Abstract
Copper amine oxidases (CAOs) have been proposed to be involved in the metabolism of xenobiotic and biogenic amines. The requirement for copper is absolute for their activity. In the fission yeast Schizosaccharomyces pombe, cao1+ and cao2+ genes are predicted to encode members of the CAO family. While both genes are expressed in wild-type cells, we determined that the expression of only cao1+ but not cao2+ results in the production of an active enzyme. Site-directed mutagenesis identified three histidine residues within the C-terminal region of Cao1 that are necessary for amine oxidase activity. By use of a cao1+-GFP allele that retained wild-type function, Cao1-GFP was localized in the cytosol (GFP is green fluorescent protein). Under copper-limiting conditions, disruption of ctr4+, ctr5+, and cuf1+ produced a defect in amine oxidase activity, indicating that a functionally active Cao1 requires Ctr4/5-mediated copper transport and the transcription factor Cuf1. Likewise, atx1 null cells exhibited substantially decreased levels of amine oxidase activity. In contrast, deletion of ccc2, cox17, and pccs had no significant effect on Cao1 activity. Residual amine oxidase activity in cells lacking atx1+ can be restored to normal levels by returning an atx1+ allele, underscoring the critical importance of the presence of Atx1 in cells. Using two-hybrid analysis, we demonstrated that Cao1 physically interacts with Atx1 and that this association is comparable to that of Atx1 with the N-terminal region of Ccc2. Collectively, these results describe the first example of the ability of Atx1 to act as a copper carrier for a molecule other than Ccc2 and its critical role in delivering copper to Cao1.
Copper is an essential metal ion cofactor that is required for many biological processes, including respiration, iron transport, superoxide anion detoxification, and assimilation of carbon and nitrogen sources (29). In excess, however, copper can participate in redox reactions that generate highly reactive oxygen species that cause damage to the cellular membrane, proteins, DNA, and RNA molecules (21). Consequently, organisms have evolved with multiple mechanisms to obtain sufficient levels of copper for incorporation into cuproproteins, while tightly controlling intracellular copper to avoid toxicity.
Copper amine oxidases (CAOs) have been identified in bacteria, yeasts, plants, and animals (39). Although little is known about their precise biological roles, CAOs from prokaryotes and lower eukaryotes can catalyze the oxidative deamination of several amine substrates to their corresponding aldehydes, providing carbon and nitrogen sources for cellular growth (7, 49). In higher eukaryotes, the situation is less well defined. Roles in wound healing, regulation of glucose uptake, metabolite signaling, and cell-cell adhesion and recognition, as well as detoxification of endogenous and xenobiotic amines, have been suggested for CAOs (42, 54). CAOs are dimeric proteins with molecular masses of ∼140 to 190 kDa. CAOs have been shown to contain a single copper ion per monomer. Each copper ion is coordinated by the imidazole groups of three highly conserved histidine residues located in the C-terminal half of each monomer (43). In addition to the copper ion, each monomer contains a covalently bound cofactor, 2,4,5-trihydroxyphenylalanine quinone (TPQ), which is generated by posttranslational modification of the first conserved tyrosine residue (indicated in bold) in the consensus sequence Asn-Tyr-(Glu/Asp)-Tyr (9, 12, 28, 36). The copper ion and oxygen are required for tyrosine modification into TPQ (10). An active CAO protein therefore has the capacity to convert a primary amine and molecular oxygen into the corresponding aldehyde, ammonia, and hydrogen peroxide. Currently, the pathways by which copper is delivered to CAOs are not well understood.
In the fission yeast Schizosaccharomyces pombe, the high-affinity copper uptake process occurs via a two-component copper transporting complex at the cell surface. This heteroprotein complex is composed of the Ctr4 and Ctr5 proteins, which are structurally related to each other and to the Ctr family of copper transporters (4, 46, 55). Ctr4 and Ctr5 are physically associated in vivo (55). Coexpression of both proteins is necessary for the proper function and localization of the heteroprotein complex at the plasma membrane (4, 55). Like most Ctr family members, the N-terminal hydrophilic domains of Ctr4 and Ctr5 are rich in methionine residues (32, 55). At the cell surface, these domains function independently to transport copper; however, both domains are required for optimal cell growth under copper-deficient conditions (4). The copper-sensing transcription factor Cuf1 plays an essential role in coordinating the copper-regulated transcription of copper transporter genes in S. pombe (2). Cuf1 activates transcription of the ctr4+ and ctr5+ genes under copper-deficient conditions and represses their expression under copper-replete conditions (5).
Once inside the cell, free-copper ions are virtually undetectable (47). One strategy by which cells transport copper to copper-dependent proteins within the cytoplasm involves the use of small soluble cytoplasmic copper carriers known as copper chaperones (19). In Saccharomyces cerevisiae, three small copper-binding proteins, Atx1 (37), Ccs1 (16), and Cox17 (20), have been identified as chaperones to deliver copper to specific intracellular localizations. Atx1 shuttles copper from the cytosol to post-Golgi vesicles by docking specifically with the Ccc2 copper-transporting P-type ATPase (45). Once transferred into the Golgi apparatus, it is thought that copper is loaded onto proteins as part of their maturation. In the mitochondrion, copper is stored mainly within the matrix, and from there copper is delivered by an as-yet-unidentified intracellular ligand in the mitochondrial intermembrane space. Once transported within the mitochondrial intermembrane space, copper is bound by Cox17, which in turn transfers copper into cytochrome c oxidase with the aid of Sco1 and Cox11 (13, 14, 24). A third copper chaperone, Ccs1, donates copper specifically to the cytosolic copper zinc-superoxide dismutase (17). For the fission yeast S. pombe, although candidate molecules for copper trafficking into cells have been inferred from sequence homology to the S. cerevisiae chaperones (31), only one has been characterized experimentally, namely, Pccs (34). This protein is orthologous to the S. cerevisiae Ccs1 cytosolic copper chaperone. In addition to its specific function to deliver copper to copper zinc-superoxide dismutase, Pccs is important for the survival of fission yeast cells in the continued presence of elevated concentrations of copper and cadmium (34). The latter observation indicates that some functional differences in copper chaperones may exist between the two species of yeast. Examination of the S. pombe genome database suggests that the open reading frame (ORF) SPBC1709.10c encodes a putative ortholog of S. cerevisiae Atx1. This putative ortholog bears 56% identity to Atx1, and the copper-binding Met-X-Cys-X-X-Cys motif is conserved between the two proteins, suggesting a role for this protein in delivering copper within the cell. The Cox17 chaperone from S. cerevisiae has a related protein in S. pombe encoded by SPBC26H8.14c. This ortholog exhibits 38% identity to the baker's yeast Cox17, with all of the functionally important cysteine residues well conserved between the two proteins. Thus, it is likely that the Cox17-like protein from fission yeast might function in delivering copper to the mitochondrial cytochrome c oxidase.
In contrast to S. pombe, S. cerevisiae does not have an endogenous CAO (9, 33, 35). However, it has been shown that heterologous expression of a CAO from another organism in S. cerevisiae generates a functional enzyme (8, 33). We showed that, when an active CAO is expressed heterologously in S. cerevisiae, its production requires CTR-mediated copper transport and the transcription factor Mac1, which is essential for the expression of the high-affinity copper transport genes (33). Furthermore, we found that Atx1 and, to a lesser extent, Ccc2 are required for the production of an active recombinant CAO in S. cerevisiae cells (33). While the results obtained with S. cerevisiae strongly suggest that the Atx1 copper chaperone is important for delivering copper to cytosolic CAO, it remains to be established whether the S. pombe SPBC1709.10c gene, which encodes a putative ortholog of Atx1, fulfills the same function in fission yeast. Given this point, we sought to examine the requirement of Atx1 for CAO activity in S. pombe.
For fission yeast, two CAO molecules, SPAC2E1P3.04 and SPBC1289.16c, have been annotated from the S. pombe Genome Project (52a). These proteins were initially designated SPAO1 and SPAO2 (33); however, recently they have been renamed Cao1 and Cao2, respectively, per the S. pombe Gene Naming Committee (http://www.genedb.org/genedb/pombe/geneRegistry.jsp). In this study, we created S. pombe strains with single or double deletions of the cao1 and cao2 genes. We found that only the expression of cao1+ resulted in the production of an active enzyme capable of catalyzing the oxidative deamination of primary amines. Cao2 played no apparent role in amine oxidase activity. Specific mutation of the His458, His460, and His627 residues to alanine within the C-terminal region of Cao1 resulted in the complete loss of CAO activity. We determined that Cao1 is a cytosolic protein which requires expression of genes involved in high-affinity copper transport to be active. Likewise, we determined that the presence of S. pombe Atx1 conferred ∼70 to 80% of the amine oxidase activity. By use of two-hybrid analysis, Atx1 was shown to interact with Cao1. Collectively, these results demonstrate that the fission yeast Atx1 protein can function as a copper chaperone for a molecule other than Ccc2, participating in the delivery of copper to Cao1.
MATERIALS AND METHODS
Yeast strains and growth media.
The S. pombe strains used in this study were all isogenic derivatives of FY435 (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210) (6) and included cuf1Δ (5), cao1Δ (FY435 plus cao1Δ::KANr), cao2Δ (FY435 plus cao2Δ::KANr), cao1Δ cao2Δ (FY435 plus cao1Δ::loxP cao2Δ::KANr), ctr4Δ ctr5Δ (4), atx1Δ (FY435 plus atx1Δ::KANr), ccc2Δ (FY435 plus ccc2Δ::KANr), cox17Δ (FY435 plus cox17Δ::KANr), and pccsΔ (34) disruption strains. To ascertain that the results observed were not specific to the S. pombe strain FY435, identical experiments were carried out with the strain FY254 (h− can1-1 leu1-32 ura4-Δ18 ade6-M210) and its isogenic derivatives cuf1Δ (32), cao1Δ (FY254 plus cao1Δ::KANr), cao2Δ (FY254 plus cao2Δ::KANr), cao1Δ cao2Δ (FY254 plus cao1Δ::loxP cao2Δ::KANr), and atx1Δ (FY254 plus atx1Δ::KANr). Double mutant gene disruptions were created by using a recyclable loxP-KANMX2-loxP cassette through homologous recombination, as described previously (26). S. pombe cells were cultured in yeast extract plus supplements, which contains 3% glucose and 225 mg/liter of adenine, histidine, leucine, uracil, and lysine (1). To maintain plasmids in various strains, synthetic Edinburgh minimal medium (1) with 225 mg/liter of the required amino acids was used; unsupplemented Edinburgh minimal medium contains 160 nM copper. Conditions of copper deprivation or copper repletion were generated by adding the indicated concentration of ammonium tetrathiomolybdate (TTM) (323446; Sigma-Aldrich) or CuSO4 to cells grown to the mid-logarithmic exponential phase (A600 of ∼0.5). After treatment for 8 h, 20-ml samples were withdrawn from the cultures for subsequent detection of CAO activity or steady-state mRNA or protein analysis.
