Abstract
The hyphae of filamentous fungi are compartmentalized by septa that have a central pore. The fungal septa and septum-associated structures play an important role in maintaining cellular and intrahyphal homeostasis. The dolipore septa in the higher Basidiomycota (i.e., Agaricomycotina) are associated with septal pore caps. Although the ultrastructure of the septal pore caps has been studied extensively, neither the biochemical composition nor the function of these organelles is known. Here, we report the identification of the glycoprotein SPC18 that was found in the septal pore cap-enriched fraction of the basidiomycetous fungus Rhizoctonia solani. Based on its N-terminal sequence, the SPC18 gene was isolated. SPC18 encodes a protein of 158 amino acid residues, which contains a hydrophobic signal peptide for targeting to the endoplasmic reticulum and has an N-glycosylation motif. Immunolocalization showed that SPC18 is present in the septal pore caps. Surprisingly, we also observed SPC18 being localized in some plugs. The data reported here strongly support the hypothesis that septal pore caps are derived from endoplasmic reticulum and are involved in dolipore plugging and, thus, contribute to hyphal homeostasis in basidiomycetous fungi.
Compartmentalization of fungal hyphae dates back to the origin of the Dikarya lineages (i.e., Ascomycota and Basidiomycota), which occurred approximately 500 to 600 million years ago (5, 22, 23). Septa play an important role in the biology of filamentous ascomycetous and basidiomycetous fungi, as they separate the fungal hyphae into functional cellular compartments and give the hyphal filaments rigidity. In addition, they are involved in mycelial differentiation (19). The regularly formed septa have a central pore through which protoplasm streams continuously and organelles like mitochondria can pass (35). Consequently, mycelia act as functional coenocytic systems and need some mechanism to restrict the effects of cellular damage within the system. Therefore, a damaged hyphal compartment needs to be isolated from the rest of the mycelial body to prevent cell lysis. In the filamentous Ascomycota group (Pezizomycotina), Woronin bodies, which are closely associated with the septum, seal the septal pores upon damage (24, 32, 56). In Neurospora crassa, it was shown that Woronin bodies confine the peroxisomal HEX-1 protein (24). Homologues of this protein have been found in other Pezizomycotina species like Aspergillus nidulans, Aspergillus oryzae, Botrytis cinerea, Trichoderma reesei, and Magnaporthe grisea (13, 24, 26, 44, 49), but not in ascomycetous yeasts (Saccharomycotina) nor in basidiomycetous fungi (24, 49).
Within the three main lineages of the phylum Basidiomycota, i.e., the Pucciniomycotina (equivalent to Urediniomycetes [47]), Ustilaginomycotina (equivalent to Ustilaginomycetes [47]), and Agaricomycotina (equivalent to Hymenomycetes [47]), the septum morphology and associated structures clearly differ. Members of the Pucciniomycotina and the Ustilaginomycotina have hyphal septa with a central pore. The septum is tapered toward the pore, like the septa in ascomycetous fungi, but lacks Woronin bodies and elaborate septal pore caps (SPCs) (3, 48). The Agaricomycotina fungi, on the other hand, are characterized by septa that are flared toward the pore, forming a barrel-shaped structure, the dolipore, and are associated with SPCs (9, 17, 21, 35, 37, 39). The endoplasmic reticulum (ER) is connected at the base of the SPC, thus suggesting that the SPC is a subdomain of the ER (9, 18, 36). The ultrastructure of SPCs has been studied extensively, and based on these observations, several functions of the SPC have been proposed. SPCs may act as a sieve to discriminate between organelles that may pass through the septal pore (55), they may guide cytoplasmic streams toward the pore (41), and they may function in protoplasmic streaming to protect the dolipore region in such way that protoplasmic streaming is not disturbed by organelles (10). Alternatively, they may be involved in dolipore plugging (1, 31, 38, 50).
In this study, we report for the first time the identification of an SPC protein. The glycoprotein SPC18 was found in the SPC-enriched fraction of the filamentous basidiomycetous fungus Rhizoctonia solani. We show that SPC18 is an ER protein that is located in both the SPCs and the pore-plugging material. The presented data clearly support the hypothesis that SPCs are part of the ER and are involved in dolipore plugging in hyphae of the higher Basidiomycota fungi.
MATERIALS AND METHODS
Preparation of an SPC-enriched fraction.
SPCs were enriched from R. solani (CBS 346.84) by isopycnic centrifugation, as described by van Driel et al. (53). R. solani was grown for 3 days in complete medium (20 g glucose, 2 g peptone L37 [Oxoid, Hampshire, United Kingdom], 2 g yeast extract [Difco, Detroit, MI], 0.5 g MgSO4·7H2O, 0.46 g KH2PO4, 1.0 g K2HPO4 per liter) with agitation at 175 rpm at 25°C. Mycelium was harvested and subsequently disrupted by using a French press (American Instrument Company, Silver Spring, MD). The cell extract was subjected to isopycnic centrifugation on a density gradient of 30 to 50 to 70% (wt/vol) sucrose. The SPC fraction on top of the 70% sucrose layer was treated with 2% (wt/vol) Triton X-100 (GE Healthcare, Uppsala, Sweden), followed by centrifugation at 5000 × g for 1 h. SPCs were enriched in the resulting pellet.
