 3-AF and then metabolized to flaviolin and its known products, e.g., 3,3′-biflaviolin, 4-HS, 5-HS, and 2-HJ. Strong supporting evidence for AT4HN as a precursor to D2HN melanin was obtained from anaerobic studies with cell-free homogenates of the albino. These experiments showed that D2HN was made in the same manner from either AT4HN or T4HN and confirmed that T4HN was a deacetylation product of AT4HN. In addition, the albino homogenate studies with 3-AF established that 3-AF is deacetylated to flaviolin, not T4HN, which resulted in the homogenates accumulating the same products as those made from flaviolin.
A somewhat unexpected result in the present study was our finding that AT4HN was metabolized to compounds in addition to T4HN (Fig. 1). For example, in the Wdyg1Δ cultures, AT4HN was converted to 3-AF and other compounds, some with UV-visible spectra similar to that of AT4HN. In addition, a portion of the available AT4HN in W. dermatitidis may have been incorporated as part of the melanin polymer before it was deacetylated to T4HN, as indicated by the appearance of a brown pigment in the Wdyg1Δ strains when they were grown in CDYB or on CDYA. The latter suggestion means that AT4HN in W. dermatitidis and YWA1 in A. fumigatus may be used similarly as melanin precursors because in A. fumigatus some YWA1 is believed to be incorporated directly into melanin before being converted into T4HN (39).
It is important that although the fungi studied produce T4HN differently, each PKS, along with WAp of A. nidulans (42), is known to share a high degree of sequence similarity based on BLAST analysis (8, 14). CLUSTAL W analysis, which was found to be more selective than BLAST analysis, showed that the PKSs of W. dermatitidis and C. lagenarium and those of A. fumigatus and A. nidulans fall into two different groups based on their deduced amino acid relationships (8). Based on sequence similarity, W. dermatitidis and A. fumigatus are now known to have similar enzymes for converting AT4HN and YWA1, respectively, to T4HN (amino acid identity, 39%; similarity, 55%). These results suggested the (highly) different substrate specificities of WdYG1p and AYG1p. In the present study, WdYg1p poorly converted YWA1 to T4HN, as indicated by the fact that YWA1 was not appreciably metabolized to T4HN when it was fed to albino cultures or used to treat cell-free homogenates. In contrast, in an earlier in vitro study, AYG1p from A. fumigatus converted YWA1 to T4HN very efficiently (17).
Our demonstration in this study that melanin biosynthesis in W. dermatitidis requires AT4HN as a precursor of T4HN is of special interest because fungi are now known to make T4HN in a third manner. The two previous ways are as follows: (i) the bluish-green fungus A. fumigatus makes T4HN from the heptaketide precursor YWA1 by loss of acetoacetic acid (17, 40), and (ii) the brown fungus C. lagenarium makes T4HN directly as a pentaketide from malonyl-CoA without using either AT4HN or YWA1 as a precursor (16). Although we presently know that a large number of other plant- and human-pathogenic fungi make melanin from D2HN via T4HN, we do not know which of the three pathways they use. Additional studies with important plant pathogens, such as V. dahliae and Magnaporthe grisea, and melanized human pathogens, such as Sporothrix schenckii (31), Fonsecaea pedrosoi (11, 15), and Hortaea werneckii (25, 36), need to be carried out to determine how they make T4HN. Specifically, it would be helpful to know if they make AT4HN by PKS in a manner similar to that for WdPks1p in W. dermatitidis (14), YWA1 via a PKS in a manner similar to that for Alb1p in A. fumigatus (38), or T4HN directly by an enzyme, as reported for PKS in C. lagenarium (16). Additional studies with important plant pathogens such as V. dahliae and Magnaporthe grisea need to be carried out to determine if they make T4HN directly or from AT4HN or YWA1. Similar studies should also be carried out with the many human-pathogenic fungi known to synthesize D2HN to see if they make melanin in the same manner as W. dermatitidis. Specifically, do all these different types of fungi make AT4HN via the PKS referred to as WdPks1p (14), do some also make it in the manner in which A. fumigatus makes YWA1, via the PKS known as Alb1p (38), and do others produce T4HN directly by its PKS in the manner of C. lagenarium (16)? Information about the different types of upstream precursors involved in melanin biosynthesis will help to confirm the relationships among the melanized fungi and should provide additional information that is useful in future studies of host-parasite interactions.