Plasmids.
Plasmid pSP1cao1+ was constructed through a three-piece ligation by simultaneously introducing a 1,934-bp SpeI-SphI PCR-amplified fragment containing the cao1+ locus starting at position −1420 from the translational start codon up to position +514 after the initiator codon and a 2,060-bp SphI-ApaI PCR-amplified fragment bearing the cao1+ ORF starting at position +515 from the translational start codon up to position +436 after the stop codon into the SpeI-ApaI-digested pSP1 vector (15). To generate the cao1+ StuI-BspEI allele, a 12-bp StuI-BspEI linker was inserted in frame to and downstream of the last codon of the cao1+ gene by an overlap extension method (22). The insertion created four extra amino acid residues after the phenylalanine at position 712 (Phe712-Arg-Pro-Ser-Gly-stop codon) of Cao1. This allele was found to be functional because of its ability to fully restore Cao activity in vivo. We used the restriction sites StuI and BspEI created within cao1+ to insert a copy of the GFP gene (30) (GFP is green fluorescent protein). The resulting plasmid was designated pSP1cao1+-GFP. To create the pBPade6+cao1+-GFP plasmid, pSP1cao1+-GFP was digested with SpeI and ApaI, allowing the purification of a DNA fragment containing the cao1+-GFP gene with its own promoter and terminator. Once purified, the DNA fragment was cloned into the SpeI-ApaI-digested pBPade6+ vector (4). For integration, pBPade6+cao1+-GFP was linearized with AatII. The cao1 mutant alleles containing site-specific mutations (H456A, H458A, H460A, H621A, and H627A) were created by a PCR overlap extension method (22). For H456A, H458A, and H460A mutants, the DNA sequence from each respective PCR-amplified fragment was digested with NruI and XhoI and exchanged with a corresponding DNA region into the pSP1cao1+ plasmid. Likewise, for H621A and H627A mutants, the DNA sequence from each respective PCR-amplified fragment was digested with XhoI and BspEI and then used to replace the corresponding fragment from the plasmid pSP1cao1+. All nucleotide changes that gave rise to mutations were verified by dideoxy sequencing. The atx1+ gene (SPBC1709.10c) was isolated by PCR amplification using primers corresponding to the start and stop codons of the ORF from an S. pombe cDNA library (ATCC 87284; deposited by S. Elledge) (kind gift of Dennis J. Thiele, Duke University). Because the primers contained EcoRI and SalI restriction sites, the purified DNA fragment was digested with these restriction enzymes and cloned into the corresponding sites of pBluescript SK vector (Stratagene, La Jolla, CA). The resulting plasmid was named pSKatx1+. Subsequently, the S. pombe atx1+ promoter up to position −1336 from the start codon of the atx1+ gene was isolated by PCR amplification and then inserted into pSKatx1+ at the NotI and PstI sites. This pSKatx1+ derivative was designated pSKprom-atx1+. The NotI-SalI DNA fragment was isolated from pSKprom-atx1+ and then inserted into the NotI-SalI-cut pSP1 plasmid, creating the pSP1atx1+ plasmid.
Analysis of gene expression.
Total RNA was extracted by a hot phenol method as described previously (11). RNA samples were quantified spectrophotometrically, and 15 μg of RNA per sample was used for an RNase protection assay, which was carried out as described previously (41). Plasmid pSKcao1+ was created by inserting a 172-bp NotI-EcoRI fragment from the cao1+ gene into the same sites of pBluescript SK (Stratagene, La Jolla, CA). The antisense RNA hybridizes to the region between positions +4 and +176 downstream from the initiator codon of cao1+. To generate pSKcao2+, a 191-bp fragment from the cao2+ gene (corresponding to the coding region between positions +2083 and +2274) was amplified and cloned into the BamHI-EcoRI sites of pBluescript SK. Plasmid pSKact1+, for measuring act1+ mRNA levels, was constructed by inserting a 151-bp BamHI-EcoRI fragment of the act1+ gene into the same restriction sites of pBluescript SK. The antisense RNA pairs to the region between positions +334 and +485 downstream from the A of the start codon of act1+. Except for pSKcao1+, which was linearized with NotI, all the above-mentioned constructs were digested with BamHI for subsequent labeling with [α-32P]UTP and T7 RNA polymerase.
CAO activity assays.
A chemiluminescence in-gel assay for CAO activity was carried out as described previously (33). Quantitative assessments of CAO activity were conducted as described by Holt and Palcic (23), with the following modifications. Cell lysates were quantified using the Bradford assay (6a), and equal amounts of cellular protein were subjected to a colorimetric assay. A “physiological HEPES” 2× stock solution (100 mM HEPES, 10 mM KCl, 4 mM CaCl2, 2.8 mM MgCl2, 280 mM NaCl) was prepared and then adjusted to a pH of 7.4 with a solution of 1 N NaOH. Before each assay, 2 mM 4-aminoantipyrine (A-4382; Sigma), 4 mM vanillic acid (H-36001; Sigma-Aldrich), and 16 U/ml of horseradish peroxidase (P-6782; Sigma) were dissolved in 2× “physiological HEPES” buffer to generate a chromogenic solution. A stock solution of ethylamine (80 mM) was prepared in water and used as a substrate. Typically, reactions were performed by adding equal volumes of the chromogenic solution (40 μl), ethylamine (40 μl), and cell lysates (equal amounts). When needed, Milli-Q water was added to reach a final reaction volume of 160 μl. Reaction mixtures were incubated at 37°C for 4 to 16 h. The appearance of red dye was measured using a spectrophotometer at 498 nm. Before each 4-aminoantipyrine-vanillic acid-peroxidase-linked assay, the chromogenic solution was tested by adding a small volume of diluted hydrogen peroxide (1:1,000) to 100 μl of the chromogenic solution to confirm that the expected red dye was generated by the colorimetric assay.
Immunoblotting.
For Western blotting experiments, protein extracts were resolved on 8% sodium dodecyl sulfate-polyacrylamide gels. After electrophoresis, protein samples were electroblotted onto polyvinylidene difluoride Hybond-P membranes (Amersham Biosciences) for 1 h at 4°C. Membranes were blocked for 2 h at 4°C in 5% powdered skim milk (Difco) in TBS (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% bovine serum albumin) with 0.1% Tween 20 (TBST). After being washed in TBST, membranes were incubated with primary antibodies in 1% powdered skim milk in TBST for 16 h at 4°C. The following primary antisera were used for immunodetection: monoclonal anti-His5 antibody (penta-His; Qiagen), monoclonal anti-GFP antibody (B-2; Santa Cruz Biotechnology), and monoclonal anti-PCNA antibody (PC10; Sigma). After incubation with the primary antibodies, membranes were washed and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences), developed with ECL reagents (Amersham Biosciences), and visualized by chemiluminescence.
Microscopic analysis of Cao1 localization.
Fluorescence microscopic analysis was performed as described previously (3). Fluorescence and differential interference contrast images of the cells were obtained with an Eclipse E800 epifluorescent microscope (Nikon, Melville, NY) equipped with an Orca ER digital cooled camera (Hamamatsu, Bridgewater, NJ). The samples were analyzed using ×1,000 magnification with the following filters: 465 to 495 nm (GFP) and 340 to 380 nm (DAPI [4′,6-diamidino-2-phenylindole]). Cell fields shown in this study are representative of experiments repeated at least five times.
Two-hybrid interaction assay.
S. cerevisiae strain L40 [MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ] (51) was used for two-hybrid analysis. For growth of cells, a modified synthetic minimal medium was used. This synthetic medium contains 83 mg/liter histidine, 83 mg/liter adenine, 30 mg/liter lysine, 2% dextrose, 50 mM MES [2-(N-morpholino)ethanesulfonic acid] buffer (pH 6.1), 10 μM NH4Fe(SO4)2, and 0.67% yeast nitrogen base minus copper and iron (MP Biomedicals, Solon, OH). To study the interaction of Cao1 with Atx1, the full-length version of the cao1+ ORF was inserted downstream of and in frame to the Escherichia coli lexA gene as bait. PCR amplification of the cao1+ gene was carried out using primers corresponding to the start and stop codons of the ORF from S. pombe genomic DNA with Pfu DNA polymerase (Stratagene). To clone the PCR product into the pLexN-a vector (51), primers designed to generate EcoRI and SalI restriction sites at the upstream and downstream termini of the cao1+ gene were used. The PCR product was digested with EcoRI and SalI and cloned into the corresponding sites of pLexN-a. A similar cloning strategy was used to generate pLexN-a-CCC2-a, which contains the wild-type (WT) ccc2+ N-terminal codons 1 to 163. The prey plasmid pVP16-Atx1 was created by cloning a 207-bp BamHI-EcoRI DNA fragment containing the full-length coding region of atx1+ into the same sites of pVP16 (51). To create the atx1(R,K) allele (see last paragraph of Results), primers corresponding to the beginning and the end of the atx1+ gene were made with mutations in the sequence that generated the Arg-20Glu, Arg-24Glu, Lys-55Glu, Lys-56Glu, and Lys-59Glu substitutions at the N- and C-terminal regions of Atx1. Each L40 transformant strain harboring the indicated bait and prey plasmids was tested for the association of the two fusion proteins by a liquid β-galactosidase assay as described previously (56), except that cells were broken by vortexing in the presence of glass beads. The expression of the LexA-Cao1 and VP16-Atx1 fusion proteins was verified by immunoblot analysis using the following antisera: monoclonal anti-LexA antibody 2-12 (Santa Cruz Biotechnology) and monoclonal anti-VP16 antibody 1-21 (Santa Cruz Biotechnology). A monoclonal anti-phosphoglycerol kinase (PGK) antibody, 22C5-D8 (Molecular Probes), was used to detect PGK protein as an internal control.
RESULTS
Primary sequences of S. pombe CAOs.