Fluorescence microscopy.
ER was stained with ER-Tracker (Invitrogen, Breda, The Netherlands) or brefeldin A conjugated to BODIPY 558/568 (BODIPY-BFA) (Invitrogen) according to the manufacturer's instructions. Mycelium samples were mounted on glass slides for light microscopy using a Zeiss Axioskop microscope (Carl Zeiss AG, Oberkochen, Germany). The filter set BP365, FT395, and LP397 was used for ER-Tracker, whereas filter set BP510-560, FT580, and LP590 was used for BODIPY-BFA. Broken mycelial fragments were stained with wheat germ agglutinin (WGA) conjugated to Alexa 488 (1:40 dilution; Invitrogen). Confocal laser scanning microscopy was performed with a Leica TCS NT microscope (Leica Microsystems, Wetzlar, Germany) using an excitation wavelength of 488 nm.
Protein analysis.
Protein content was determined using the bicinchonic acid method as provided by the supplier (Pierce, Rockford, IL), using bovine serum albumin (BSA) as a standard. Protein samples were concentrated by the addition of 9 volumes of ice-cold acetone. After a 30-min incubation on ice, samples were centrifuged for 10 min at 14,000 rpm at 4°C. The pellet was air dried and subsequently dissolved in sample buffer (Pierce) containing dithiothreitol with a final concentration of 50 mM. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by the method of Laemmli (30) in a 10 to 20% gradient Tris-HCl gel (Bio-Rad Laboratories, Hercules, CA) in 25 mM Tris, 192 mM glycine, and 0.1% (wt/vol) SDS (pH 8.3). Proteins were stained with Coomassie R350 (GE Healthcare).
Deglycosylation of the protein with the EndoH enzyme (New England Biolabs, Ipswich, MA) was performed according to the manufacturer's instructions.
For N-terminal sequencing, proteins were subjected to SDS-PAGE and electroblotted onto an Immobilon-P transfer membrane (Millipore, Billerica, MA). After being stained with Coomassie R350, the protein of interest was cut out and subjected to N-terminal sequencing using an Applied Biosystems 476A sequencer (Applied Biosystems, Foster City, CA).
Isolation of the full-length SPC18 cDNA and sequence analysis.
A 3-day-old submerged culture of R. solani was snap-frozen in liquid nitrogen and subsequently ground to a fine powder in a mortar. Total RNA was isolated using TRIzol reagent (Invitrogen, Paisley, United Kingdom). The forward degenerate primer SPC18#02 (Table 1) was designed according to the N terminus of mature SPC18 and adjusted for codon usage of R. solani. Codon usage was determined by analyzing the nucleotides of the coding sequences of the laccase, gpd, ABC transporter (partial), aromatic polypeptide, and ste3-like (partial) genes of R. solani (coding sequence of Gly, GGN → GGH; Ile, ATH → ATY; and Val, GTN → GTB; where N = A, T, C, or G; H = A, C, or T; Y = C or T; and B = C, G, or T). cDNA was made by using ImProm-II reverse transcriptase (Promega, Madison, WI), recombinant RNAsin RNase inhibitor (Promega), the degenerate primer SPC18#02, and the oligo(dT) primer (reverse transcription-PCR [RT-PCR] was performed for 5 min at 25°C, 60 min at 42°C, and 15 min at 70°C; PCR amplification was performed for 4 min at 94°C, followed by 35 cycles of 20 s at 94°C, 20 s at 47°C, and 2 min at 72°C, followed by 10 min at 72°C). To obtain the full SPC18 cDNA sequence, 5′ rapid amplification of cDNA ends (RACE) was performed with a 5′/3′ RACE kit (Roche, Basel, Switzerland) using the primers SPC18_SP1, SPC18_SP2, and SPC18_SP3 (Table 1) according to the manufacturer's instructions. The genomic sequence of SPC18 was obtained by PCR with the primers SPC18-fw1 and SPC18-rv1 (Table 1) using genomic DNA as a template (for 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 54°C, and 2 min at 72°C, followed by 7 min at 72°C). PCR products were cloned into the pGEM-T vector (Promega) and sequenced using BigDye version 3.1 chemistry (Applied Biosystems) and analyzed on an ABI 730XL DNA analyzer (Applied Biosystems) according to the manufacturer's instructions. Hydropathy analysis of the protein was done according to Kyte and Doolittle (29), using a window size of 11 amino acids (29) using ProtScale (16). ProtParam (16) was used to calculate molecular weights, SignalP version 3.0 (4) was used to predict signal sequence, and ScanProsite (15) was used to predict N-glycosylation sites. Similarity searches were done with BLAST software against sequences in GenBank and Swiss-Prot (http://www.ncbi.nlm.nih.gov/BLAST) and in fungal genome databases of the Broad Institute (http://www.broad.mit.edu/annotation/fgi/), the DOE Joint Genome Institute (http://www.jgi.doe.gov/), and the J. Craig Venter Institute (http://www.tigr.org/tdb/fungal/). DNA hybridization (Southern) was done according to Schuren et al. (43). Genomic DNA of R. solani was digested with HindIII and hybridized with the probe that was derived by PCR with the SPC18-fw1 and SPC18-rv1 primers (Table 1), using genomic DNA as a template.