Acknowledgments
We thank Li Zheng and Qiang Cheng, who, while members of the Szaniszlo laboratory at the University of Texas at Austin, identified and isolated the WdBrm1 strain and cloned and sequenced the WdYG1 gene, respectively; both Robert D. Stipanovic at the Cotton Pathology Research Unit with USDA/ARS and Shane E. Tichy at the LBMS laboratory at Texas A&M University, for obtaining and helping to interpret mass spectra; and Lucile Young for technical assistance.
This research was supported in part by a grant to P.J.S. from the National Institute of Allergy and Infectious Diseases (AI 33049). A part of this work was also supported by a grant-in-aid for scientific research (B) (no. 19310139) to I.F. from the Japan Society for the Promotion of Science and by a grant-in-aid for scientific research on priority areas (applied genomics) to I.F. from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 3.Becher, D., C. Djerassi, R. E. Moore, H. Singh, and P. J. Scheuer. 1966. Mass spectrometry in structural and stereochemical problems. CXI. The mass spectrometric fragmentation of substituted naphthoquinones and its application to structural elucidation of Echinoderm pigments. J. Org. Chem. 313650-3660. [Google Scholar]
- 4.Bell, A. A., R. D. Stipanovic, and J. E. Puhalla. 1976. Pentaketide metabolites of Verticillium dahliae. Identification of (+) scytalone as a natural precursor to melanin. Tetrahedron 321353-1356. [Google Scholar]
- 5.Bell, A. A., and M. H. Wheeler. 1986. Biosynthesis and function of fungal melanins. Annu. Rev. Phytopathol. 24411-451. [Google Scholar]
- 6.Brakhage, A. A., and B. Liebmann. 2005. Aspergillus fumigatus conidial pigment and cAMP signal transduction: significance for virulence. Med. Mycol. 43(Suppl. 1)S75-S82. [DOI] [PubMed] [Google Scholar]
- 7.Butler, M. J., and A. W. Day. 1998. Fungal melanins: a review. Can. J. Microbiol. 441115-1136. [Google Scholar]
- 8.Cheng, Q., K. A. Kinney, C. P. Whitman, and P. J. Szaniszlo. 2004. Characterization of two polyketide synthase genes in Exophiala lecanii-corni, a melanized fungus with bioremediation potential. Bioorg. Chem. 3292-108. [DOI] [PubMed] [Google Scholar]
- 9.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 862172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, Jr., C. R., and P. J. Szaniszlo. 1997. Melanin as a virulence factor in dematiaceous pathogenic fungi, p. 81-93. In H. V. Bossche, D. A. Stevens, and F. C. Odds (ed.), Host-fungus interplay. National Foundation for Infectious Diseases, Bethesda, MD.
- 11.Cunha, M. M., A. J. Franzen, D. S. Alviano, E. Zanardi, C. S. Alviano, W. De Souza, and S. Rozental. 2005. Inhibition of melanin synthesis pathway by tricyclazole increases susceptibility of Fonsecaea pedrosoi against mouse macrophages. Microsc. Res. Tech. 68377-384. [DOI] [PubMed] [Google Scholar]
- 12.Dixon, D. M., P. J. Szaniszlo, and A. Polak. 1991. Dihydroxynaphthalene (DHN) melanin and its relationship with virulence in the early stages of phaeohyphomycosis, p. 297-318. In G. T. Cole and H. C. Hoch (ed.), The fungal spore and disease initiation in plants. Plenum Press, New York, NY.