In S. pombe, there are two genes that encode proteins related to the CAO family of enzymes. We initially designated the SPAC2E1P3.04- and SPBC1289.16c-encoded proteins SPAO1 and SPAO2, respectively (33). However, because of potential name conflicts brought up by the S. pombe Gene Naming Committee, SPAC2E1P3.04 and SPBC1289.16c genes were renamed cao1+ and cao2+, respectively. The amino acid sequences of Cao1 and Cao2 are 57% and 40% identical, respectively, to that of the Hansenula polymorpha methylamine oxidase HPAO (Fig. 1). These represent the highest percentages of identity when Cao1 and Cao2 were compared to other prokaryotic and eukaryotic CAOs (C. Peter, J. Laliberté, and S. Labbé, unpublished data). Like HPAO and the other microbial and metazoan CAOs, both Cao1 and Cao2 have a tyrosine residue (Tyr407 for Cao1; Tyr394 for Cao2) that is contained within the highly conserved Asn-Tyr-Glu/Asp-Tyr sequence (Fig. 1). The first tyrosine within this sequence (indicated in bold) is known to be posttranslationally converted to TPQ by an autocatalytic mechanism (9). The conversion of the precursor tyrosine to TPQ requires the presence of molecular oxygen and a mononuclear copper center. Based on the sequence alignment of Cao1 and Cao2 with HPAO, histidine residues (His456, His458, His460, His621, and His627 for Cao1; His445, His447, His598, and His604 for Cao2) may coordinate a single copper atom. Furthermore, the two amino acids (Tyr307 and Asp321) in Cao1 corresponding to HPAO Tyr305 and Asp319 are also conserved and might, by analogy with HPAO, serve to promote the active conformation of TPQ during the enzymatic reaction. In Cao2, however, a Phe residue is found at position 295 instead of Tyr. Despite this difference found in Cao2, amino acid alignments of Cao1 and Cao2 with the H. polymorpha protein HPAO and with CAOs from other organisms (data not shown) suggested that both Cao1 and Cao2 possess conserved motifs thought to be essential for CAO activity.
FIG. 1.
Amino acid alignment of S. pombe Cao1 and Cao2 with H. polymorpha HPAO. Amino acid residues identical in the compared proteins are shown in inverse highlighting. The rectangle shown in the middle portion of the protein sequences indicates the location of the conserved N-Y-E-Y motif in which the first peptidyl tyrosine residue (black circle) is predicted to serve as a precursor for TPQ. Arrowheads indicate putative amino acids that may promote the active conformation of TPQ during the enzymatic reaction. Asterisks show putative histidine residues located in the C-terminal halves of the proteins that are potentially involved in the coordination of one copper atom. The amino acid sequence numbers refer to the position relative to the first amino acid of each protein.
Expression of cao1+ and cao2+ is copper independent and is not regulated by Cuf1.
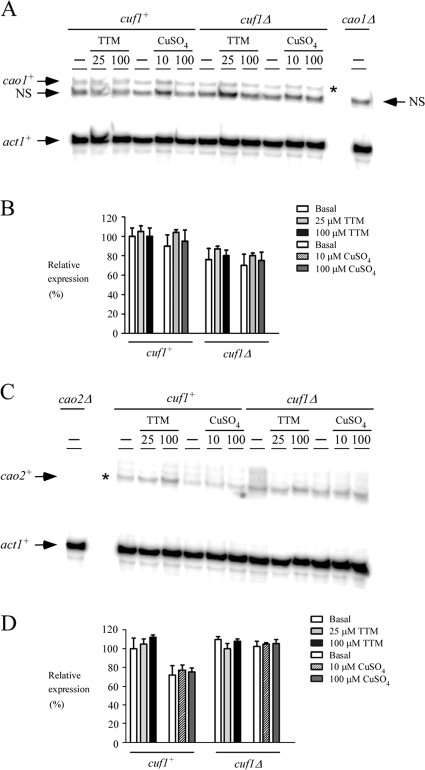
As determined by RNase protection analyses, the steady-state mRNA levels of cao1+ and cao2+ in the WT strain FY435 were not regulated by either the copper chelator TTM (25 and 100 μM) or exogenous CuSO4 (10 and 100 μM) (Fig. 2). There were no significant changes in levels of cao1+ and cao2+ gene expression in treated cells compared to the basal levels in untreated cells (Fig. 2). As controls for signal specificity, cao1+ and cao2+ mRNAs were absent when RNA samples were isolated from cao1Δ and cao2Δ mutant cells, respectively (Fig. 2A and C). RNA levels in each experiment were quantitated (Fig. 2B and D). To further examine whether cao1+ and cao2+ transcription is controlled by the S. pombe regulatory transcription factor Cuf1, a cuf1Δ mutant strain was grown in the absence or presence of 25 and 100 μM TTM or 10 and 100 μM CuSO4. As shown in Fig. 2, the cuf1Δ mutant had no significant effect on the expression of the cao1+ and cao2+ genes. We therefore concluded that Cuf1, a transcription factor that is required for expression of genes involved in copper transport, is not required for constitutive transcription of the cao1+ and cao2+ genes.
FIG. 2.
cao1+ and cao2+ mRNA levels are constitutively expressed and present in cuf1Δ cells. (A) The indicated isogenic strains were grown to logarithmic phase in yeast extract plus supplements. Cultures were incubated in the absence (−) or presence of TTM (25 and 100 μM) or CuSO4 (10 and 100 μM) for 1 h at 30°C. Total RNA was isolated and analyzed by RNase protection assays. Steady-state mRNA levels of cao1+ and act1+ (indicated with arrows) were analyzed in strains expressing (cuf1+) or lacking (cuf1Δ) the cuf1+ allele. As a control, the cao1+ mRNA was not detected in the isogenic cao1Δ null strain. NS, nonspecific signal. The asterisk indicates the cao1+ mRNA. (B) Quantification of results of three independent RNase protection assays, including results shown in panel A. (C) Aliquots of the cultures described for panel A were examined by RNase protection assays for steady-state levels of cao2+ mRNA. The arrows indicate signals corresponding to cao2+ and act1+ steady-state mRNA levels. RNA isolated and analyzed from the isogenic cao2Δ null strain was used as a control. The asterisk indicates the cao2+ mRNA. (D) Quantification of cao2+ mRNA levels after the treatments. The values are the averages from triplicate determinations ± standard deviations.
Cao1 is necessary for CAO activity in S. pombe.
Our previous studies of Cao1 and Cao2 took advantage of the fact that, although S. cerevisiae does not have an endogenous CAO, it can serve as an excellent host for the expression and characterization of genes encoding CAOs from other organisms (8, 33). We showed that, when heterologously expressed in S. cerevisiae, only cao1+ but not cao2+ resulted in the production of an active enzyme capable of catalyzing the oxidative deamination of ethylamine (33). To assess the amine oxidase activities of Cao1 and Cao2 in S. pombe, we used isogenic strains harboring WT cao1+ and cao2+ genes or insertionally inactivated cao1Δ, cao2Δ, or cao1Δ cao2Δ double mutant genes. In the WT strain, we detected a strong chemiluminescent signal indicating the presence of an active amine oxidase (Fig. 3A). In contrast, no detectable CAO activity was observed with the cao1Δ single mutant (Fig. 3A). The cao2Δ mutant behaved similarly to the WT strain and was equally competent in catalyzing the oxidative deamination of ethylamine. As expected, no CAO activity was observed with the cao1Δ cao2Δ double mutant strain (Fig. 3A). To test whether transcripts of both cao1+ and cao2+ were present in WT or single or double deletion strains, the steady-state mRNA levels of cao1+, cao2+, and act1+ were analyzed by RNase protection assays. The results shown in Fig. 3B indicate that, although Cao2 was inactive, its transcript was clearly detected in WT and cao1Δ strains. In contrast, no cao2+ transcript was observed with the cao2Δ single mutant and cao1Δ cao2Δ double mutant strains (Fig. 3B). Further analysis showed that both the WT and the cao2Δ mutant strains expressed cao1+, while the cao1Δ and cao1Δ cao2Δ mutants exhibited no cao1+ transcripts (Fig. 3B). Taken together, our analysis of mutants defective in cao1+, cao2+, or both genes revealed that cao1+ plays a unique role in producing amine oxidase activity in S. pombe.
FIG. 3.
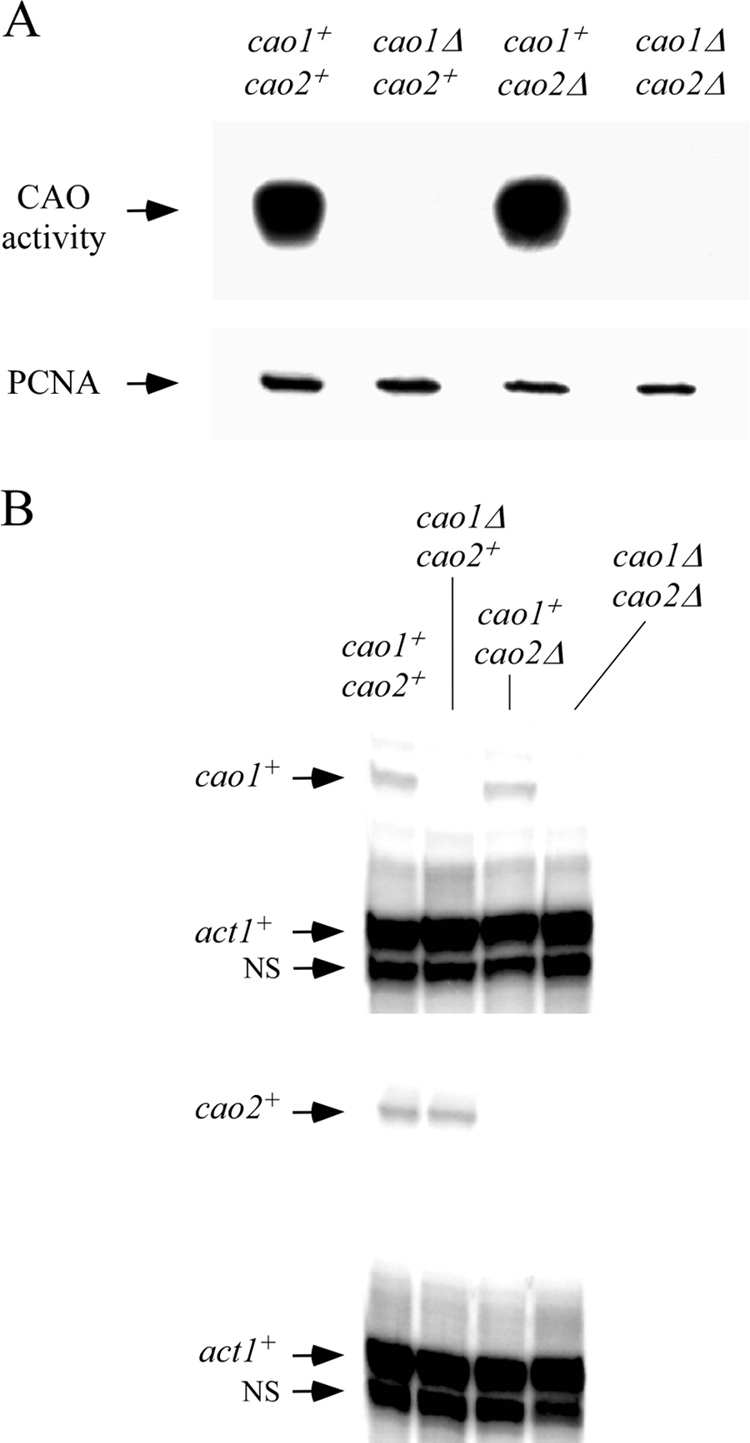
Cao1 but not Cao2 catalyzes the oxidative deamination of ethylamine. (A) Four isogenic fission yeast strains (cao1+ cao2+, cao1Δ cao2+, cao1+ cao2Δ, and cao1Δ cao2Δ strains) were analyzed for the presence of CAO activity by use of a chemiluminescence activity assay with ethylamine (10 mM) as a substrate (top). As an internal control, aliquots of total protein extracts were analyzed by immunoblotting using an anti-PCNA antibody (bottom). (B) Total RNA was prepared from aliquots of cultures used in the experiment described for panel A. Representative RNase protection assays of cao1+ (top) and cao2+ (bottom) are shown, indicating steady-state mRNA levels. Actin (act1+) mRNA levels were probed as an internal control. NS, nonspecific signal.
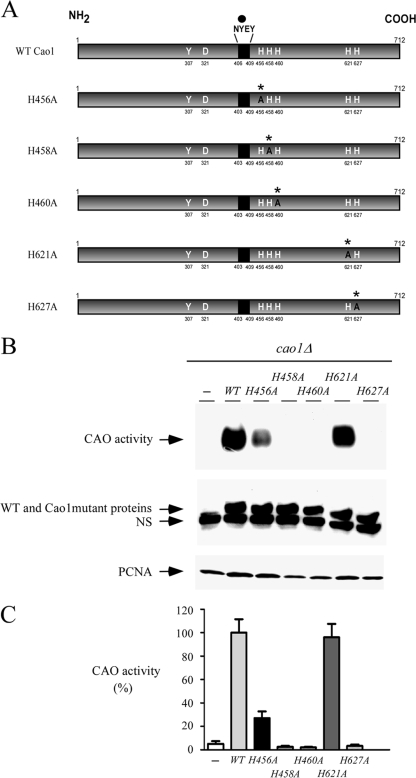
In vivo mapping of critical histidine residues required for Cao1 function.
Within the C-terminal half of Cao1, several His residues are conserved and may act as potential copper ligands. To ascertain their role in Cao1 activity, five His residues, His456, His458, His460, His621, and His627, were individually replaced with Ala (Fig. 4A). These mutations had no effect on the steady-state levels of Cao1 protein (Fig. 4B, middle). The WT and His mutant proteins were detected by immunoblotting using an anti-His5 antibody, due to the presence of an endogenous cluster that contains 5 His residues located within the N-terminal residues 10 to 14 of Cao1 (Fig. 1). cao1Δ cells transformed with the vector alone exhibited no significant CAO activity (Fig. 4B and C). cao1Δ cells were transformed with WT cao1+ or cao1+ with mutations in various His residues and treated with 10 μM CuSO4, 20 μM TTM, and 100 μM TTM. Figure 4B and C show results obtained for cells treated with 10 μM CuSO4. Identical results were obtained when cells were treated with 20 μM TTM or 100 μM TTM (data not shown). In these experiments, expression of WT Cao1 restored CAO activity (Fig. 4B and C). In contrast, expression of the mutant alleles [cao1(H458A), cao1(H460A), and cao1(H627A)] in the cao1Δ cells failed to restore active CAO protein (Fig. 4B and C). On the other hand, we observed that cao1Δ cells expressing the cao1(H621A) allele displayed a CAO activity at a level similar to that of the WT cao1+ allele expressed under the same conditions (Fig. 4B and C). Singularly, we noted that the Cao1 H456A mutant exhibited a much lower CAO activity, which is 5.9 times weaker than that of WT Cao1 (Fig. 4B and C). Taken together, these data reveal that the His458, His460, and His627 amino acid residues are necessary for Cao1 function, while replacement of the His456 amino acid residue with an Ala residue resulted only in a partially functional Cao1 protein.
FIG. 4.
Functional dissection of critical His residues in Cao1. (A) Schematic representation of the WT Cao1, Cao1 H456A, Cao1 H458A, Cao1 H460A, Cao1 H621A, and Cao1 H627A mutant proteins. The point mutations are marked with asterisks and an A (instead of the WT H residues). The black region indicates the location of the highly conserved consensus sequence NYEY, in which the first peptidyl tyrosine (Y) residue (black circle) serves as a putative precursor for TPQ formation. The active conformation of TPQ is presumably stabilized through interactions with Y307 and D321 in Cao1. The amino acid sequence numbers refer to the position relative to the first amino acid of the protein. (B) CAO activity was determined for cao1Δ cells that were transformed with a plasmid alone (−), WT cao1+, or the indicated mutant alleles of cao1 (top). Protein extracts were prepared from aliquots of cultures and then analyzed by immunoblotting using either anti-His5 or anti-PCNA (as an internal control) antibody. NS, nonspecific signal. (C) Total extracts from cells transformed as described for panel B were assayed for CAO activity using a spectrophotometric method with 4-aminoantipyrine and vanillic acid (23). The CAO activities reported represent the means from three separate determinations ± standard deviations.
Cellular localization of Cao1.
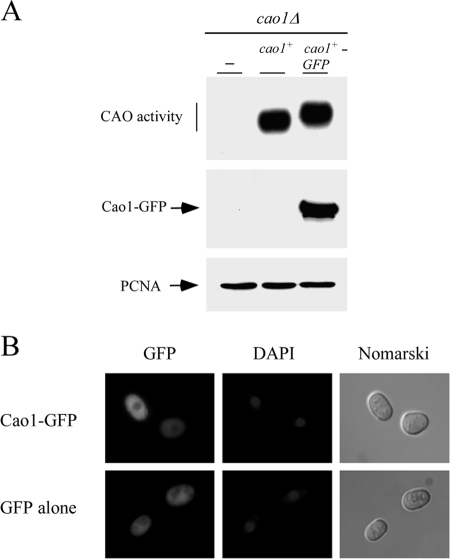
To understand how Cao1 functions in S. pombe copper metabolism, we examined its cellular localization in living cells by fusing GFP to the C terminus of Cao1. A plasmid expressing the Cao1-GFP fusion protein was transformed into a cao1Δ mutant strain. Transformants were analyzed to determine the ability of the Cao1-GFP fusion protein to catalyze the oxidative deamination of ethylamine compared to the ability of the WT strain by use of the chemiluminescence activity assay. The results in Fig. 5A show that the Cao1-GFP fusion protein, expressed from its own promoter, retained CAO activity comparable to that of the WT, untagged Cao1 protein. By use of direct fluorescence microscopy, Cao1-GFP exhibited uniform fluorescent staining throughout the cytosol. Costaining with DAPI indicated that Cao1-GFP was excluded from the nucleus (Fig. 5B, top). As a control, GFP alone produced from a recombinant construct showed a pattern of fluorescence throughout the cytoplasm and the nucleus (Fig. 5B, bottom). Thus, the absence of Cao1 from the nucleus strongly suggests that the protein is localized primarily in the cytoplasm when expressed in fission yeast.
FIG. 5.
Cytosolic localization of a functional Cao1-GFP fusion protein. (A) S. pombe cells carrying a disrupted cao1Δ allele were transformed with an empty plasmid (−) or a plasmid expressing cao1+ or cao1+-GFP and then tested for the ability to mediate the oxidative deamination of ethylamine using a chemiluminescence activity assay (top). Extracts used to detect CAO activity were also probed with anti-GFP (middle) or anti-PCNA (bottom) antibody. (B) Representative cao1Δ cells expressing Cao1-GFP and GFP alone (as a control) are shown. Cells were grown to exponential phase and then analyzed by direct fluorescence microscopy for GFP. DAPI staining visualized nuclear DNA, and Nomarski optics were used to examine cell morphology.
Active Cao1 in S. pombe requires Ctr4/5-mediated copper transport and the transcription factor Cuf1.