- 13.Dixon, D. M., J. Migliozzi, C. R. Cooper, Jr., O. Solis, B. Breslin, and P. J. Szaniszlo. 1992. Melanized and non-melanized multicellular-form mutants of Wangiella dermatitidis in mice: mortality and histopathology studies. Mycoses 3517-21. [DOI] [PubMed] [Google Scholar]
- 14.Feng, B., X. Wang, M. Hauser, S. Kaufmann, S. Jentsch, G. Haase, J. M. Becker, and P. J. Szaniszlo. 2001. Molecular cloning and characterization of WdPKS1, a gene involved in dihydroxynaphthalene melanin biosynthesis and virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 691781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franzen, A. J., M. M. L. Cunha, E. J. O. Batista, S. H. Seabra, W. De Souza, and S. Rozental. 2006. Effects of tricyclazole (5-methyl-1,2,4-triazol[3,4] benzothiazole), a specific DHN-melanin inhibitor, on the morphology of Fonsecaea pedrosoi conidia and sclerotic cells. Microsc. Res. Tech. 69729-737. [DOI] [PubMed] [Google Scholar]
- 16.Fujii, I., Y. Mori, A. Watanabe, Y. Kubo, G. Tsuji, and Y. Ebizuka. 2000. Enzymatic synthesis of 1,3,6,8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase PKS1. Biochemistry 398853-8858. [DOI] [PubMed] [Google Scholar]
- 17.Fujii, I., Y. Yasuoka, H.-F. Tsai, Y. C. Chang, K. J. Kwon-Chung, and Y. Ebizuka. 2004. Hydrolytic polyketide shortening by Ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J. Biol. Chem. 27944613-44620. [DOI] [PubMed] [Google Scholar]
- 18.Fujii, I., N. Yoshida, S. Shimomak, H. Oikawa, and Y. Ebizuka. 2005. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem. Biol. 121301-1309. [DOI] [PubMed] [Google Scholar]
- 19.Fujii, T., H. Yamaoka, K. Gomi, K. Kitamoto, and C. Kumagai. 1995. Cloning and nucleotide sequence of the ribonuclease T1 gene (rntA) from Aspergillus oryzae and its expression in Saccharomyces cerevisiae and Aspergillus oryzae. Biosci. Biotechnol. Biochem. 591869-1874. [DOI] [PubMed] [Google Scholar]
- 20.Geis, P. A., M. H. Wheeler, and P. J. Szaniszlo. 1984. Pentaketide metabolites of melanin synthesis in the dematiaceous fungus Wangiella dermatitides. Arch. Microbiol. 137324-328. [DOI] [PubMed] [Google Scholar]
- 21.Gomi, K., Y. Imamura, and S. Hara. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 512549-2555. [Google Scholar]
- 22.Henson, J. M., M. J. Butler, and A. W. Day. 1999. The dark side of the mycelium: melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37447-471. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, E. S. 2000. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karuppayil, S. M., and P. J. Szaniszlo. 1997. Importance of calcium to the regulation of polymorphism in Wangiella dermatitidis. J. Med. Vet. Mycol. 35379-388. [DOI] [PubMed] [Google Scholar]
- 25.Kogej, T., M. H. Wheeler, T. L. Rizner, and N. Gunde-Cimerman. 2004. Evidence for 1,8-dihydroxynaphthalene melanin in three halophilic black yeasts grown under saline and non-saline conditions. FEMS Microbiol. Lett. 232203-209. [DOI] [PubMed] [Google Scholar]
- 26.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38143-158. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2004. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperatures of infection. Eukaryot. Cell 340-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, R. E., H. Singh, W. J. Clifford, W. J. Chang, and P. J. Scheuer. 1966. Sodium borohydride reduction of spinochrome A. Removal of phenolic hydroxyls in the naphthazarins system. J. Org. Chem. 313638-3645. [Google Scholar]
- 29.Nosanchuk, J. D., and A. Casadevall. 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 503519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paolo, W. F., Jr., E. Dadachova, P. Mandal, A. Casadevall, P. J. Szaniszlo, and J. D. Nosanchuck. 2006. Effects of disrupting the polyketide synthase gene WDPKS1 in Wangiella (Exophiala) dermatitidis on melanin production and resistance to killing by antifungal compounds, enzymatic degradation, and extremes in temperature. BMC Microbiol. 655. http://www.biomedcentral.com/1471-2180-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero-Martinez, R., M. Wheeler, A. Guerrero-Plata, G. Rico, and H. Torres-Guerrero. 2000. Biosynthesis and functions of melanin in Sporothrix schenckii. Infect. Immun. 683696-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 33.Schnitzler, N., H. Peltroche-Llacsahuanga, N. Bestier, J. Zundorf, R. Lutticken, and G. Haase. 1998. Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidis on phagocytosis, oxidative burst, and killing by human neutrophils. Infect. Immun. 6794-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szaniszlo, P. J. 2002. Molecular genetic studies of the model dematiaceous pathogen Wangiella dermatitidis. Int. J. Med. Microbiol. 292381-390. [DOI] [PubMed] [Google Scholar]
- 35.Szaniszlo, P. J. 2006. Virulence factors in black molds with emphasis on melanin, chitin, and Wangiella as a molecularly tractable model, p. 407-428. In J. Heitman, S. Filler, A. Mitchell, and J. Edwards (ed.), Molecular principles of fungal pathogenesis. American Society for Microbiology, Washington, DC.