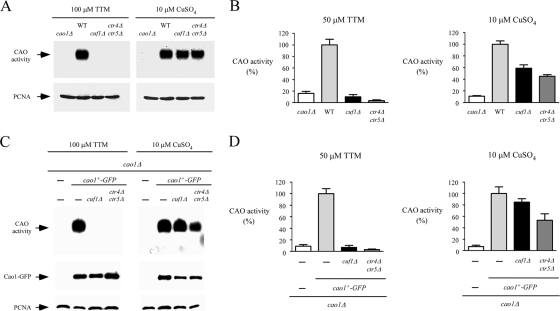
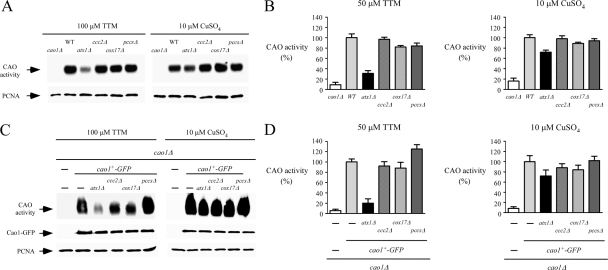
Cellular management of copper requires transport from the environment through the cellular membrane for delivery to copper-requiring enzymes. In S. pombe, ctr4+ and ctr5+ genes are known to encode two transmembrane proteins that form a tight complex localized at the cell surface (4, 55). Once assembled, this heteroprotein complex is required for high-affinity copper transport (55). The ctr4+ and ctr5+ genes are regulated by the copper-sensing transcription factor Cuf1 (2, 32). They are activated in response to copper deficiency and repressed under conditions of copper sufficiency. To ascertain the necessity of Cuf1 and the heteroprotein complex Ctr4/Ctr5 in supplying copper to Cao1, CAO activity was assayed with whole-cell extracts from WT, cuf1Δ, and ctr4Δ ctr5Δ cells. Under conditions of low copper supply, in the presence of 100 μM TTM in the growth medium, cuf1Δ or ctr4Δ ctr5Δ mutant cells exhibited no detectable CAO activity by use of an in-gel assay to detect enzymatic activity (Fig. 6A, left). In contrast, CAO activity was restored by supplementing the growth medium with 10 μM of exogenous copper (Fig. 6A, right). This is likely due to the accumulation of intracellular copper ions via a low-affinity copper uptake system, which bypasses the requirement for the S. pombe high-affinity Ctr4/Ctr5 copper transport system. To further confirm these results, we used a quantitative spectrophotometric method with 4-aminoantipyrine and vanillic acid (23) to determine whether deletions of cuf1+ and ctr4+ ctr5+ genes dramatically reduced CAO activity in the presence of 50 μM TTM (Fig. 6B, left). Consistent with the restoration of CAO activity by the addition of exogenous copper (10 μM CuSO4), we observed that copper-replete cuf1Δ and ctr4Δ ctr5Δ cells displayed an increase in CAO activity to 59 and 45% of the level observed for the WT strain, respectively (Fig. 6B, right). To ensure that the absence of CAO activity in the cuf1Δ and ctr4Δ ctr5Δ mutant strains grown under conditions of copper deprivation was not due to a defect in Cao1 expression, we integrated a functional cao1+-GFP fusion allele into S. pombe cao1Δ, cao1Δ cuf1Δ, and cao1Δ ctr4Δ ctr5Δ strains. As shown by two distinct CAO assays (Fig. 6C and D, left), the cao1Δ cuf1Δ and cao1Δ ctr4Δ ctr5Δ mutant strains failed to display significant measurable CAO activity when cells were grown in the presence of the copper chelator TTM (50 or 100 μM). In contrast, when a cao1Δ disruptant was transformed with the GFP epitope-tagged cao1+ allele, high CAO activity was restored (Fig. 6C and D). Likewise, CAO activity was restored by the addition of 10 μM CuSO4 to the growth medium (Fig. 6C and D, right). To ascertain that the Cao1-GFP fusion protein was expressed in the cao1Δ, cao1Δ cuf1Δ, and cao1Δ ctr4Δ ctr5Δ cells, total protein extracts from transformed cells were analyzed by immunoblotting (Fig. 6C, middle). These results demonstrated that Cao1-GFP was produced in the cao1Δ cuf1Δ and cao1Δ ctr4Δ ctr5Δ strains, indicating that the absence of CAO activity in these mutant strains was not due to the lack of Cao1-GFP expression. Collectively, these data indicate that under low-copper conditions, the production of active Cao1 in S. pombe requires the plasma membrane-associated high-affinity copper transporters Ctr4 and Ctr5, as well as the transcription factor Cuf1, which is necessary for the expression of the high-affinity copper uptake genes.
FIG. 6.
Cao1 activity requires expression of the Ctr4/Ctr5 copper transporting complex at the cell surface and the transcription factor Cuf1. (A) The WT and the isogenic cao1Δ, cuf1Δ, and ctr4Δ ctr5Δ mutant strains were incubated in the presence of TTM (100 μM) or CuSO4 (10 μM) and analyzed using an in-gel assay for CAO activity (top). Aliquots of total-extract preparations were assayed by immunoblotting using anti-PCNA antibody (bottom). (B) For a quantitative assay of CAO activity, hydrogen peroxide released from a reaction catalyzed by amine oxidase was determined with a spectrophotometric method using 4-aminoantipyrine and vanillic acid to generate a quinoneimine dye. Each sample was assayed in triplicate. (C) Isogenic strains harboring insertionally inactivated cao1Δ, cuf1Δ cao1Δ, or ctr4Δ ctr5Δ cao1Δ genes were transformed with the GFP-tagged cao1+ allele. After treatment with TTM (100 μM) or CuSO4 (10 μM), whole-cell extracts were prepared and analyzed using an in-gel assay for CAO activity (top). Aliquots of lysates were also analyzed by immunoblotting to verify the presence of Cao1-GFP (middle). Furthermore, all samples were subjected to immunoblotting with anti-PCNA antibody (bottom). As a control, an S. pombe strain bearing a cao1 deletion was transformed with an empty vector (−). (D) Extract preparations from strains described for panel C were analyzed using the spectrophotometric method described for panel B. The reported values of CAO activities are the means from three replicates ± standard deviations.
S. pombe Atx1-like protein is required for the synthesis of fully active Cao1.
Copper is an essential trace element that is also toxic due to its proclivity to engage in redox reactions that generate detrimental reactive oxygen species (21). Consequently, organisms have evolved with cellular components that function to acquire and distribute copper in a carefully controlled fashion. The discovery of copper chaperones that are involved in intracellular distribution of copper is a typical example of this fine control (19). For S. cerevisiae, three distinct copper chaperones, Atx1 (37), Ccs1 (16), and Cox17 (20), have been identified and shown to deliver copper to distinct pathways or organelles. Based on the S. pombe genome database and published data, it has been hypothesized or proven that the SPBC1709.10c (data not shown), pccs+ (SPAC22E12.04) (34), and SPBC26H8.14c genes encode chaperones that are orthologous to S. cerevisiae Atx1, Ccs1, and Cox17, respectively. Furthermore, S. pombe atx1Δ (SPBC1709.10cΔ) mutant cells were defective for iron acquisition (data not shown), presumably because of a lack of copper incorporation into the ferroxidase Fio1 (31), which would prevent Fio1-Fip1 high-affinity iron transport activity at the plasma membrane. This result strongly suggests that the product of the fission yeast gene SPBC1709.10c is a functional homolog of S. cerevisiae Atx1. To determine if one of the copper chaperones Atx1, Ccs1, and Cox17 was involved in delivering copper to Cao1, we generated mutants lacking each of the copper chaperone genes, as well as a mutant lacking a fourth gene, ccc2 (SPBC29A3.01), which is predicted to transport copper across intracellular membranes into the secretory pathway. The WT strain and the single mutants were tested for CAO activity. Strains were grown under both copper-limiting (50 or 100 μM TTM) and copper-replete (10 μM CuSO4) conditions. As shown in Fig. 7A and B (left), in the presence of TTM, atx1Δ cells exhibited a CAO activity much lower than those of WT cells and the other mutant cells. Using a spectrophotometric CAO assay, we determined that cells bearing an atx1 deletion displayed an activity ∼2.6 to 3.2 times weaker than those of the WT cells or cells that harbored a ccc2, cox17, or pccs deletion (Fig. 7B). As expected, the decrease in CAO activity observed with the atx1Δ mutant cells was largely restored by the addition of 10 μM CuSO4 to the growth medium (Fig. 7A and B, right). To ensure that the diminution of CAO activity in an atx1Δ mutant was not due to the lack of expression of the Cao1 protein, we generated cao1Δ atx1Δ, cao1Δ ccc2Δ, cao1Δ cox17Δ, and cao1Δ pccsΔ double mutants. Subsequently, we integrated a functional cao1+-GFP allele expressed under the control of the cao1+ promoter into the double disruption strains, as well as the cao1Δ single disruptant. The transformed strains were first grown in low-copper medium (50 or 100 μM TTM). Peroxidase-catalyzed chemiluminescence assays showed that the cao1Δ atx1Δ double mutant strain expressing cao1+-GFP appeared to have much less CAO activity than the other double mutants, even though the protein levels were comparable (Fig. 7C, left). The reduced level of CAO activity observed for cao1Δ atx1Δ cells was reversed by the addition of exogenous CuSO4 (10 μM) to the medium (Fig. 7C, right). These results were recapitulated by a second method that used a peroxidase-linked spectrophotometric CAO activity assay. As shown in Fig. 7D (left), cao1Δ atx1Δ double mutant cells expressing cao1+-GFP displayed ∼70 to 80% less CAO activity under copper-limiting conditions. As expected, the mutant phenotype was largely corrected by the addition of copper to the growth medium (Fig. 7D, right).
FIG. 7.
Production of fully active Cao1 requires functional Atx1. (A) Logarithmic-phase cultures of isogenic FY435 (WT), cao1Δ, atx1Δ, ccc2Δ, cox17Δ, and pccsΔ strains were treated in the presence of TTM (100 μM) or CuSO4 (10 μM) at 30°C. After 8 h of treatment, cell lysates were prepared from each culture and analyzed using an in-gel peroxidase-catalyzed chemiluminescence assay for CAO activity (top). As an internal control, aliquots of cell lysates were probed by immunoblotting using anti-PCNA antibody (bottom). (B) CAO activity was quantitated from the cell lysates described for panel A by use of a spectrophotometric method with 4-aminoantipyrine and vanillic acid. Error bars indicate the standard deviation of activities from samples analyzed in triplicate. (C) Isogenic S. pombe strains bearing a single deletion (cao1Δ) or a double deletion (atx1Δ cao1Δ, ccc2Δ cao1Δ, cox17Δ cao1Δ, or pccsΔ cao1Δ) were transformed with an integrative plasmid expressing a functional Cao1-GFP fusion protein. Whole-cell extracts from each transformant grown in the presence of TTM (100 μM) or CuSO4 (10 μM) were prepared and analyzed for CAO activity using an in-gel assay (top). Aliquots of total-extract preparations were examined by immunoblotting using either anti-GFP (middle) or anti-PCNA (bottom) antibody. As a control, an S. pombe strain bearing a cao1 deletion was transformed with an empty integrative vector (−). (D) Total-extract preparations from strains described for panel C were analyzed by a spectrophotometric method using 4-aminoantipyrine/vanillic acid. The values of CAO activities are the means from three replicates ± standard deviations.
Because the loss of function of the Atx1-like protein through gene inactivation resulted in less CAO activity, we reasoned that introduction of the WT atx1+ locus on an episomal plasmid should restore CAO activity to WT levels. A cDNA fragment encompassing the atx1+ gene was cloned into the pSP1 vector and transformed into two different atx1Δ mutant strains. While atx1Δ mutant strains carrying empty pSP1 exhibited low CAO activity under conditions of copper deprivation, the same strains expressing atx1+ from the pSP1 plasmid displayed elevated levels of CAO activity that were comparable to those found in cells expressing an endogenous atx1+ gene (Fig. 8). Taken together, these results reveal that optimal activity of the Cao1 protein requires a functional atx1+-like gene in fission yeast.
FIG. 8.