- 36.Taylor, B. E., M. H. Wheeler, and P. J. Szaniszlo. 1987. Evidence for pentaketide biosynthesis in dematiaceous human pathogenic fungi. Mycologia 79320-322. [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, H.-F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 1803031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, H.-F., M. H. Wheeler, Y. C. Chang, Y. C., and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 1816469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, H.-F., I. Fujii, A. Watanabe, M. H. Wheeler, Y. C. Chang, Y. Yasuoka, Y. Ebizuka, and K. J. Kwon-Chung. 2001. Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J. Biol. Chem. 27629292-29298. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Z., L. Zheng, M. Hauser, J. M. Becker, and P. J. Szaniszlo. 1999. WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support the growth of Wangiella (Exophiala) dermatitidis at temperature of infection. Infect. Immun. 676619-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, A., I. Fujii, U. Sankawa, M. E. Mayorga, W. E. Timberlake, and Y. Ebizuka. 1999. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 4091-94. [Google Scholar]
- 43.Watanabe, A., and Y. Ebizuka. 2002. A novel hexaketide naphthalene synthesized by a chimeric polyketide synthase composed of fungal pentaketide and heptaketide synthases. Tetrahedron Lett. 43843-846. [Google Scholar]
- 44.Wheeler, M. H., and R. D. Stipanovic. 1985. Melanin biosynthesis and the metabolism of flaviolin and 2-hydroxyjuglone in Wangiella dermatitidis. Arch. Microbiol. 142234-241. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler, M. H., and A. A. Bell. 1988. Melanins and their importance in pathogenic fungi, p. 338-387. In M. R. McGinnis (ed.), Current topics in medical mycology, vol. 2. Springer-Verlag, New York, NY. [DOI] [PubMed] [Google Scholar]
- 46.Wheeler, M. H., B. D. Bruton, L. S. Puckhaber, J. Zhang, and R. D. Stipanovic. 2004. Identification of 1,8-dihydroxynaphthalene melanin in Monosporascus cannonballus and the analysis of hexaketide and pentaketide compounds produced by wild-type and pigmented isolates of the fungus. J. Agric. Food Chem. 524113-4120. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler, M. H., Q. Cheng, L. S. Puckhaber, and P. J. Szaniszlo. 2005. Evidence for heptaketide melanin biosynthesis and chain-shortening of the melanin precursor YWA1 to 1,3,6,8-tetrahydroxynaphthalene in Wangiella (Exophiala) dermatitidis, abstr. F-036, p. 266. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 48.Zheng, L. 1997. Establishment of genetic transformation systems in and molecular cloning of the chitin synthase 2 (WdCHS2) gene, and the characterization of the WdCHS1 and WdCHS2 genes of Wangiella dermatitidis. Ph.D. thesis. The University of Texas at Austin, Austin.
- 49.Zheng, L., and P. J. Szaniszlo. 1999. Cloning and use of the WdURA5 gene as a hisG cassette selection marker for potentially disrupting multiple genes in Wangiella dermatitidis. Med. Mycol. 3785-96. [PubMed] [Google Scholar]