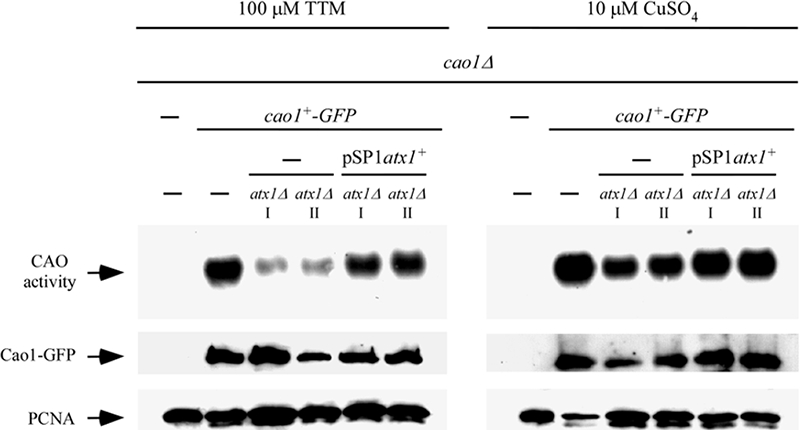
Loss of CAO activity in atx1Δ cells is restored by exogenous copper or a plasmid-borne copy of the WT atx1+ gene. An integrative plasmid expressing a functional GFP-tagged cao1+ allele was transformed into cao1Δ and cao1Δ atx1Δ cells. Disruption of the atx1+ gene decreased CAO activity levels. The CAO activity defect was corrected either by returning a WT copy of the atx1+ gene expressed from a plasmid or by adding CuSO4 (10 μM) to the growth medium. Roman numerals (I and II) indicate two separate cao1Δ atx1Δ strains in which cao1+-GFP was returned by integration.
Cao1 associates with the Atx1 chaperone.
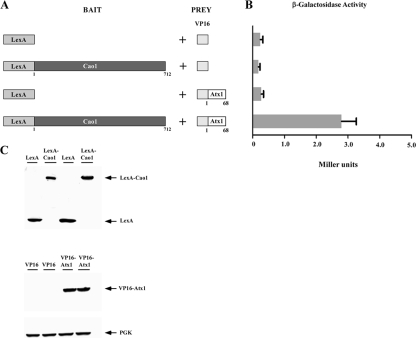
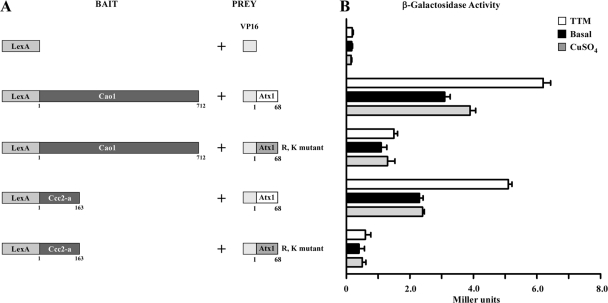
Given the results that Cao1 requires the presence of the Atx1 protein to be fully active, we tested the possibility that Cao1 physically interacts with Atx1. We carried out two-hybrid experiments using the entire 712-amino-acid coding region of Cao1 fused in frame to that of the DNA-binding domain of LexA as bait, whereas the coding region for the VP16 activation domain was fused to the 68-amino-acid coding region of Atx1 as prey (Fig. 9A). As shown in Fig. 9B, when cotransformed S. cerevisiae L40 cells were grown in low-copper synthetic minimal medium, a weak but reproducible interaction between LexA-Cao1 and VP16-Atx1 was observed, indicating a physical interaction between these proteins. The strength of the signal was low (2.84 Miller units) but significantly higher than those for negative controls (which consist of vectors only) (0.24 Miller units), the chimeric LexA-Cao1 molecule with the VP16 polypeptide alone (0.17 Miller units), and the DNA binding domain of LexA alone with the chimeric VP16-Atx1 molecule (0.27 Miller units). To ensure that the fusion proteins were expressed in the transformed cells, immunoblot analyses of protein extracts were carried out using anti-LexA and anti-VP16 antibodies (Fig. 9C). Although the VP16-Atx1 fusion protein used in this work was consistently detected by immunoblotting, we were unable to detect the VP16 polypeptide alone, perhaps owing to its low predicted molecular mass of ∼8 kDa. It has been proposed that copper chaperones are required under conditions where copper is not readily available for copper-dependent enzymes (44, 50). Furthermore, previous studies have reported that the N-terminal cytoplasmic region of S. cerevisiae Ccc2 interacts with Atx1 in a copper-dependent manner (44, 45, 50). It has also been observed that either very high or very low concentrations of copper result in a decrease in the interaction of Atx1 with Ccc2 (44, 45, 50). Given these observations, we tested whether high-copper treatment or the presence of the copper chelator TTM had an effect on the interaction of Atx1 with Cao1. As shown in Fig. 10, we detected an interaction between LexA-Cao1 and VP16-Atx1 in the presence of 1 mM TTM (6.20 Miller units) slightly stronger than that observed in the presence of exogenous copper (3.91 Miller units) or under basal conditions (3.10 Miller units). In a control experiment, coexpression of the N-terminal cytoplasmic region of S. pombe Ccc2 (amino acids 1 to 163) (designated Ccc2-a) fused to LexA with the VP16-Atx1 fusion protein produced weak but reproducible levels of β-galactosidase activity in the presence of TTM (5.12 Miller units). Analogous to the interaction between Cao1 and Atx1, untreated cultures (2.33 Miller units) or cultures grown in high copper concentrations (2.42 Miller units) exhibited a decreased physical interaction between the N-terminal region of Ccc2 and Atx1 (Fig. 10). To begin to characterize the mechanism whereby Atx1 interacts with Cao1, we examined the role of putative arginine and lysine patches represented by Arg-20, Arg-24, Lys-55, Lys-56, and Lys-59 of S. pombe Atx1, which are thought to be important for physical interaction with a partner copper-requiring enzyme. Replacing the five arginine and lysine residues with acidic glutamates resulted in an Atx1 molecule that was severely inhibited in its ability to interact with Cao1 [the Atx1(R,K) mutant] (Fig. 10). As a control, we monitored the physical interaction between the VP16-Atx1(R,K) mutant and LexA-Ccc2-a and found an ∼84% decrease in the activity of the reporter gene product compared with those of the unaltered VP16-Atx1 and LexA-Ccc2-a fusion proteins (Fig. 10). Using Western blot analysis, we observed equivalent expression levels of the LexA-Cao1, LexA-Ccc2-a, VP16-Atx1, and VP16-Atx1(R,K) mutant fusion proteins regardless of the presence of copper or a copper chelator in the growth media (data not shown). Taken together, these results strongly suggest that Atx1 can act as a copper carrier for a molecule other than Ccc2, participating in the delivery of copper to Cao1.
FIG. 9.
S. pombe Atx1 interacts with Cao1 by two-hybrid assay. (A) Schematic representation of the LexA DNA binding domain alone or fused upstream of and in frame to the full-length Cao1 coding region. The indicated bait molecule was coexpressed with the VP16 activation domain or the VP16-Atx1 fusion protein. (B) Interactions between the proteins were detected by liquid β-galactosidase assays. The values are the averages from triplicate determinations ± standard deviations. (C) Total-cell-extract preparations from aliquots of cultures used in the assays described for panel B were analyzed by immunoblotting with anti-LexA, anti-VP16, or anti-PGK (as an internal control) antibody.
FIG. 10.
The interaction between Cao1 and Atx1 is similar to that of Ccc2-a and Atx1 and is enhanced under low-copper conditions. (A) Schematic diagrams of the LexA DNA binding domain and fusions with Cao1 and the putative N-terminal cytosolic domain of the S. pombe Ccc2 polypeptide (Ccc2-a) are depicted at left. The VP16 and chimeric VP16-Atx1 and VP16-Atx1(R,K) molecules used as prey are shown at right. (B) Mid-logarithmic-phase cells cotransformed with the indicated plasmids were grown under basal or copper-deficient (1 mM TTM) conditions or with excess copper (100 μM CuSO4) for 5 h. Protein-protein interactions were detected by liquid β-galactosidase assays, and results are indicated in Miller units. Error bars indicate the standard deviations of samples analyzed in triplicate.
DISCUSSION
Global analysis of the fission yeast genome indicates the presence of two candidate copper-containing amine oxidase genes, cao1+ and cao2+, in S. pombe. Consistent with a role for Cao1 in the metabolism of primary amines as alternate sources of nitrogen to support growth, we have shown previously that the inability of S. cerevisiae to utilize ethylamine as the sole nitrogen source can be corrected by the heterologous expression of Cao1 from S. pombe in S. cerevisiae (33). Furthermore, we have observed that S. pombe cells harboring an inactivated cao1+ gene (cao1Δ) failed to grow in medium containing ethylamine as the sole source of nitrogen (Peter, Laliberté, and Labbé, unpublished). On the contrary, cao1Δ cells in which a WT cao1+ allele was reintegrated regained the capacity to utilize ethylamine as a nitrogen source (Peter, Laliberté, and Labbé, unpublished). Interestingly, it has been shown that the transcription of cao1+ is increased under nitrogen-limiting conditions (52). Given the fact that CAOs convert primary amines into aldehydes, also generating hydrogen peroxide and ammonia, one can envision that under low-nitrogen conditions, Cao1 may enable S. pombe cells to utilize primary amines as sources of nitrogen. The fact that S. cerevisiae does not express an endogenous protein homologous to Cao1 or to any of the members of the CAO family may reflect an important degree of divergence between the two yeasts with respect to environmental challenges they may have to cope with in their respective habitats. Alternatively, it may simply reflect a distinction between the biochemical pathways that are employed by these two yeasts to utilize carbon and nitrogen sources.
Although we detected CAO activity only in S. pombe cells expressing Cao1, we determined that both cao1+ and cao2+ genes were constitutively expressed under both copper-limiting and copper-replete conditions. Furthermore, we found that their expression was independent of cuf1+, a gene encoding the nutritional copper sensing trans inducer of the copper transport genes ctr4+, ctr5+, and ctr6+ in fission yeast (5). Interestingly, cao1+ and cao2+ mRNA steady-state levels were reportedly induced by hydrogen peroxide (11). Because one of the by-products of CAO catalysis is hydrogen peroxide, this observation may suggest a mechanism for the positive transcriptional autoregulation of these genes, thereby ensuring their continuous expression.
The role of Cao2 in S. pombe is unclear because of the lack of CAO activity and the phenotype of the cao2Δ mutant. The lack of activity may be due to the substitution of Tyr for Phe295 in the Cao2 protein. Based on the X-ray crystal structures of HPAO (36) and E. coli CAO (43), this conserved peptidyl Tyr residue is critical for the proper orientation of TPQ for optimal catalysis. Furthermore, we noticed that Cao2 contains a Cys354 → Ser amino acid modification. This amino acid substitution would prevent the formation of an important disulfide bridge that is highly conserved in all known eukaryotic CAOs (36), thereby potentially interfering with the correct topology of Cao2. Thus, these amino acid changes might render the Cao2 enzyme incapable of catalyzing ethylamine oxidation. On the other hand, perhaps we have not identified the appropriate amine substrate for Cao2. In this study, the primary amine substrates that we tested included monoamines (e.g., ethylamine), aromatic monoamines (e.g., benzylamine), and diamines (e.g., putrescine and 1,8-diaminooctane). None of these substrates were oxidized by Cao2 (Peter, Laliberté, and Labbé, unpublished). In contrast, Cao1 catalyzed the oxidative deamination of all of the primary amines that we have tested. The best substrates for Cao1 were ethylamine, putrescine, and 1,8-diaminooctane, while the aromatic monoamine benzylamine was oxidized less efficiently (Peter, Laliberté, and Labbé, unpublished). Collectively, our data reveal that CAO activity was found in S. pombe cells expressing cao1+ but not in cells expressing only cao2+. It is possible, however, that the expression of cao2+ may make a contribution under conditions not tested in our study. Further molecular and biochemical studies are needed to better define the role of Cao2.
Given the requirement of copper for TPQ formation in active CAOs, we sought to identify the cellular proteins involved in copper delivery to Cao1. The analysis of fission yeast cells in which both ctr4+ and ctr5+ genes have been inactivated demonstrated the loss of CAO activity under copper-limiting conditions. Likewise, the lack of Cuf1, the transcription factor that directs expression of the ctr4+ and ctr5+ genes, also results in the loss of CAO activity. As expected, the requirement of Ctr4/Ctr5 or Cuf1 for CAO activity was bypassed by the addition of exogenous copper at concentrations equivalent to or exceeding 10 μM to the growth medium. The lack of CAO activity in these mutant strains underscores the requirement of copper delivered by the high-affinity copper uptake machinery for the synthesis of an active CAO protein.
To further explore the pathway by which copper is distributed to Cao1, we examined the effects of deleting the atx1+, ccc2+, cox17+, and pccs+ genes, which are predicted (or proven, for the pccs+ gene [34]) to be involved in delivering intracellular copper, on Cao1 activity. Under conditions of copper starvation, cells lacking Atx1 exhibited a substantial decrease (∼70 to 80%) in CAO activity. In contrast, under the same conditions, the deletions of the other genes failed to show any significant inhibition of CAO activity. We also noticed that while Atx1 can be bypassed even under low-copper conditions (with ∼20 to 30% CAO activity), a defect in atx1 affects Cao1 activation under both low-copper (100 μM TTM) and high-copper (10 μM CuSO4) conditions. We believe that the latter observation (independent of the presence of TTM) further supports a specific function for Atx1 in the activation of Cao1. A previous study of global analysis of protein localization in the fission yeast S. pombe revealed that Atx1 is localized throughout the cytoplasm and the nucleus (40). Because both Cao1 and Atx1 can colocalize in the cytoplasm, we tested the ability of Cao1 to physically associate with Atx1. Using yeast two-hybrid analysis, we detected a weak but highly reproducible interaction between Cao1 and Atx1. How this might occur? X-ray crystal structures of S. cerevisiae Atx1 have revealed a region containing several Lys residues that may generate a positively charged patch on the surface of the protein (44, 48). Positively charged amino acids such as Lys and Arg have extra amino (NH2) groups on their side chains that may mimic NH2 groups of primary amines. The Lys-rich face of Atx1 has been shown to be required for physical association with Ccc2 (44). Analysis of X-ray crystal structures of CAOs, including HPAO from H. polymorpha (the closest ortholog of Cao1), indicates that they form homodimeric molecules, with an overall structure reminiscent of a mushroom cap (18, 36, 38, 43). The bulk of the homodimer has two identical active sites arranged along a molecular twofold symmetrical axis. Each active site is located in the interior of the protein and harbors one copper ion and one TPQ cofactor, both at a very close distance to each other. The active site pocket forms one open channel. Interestingly, the cavity of the channel leading to the active site contains residues that promote favorable binding for positively charged molecules, such as compounds containing amine groups. S. pombe Atx1 encodes a very small polypeptide of only 7.6 kDa. We observed that the corresponding Lys24, Lys28, Lys61, Lys62, and Lys65 residues, which generate a positively charged surface on the S. cerevisiae Atx1 protein, are either replaced by Arg (instead of Lys) for the first 2 residues or perfectly conserved for the last 3 residues in S. pombe Atx1. It is therefore possible that Atx1 docks on the cavity of the entry channel at the active site of Cao1, which is predicted to have the capacity to retain relatively large substrates, particularly those with multiple positively charged Lys residues with NH2 groups on their surface (27). In fact, it is known that peptides with exposed lysines can associate with and inhibit human Cao1/Vap1-dependent lymphocyte rolling activity (53). To support this hypothesis, using two-hybrid analysis (Fig. 10), we observed that changing the overall charge of the putative Atx1 basic region from positive to negative, as well as removing the predicted exposed NH2 groups from arginine and lysine residues, resulted in an Atx1 molecule that showed a significant decrease (∼70%) in physical association with Cao1. In addition to the Atx1 Lys-rich face, which is known to be important for the interaction between the chaperone and its target, the N terminus of Atx1 harbors a Met-X-Cys-X2-Cys copper-binding motif that is present in two copies at the N terminus of S. cerevisiae Ccc2. The current model proposes that the transfer of copper from Atx1 to Ccc2 involves a direct metal ion exchange between the conserved Met-X-Cys-X2-Cys motifs found in both proteins (25). Interestingly, S. pombe Cao1 has a Met-X-His-X2-Cys sequence (amino acids 151 to 156), which is predicted to be located at the cell surface near the entrance to the active site of the protein. This prediction was made based on the HPAO crystal structure and visualized using the 3D Molecule Viewer program (Invitrogen Corporation, San Jose, CA). A similar amino acid sequence is also present in HPAO (amino acids 148 to 153), except that the His residue is found immediately after the Met residue. In contrast, no such sequence exists in Cao2. Further characterization of Cao1 is required to ascertain whether the protein lacking the Met-X-His-X2-Cys sequence is unable to interact with Atx1 and/or is less active under conditions of copper deprivation. It is also possible that Atx1 delivers copper with the aid of an accessory protein, which would participate in the insertion of copper into Cao1; however, no such factor has yet been identified.
In S. pombe, the Atx1 metallochaperone represents an important source of copper for Cao1. However, Atx1 is not the only means by which Cao1 can be activated. As shown in this study, Cao1 can obtain copper in an Atx1-independent manner. The activity of Cao1 in yeast cells lacking Atx1 was detectable and highly reproducible and ranged from ∼20 to 30% that of the Atx1-dependent activity. We used a classical genetic approach to identify additional trans-acting proteins responsible for Atx1-independent activation of Cao1. Different copper-binding proteins were examined, including the putative copper chaperones encoded by pccs+ and cox17+ and the copper-transporting P-type ATPase Ccc2. However, deletions of these genes had no significant effect on Cao1 activity. Furthermore, the requirement for glutathione, which is known to play an important role in copper homeostasis, was also studied. We found that mutant cells defective in glutathione biogenesis showed no marked decrease in Cao1 activity (Peter, Laliberté, and Labbé, unpublished). Thus, the identity of the other molecule(s) that may be involved in delivering copper to Cao1 in fission yeast remains to be established. Because related proteins exist in pathogenic fungi, the mechanisms of copper loading on Cao1 homologs are indeed an interesting area for future study.
Acknowledgments
We are grateful to Maria M. O. Peña for critically reading the manuscript and providing valuable suggestions.
C.P. was a recipient of a studentship from the Faculty of Medicine and the Université de Sherbrooke. J.L. was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). This work was supported by Canadian Institutes of Health Research (CIHR) grant MOP-36450 to S.L. and by the Fondation de la Recherche sur les Maladies Infantiles du Québec. S.L. was supported by a senior scholarship from the Fonds de la Recherche en Santé du Québec (FRSQ).
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast, a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Beaudoin, J., and S. Labbé. 2001. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1-like recognition sequence. J. Biol. Chem. 27615472-15480. [DOI] [PubMed] [Google Scholar]
- 3.Beaudoin, J., and S. Labbé. 2006. Copper induces cytoplasmic retention of fission yeast transcription factor Cuf1. Eukaryot. Cell 5277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaudoin, J., J. Laliberté, and S. Labbé. 2006. Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology 152209-222. [DOI] [PubMed] [Google Scholar]
- 5.Beaudoin, J., A. Mercier, R. Langlois, and S. Labbé. 2003. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 27814565-14577. [DOI] [PubMed] [Google Scholar]
- 6.Bezanilla, M., S. L. Forsburg, and T. D. Pollard. 1997. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell 82693-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brazeau, B. J., B. J. Johnson, and C. M. Wilmot. 2004. Copper-containing amine oxidases. Biogenesis and catalysis; a structural perspective. Arch. Biochem. Biophys. 42822-31. [DOI] [PubMed] [Google Scholar]
- 8.Cai, D., and J. P. Klinman. 1994. Copper amine oxidase: heterologous expression, purification, and characterization of an active enzyme in Saccharomyces cerevisiae. Biochemistry 337647-7653. [DOI] [PubMed] [Google Scholar]
- 9.Cai, D., and J. P. Klinman. 1994. Evidence of a self-catalytic mechanism of 2,4,5-trihydroxyphenylalanine quinone biogenesis in yeast copper amine oxidase. J. Biol. Chem. 26932039-32042. [PubMed] [Google Scholar]
- 10.Cai, D., N. K. Williams, and J. P. Klinman. 1997. Effect of metal on 2,4,5-trihydroxyphenylalanine (topa) quinone biogenesis in the Hansenula polymorpha copper amine oxidase. J. Biol. Chem. 27219277-19281. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bähler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, Y. H., R. Matsuzaki, S. Suzuki, and K. Tanizawa. 1996. Role of conserved Asn-Tyr-Asp-Tyr sequence in bacterial copper/2,4, 5-trihydroxyphenylalanyl quinone-containing histamine oxidase. J. Biol. Chem. 27122598-22603. [DOI] [PubMed] [Google Scholar]
- 13.Cobine, P. A., F. Pierrel, M. L. Bestwick, and D. R. Winge. 2006. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 28136552-36559. [DOI] [PubMed] [Google Scholar]
- 14.Cobine, P. A., F. Pierrel, and D. R. Winge. 2006. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta 1763759-772. [DOI] [PubMed] [Google Scholar]
- 15.Cottarel, G., D. Beach, and U. Deuschle. 1993. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr. Genet. 23547-548. [DOI] [PubMed] [Google Scholar]
- 16.Culotta, V. C., L. W. Klomp, J. Strain, R. L. Casareno, B. Krems, and J. D. Gitlin. 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 27223469-23472. [DOI] [PubMed] [Google Scholar]
- 17.Culotta, V. C., M. Yang, and T. V. O'Halloran. 2006. Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta 1763747-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duff, A. P., A. E. Cohen, P. J. Ellis, J. A. Kuchar, D. B. Langley, E. M. Shepard, D. M. Dooley, H. C. Freeman, and J. M. Guss. 2003. The crystal structure of Pichia pastoris lysyl oxidase. Biochemistry 4215148-15157. [DOI] [PubMed] [Google Scholar]
- 19.Field, L. S., E. Luk, and V. C. Culotta. 2002. Copper chaperones: personal escorts for metal ions. J. Bioenerg. Biomembr. 34373-379. [DOI] [PubMed] [Google Scholar]
- 20.Glerum, D. M., A. Shtanko, and A. Tzagoloff. 1996. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 27114504-14509. [DOI] [PubMed] [Google Scholar]
- 21.Halliwell, B., and J. M. Gutteridge. 1992. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett. 307108-112. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 23.Holt, A., and M. M. Palcic. 2006. A peroxidase-coupled continuous absorbance plate-reader assay for flavin monoamine oxidases, copper-containing amine oxidases and related enzymes. Nat. Protoc. 12498-2505. [DOI] [PubMed] [Google Scholar]
- 24.Horng, Y. C., P. A. Cobine, A. B. Maxfield, H. S. Carr, and D. R. Winge. 2004. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 27935334-35340. [DOI] [PubMed] [Google Scholar]
- 25.Huffman, D. L., and T. V. O'Halloran. 2000. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J. Biol. Chem. 27518611-18614. [DOI] [PubMed] [Google Scholar]
- 26.Iwaki, T., and K. Takegawa. 2004. A set of loxP marker cassettes for Cre-mediated multiple gene disruption in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68545-550. [DOI] [PubMed] [Google Scholar]
- 27.Jakobsson, E., J. Nilsson, D. Ogg, and G. J. Kleywegt. 2005. Structure of human semicarbazide-sensitive amine oxidase/vascular adhesion protein-1. Acta Crystallogr. D 611550-1562. [DOI] [PubMed] [Google Scholar]
- 28.Janes, S. M., D. Mu, D. Wemmer, A. J. Smith, S. Kaur, D. Maltby, A. L. Burlingame, and J. P. Klinman. 1990. A new redox cofactor in eukaryotic enzymes: 6-hydroxydopa at the active site of bovine serum amine oxidase. Science 248981-987. [DOI] [PubMed] [Google Scholar]
- 29.Kim, B. E., T. Nevitt, and D. J. Thiele. 2008. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4176-185. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J., and J. P. Hirsch. 1998. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics 149795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labbé, S., J. Beaudoin, D. R. Bellemare, and B. Pelletier. 2002. Regulatory responses to copper ions in fungi, p. 571-587. In E. J. Massaro (ed.), Handbook of copper pharmacology and toxicology. Humana Press, Totowa, NJ.
- 32.Labbé, S., M. M. O. Peña, A. R. Fernandes, and D. J. Thiele. 1999. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 27436252-36260. [DOI] [PubMed] [Google Scholar]
- 33.Laliberté, J., and S. Labbé. 2006. Mechanisms of copper loading on the Schizosaccharomyces pombe copper amine oxidase 1 expressed in Saccharomyces cerevisiae. Microbiology 1522819-2830. [DOI] [PubMed] [Google Scholar]
- 34.Laliberté, J., L. J. Whitson, J. Beaudoin, S. P. Holloway, P. J. Hart, and S. Labbé. 2004. The Schizosaccharomyces pombe Pccs protein functions in both copper trafficking and metal detoxification pathways. J. Biol. Chem. 27928744-28755. [DOI] [PubMed] [Google Scholar]
- 35.Large, P. J. 1986. Degradation of organic nitrogen compounds by yeasts. Yeast 21-34. [Google Scholar]
- 36.Li, R., J. P. Klinman, and F. S. Mathews. 1998. Copper amine oxidase from Hansenula polymorpha: the crystal structure determined at 2.4 A resolution reveals the active conformation. Structure 6293-307. [DOI] [PubMed] [Google Scholar]
- 37.Lin, S. J., R. A. Pufahl, A. Dancis, T. V. O'Halloran, and V. C. Culotta. 1997. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 2729215-9220. [PubMed] [Google Scholar]
- 38.Lunelli, M., M. L. Di Paolo, M. Biadene, V. Calderone, R. Battistutta, M. Scarpa, A. Rigo, and G. Zanotti. 2005. Crystal structure of amine oxidase from bovine serum. J. Mol. Biol. 346991-1004. [DOI] [PubMed] [Google Scholar]
- 39.MacPherson, I. S., and M. E. Murphy. 2007. Type-2 copper-containing enzymes. Cell. Mol. Life Sci. 642887-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, S. Horinouchi, and M. Yoshida. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24841-847. [DOI] [PubMed] [Google Scholar]
- 41.Mercier, A., S. Watt, J. Bahler, and S. Labbé. 2008. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7493-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Sullivan, J., M. Unzeta, J. Healy, M. I. O'Sullivan, G. Davey, and K. F. Tipton. 2004. Semicarbazide-sensitive amine oxidases: enzymes with quite a lot to do. Neurotoxicology 25303-315. [DOI] [PubMed] [Google Scholar]
- 43.Parsons, M. R., M. A. Convery, C. M. Wilmot, K. D. Yadav, V. Blakeley, A. S. Corner, S. E. Phillips, M. J. McPherson, and P. F. Knowles. 1995. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure 31171-1184. [DOI] [PubMed] [Google Scholar]
- 44.Portnoy, M. E., A. C. Rosenzweig, T. Rae, D. L. Huffman, T. V. O'Halloran, and V. C. Culotta. 1999. Structure-function analyses of the Atx1 metallochaperone. J. Biol. Chem. 27415041-15045. [DOI] [PubMed] [Google Scholar]
- 45.Pufahl, R. A., C. P. Singer, K. L. Peariso, S. J. Lin, P. J. Schmidt, C. J. Fahrni, V. C. Culotta, J. E. Penner-Hahn, and T. V. O'Halloran. 1997. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278853-856. [DOI] [PubMed] [Google Scholar]
- 46.Puig, S., and D. J. Thiele. 2002. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6171-180. [DOI] [PubMed] [Google Scholar]
- 47.Rae, T. D., P. J. Schmidt, R. A. Pufahl, V. C. Culotta, and T. V. O'Halloran. 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284805-808. [DOI] [PubMed] [Google Scholar]
- 48.Rosenzweig, A. C., D. L. Huffman, M. Y. Hou, A. K. Wernimont, R. A. Pufahl, and T. V. O'Halloran. 1999. Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution. Structure 7605-617. [DOI] [PubMed] [Google Scholar]
- 49.Samuels, N. M., and J. P. Klinman. 2005. 2,4,5-Trihydroxyphenylalanine quinone biogenesis in the copper amine oxidase from Hansenula polymorpha with the alternate metal nickel. Biochemistry 4414308-14317. [DOI] [PubMed] [Google Scholar]
- 50.van Dongen, E. M., L. W. Klomp, and M. Merkx. 2004. Copper-dependent protein-protein interactions studied by yeast two-hybrid analysis. Biochem. Biophys. Res. Commun. 323789-795. [DOI] [PubMed] [Google Scholar]
- 51.Vojtek, A. B., J. A. Cooper, and S. M. Hollenberg. 1997. Searching for interacting proteins with the two-hybrid system II, p. 29-42. In P. Bartel and S. Fields (ed.), The yeast two-hybrid system: a practical approach. Oxford University Press, New York, NY.
- 52.Wilhelm, B. T., S. Marguerat, S. Watt, F. Schubert, V. Wood, I. Goodhead, C. J. Penkett, J. Rogers, and J. Bahler. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 4531239-1243. [DOI] [PubMed] [Google Scholar]
- 52a.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schäfer, S. Müller-Auer, C. Gabel, M. Fuchs, A. Düsterhöft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dréano, S. Gloux, V. Lelaure, S. Mottier, F. Galibert, S. J. Aves, Z. Xiang, C. Hunt, K. Moore, S. M. Hurst, M. Lucas, M. Rochet, C. Gaillardin, V. A. Tallada, A. Garzon, G. Thode, R. R. Daga, L. Cruzado, J. Jimenez, M. Sánchez, F. del Rey, J. Benito, A. Domínguez, J. L. Revuelta, S. Moreno, J. Armstrong, S. L. Forsburg, L. Cerutti, T. Lowe, W. R. McCombie, I. Paulsen, J. Potashkin, G. V. Shpakovski, D. Ussery, B. G. Barrell, and P. Nurse. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415871-880. [DOI] [PubMed] [Google Scholar]
- 53.Yegutkin, G. G., T. Salminen, K. Koskinen, C. Kurtis, M. J. McPherson, S. Jalkanen, and M. Salmi. 2004. A peptide inhibitor of vascular adhesion protein-1 (VAP-1) blocks leukocyte-endothelium interactions under shear stress. Eur. J. Immunol. 342276-2285. [DOI] [PubMed] [Google Scholar]
- 54.Yu, P. H., S. Wright, E. H. Fan, Z. R. Lun, and D. Gubisne-Harberle. 2003. Physiological and pathological implications of semicarbazide-sensitive amine oxidase. Biochim. Biophys. Acta 1647193-199. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, H., and D. J. Thiele. 2001. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 27620529-20535. [DOI] [PubMed] [Google Scholar]
- 56.Znaidi, S., B. Pelletier, Y. Mukai, and S. Labbé. 2004. The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J. Biol. Chem. 2799462-9474. [DOI] [PubMed] [Google Scholar]