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| SPC18#2 | GCNATGGGHGAYGTBATYGT |
| Oligo(dT) | TTAAT22(A G C) |
| SPC18_SP1 | GGTGGTCGGTGATAATGAGG |
| SPC18_SP2 | GTCGGAAGGCTTGTTGATGT |
| SPC18_SP3 | GAACAACGTAGCCCTGAGGA |
| RSFWTotalSPC18 | TCAGCATATGATCTTCCCCGTCGCC |
| RSRVSPC18 | GTTAAAGCTTAGAGTTCCTTGAAGTCCTTG |
| SPC18-fw1 | AGCCTCCTCAGGGCTACG |
| SPC18-rv1 | GGATATGCAAAAAGGCAAGC |
Overexpression of SPC18 in Escherichia coli.
The coding sequence of SPC18 was amplified by PCR with the RSFWTotalSPC18 and RSRVSPC18 primers (Table 1), using cDNA as a template (for 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by 7 min at 72°C). The cDNA fragment was cloned into pET11a (Promega). The recombinant SPC18 (recSPC18) protein was expressed in E. coli BL21 by isopropyl-β-d-thiogalactopyranoside induction. Inclusion bodies were isolated by sonication of the cells in ice-cold 10 mM Tris-HCl, 3 mM EDTA, and 1× protease inhibitor (Roche), followed by centrifugation at 4,000 rpm for 10 min. Finally, inclusion bodies were purified by centrifugation at 75,000 rpm for 1 h using a Beckman TL-100 machine and were dissolved in 20 mM Tris-HCl, 100 mM glycine, and 8 M urea.
Preparation of antibodies.
Polyclonal antibodies were raised in rabbits, using the mature SPC18 (anti-SPC18) protein or the recSPC18 (anti-recSPC18) protein that had been cut out of an SDS-polyacrylamide gel (Eurogentec, Seraing, Belgium). Immunoglobulin G was purified from the serum by using a Melon gel immunoglobulin G purification system (Pierce) according the manufacturer's instructions.
To analyze the specificity of the recSPC18 antibodies, we performed preabsorption control experiments. Protran nitrocellulose membrane strips (Whatman, Maidstone, United Kingdom) were bound with 12 μg, 60 μg, and 300 μg of recSPC18 protein. Then, the strips were washed three times for 10 min each with 0.4% Marvel milk powder, 0.1% Tween 20 in phosphate-buffered saline (PBS), followed by three washes with PBS for 10 min each. After being washed, the strips were blocked with 10% calf serum for 10 min and twice for 5 min in 1% BSA-c in PBS. Thereafter, the strips were incubated with anti-recSPC18 antibodies (3 mg/ml) diluted 1:10 in 1% BSA-c in PBS overnight at 4°C. The serum that was preabsorbed with the recSPC18 protein was used to label Lowicryl HM20 sections (as described above). Labeling of preabsorbed serum was compared with the labeling of serum that was not preabsorbed. The antigen-antibody complexes were visualized by incubation with protein A (1:80) conjugated with 10 nm gold. Sections were not contrasted with uranyl acetate.
Immunoblot analysis.
After SDS-PAGE, proteins were electroblotted onto an Immobilon-P transfer membrane. After being blocked with 3% TopBlock (Fluka, Buchs, Switzerland), the membrane was incubated with anti-SPC18 (1:250) or anti-recSPC18 (1:500) antibody. This step was followed by incubation with goat anti-rabbit antibodies conjugated with alkaline phosphatase (1:15,000) (Sigma-Aldrich). These antibodies were detected by using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium as substrate.
Chemical fixation and embedding in Spurr's resin.
Samples were fixed overnight at 4°C in 1.5% glutaraldehyde (Agar Scientific Ltd., Essex, United Kingdom) buffered with 20 mM HEPES (pH 6.8). This step was followed by postfixation in 1% (wt/vol) aqueous osmium tetroxide (EMS, Hatfield, PA) for 1 h at room temperature. Samples were gradually dehydrated in a series of ascending concentrations of acetone, followed by a gradual infiltration in Spurr's resin (45). The resin was polymerized at 65°C for 48 h in BEEM capsules (EMS).
High-pressure freezing, freeze substitution, and Lowicryl HM20 embedding.
Mycelia were high-pressure frozen and freeze-substituted (36). Sections of about 90 nm and 350 nm were contrasted with 4% (wt/vol) aqueous uranyl acetate (Merck) for 10 min and with 0.4% (wt/vol) aqueous lead citrate (Merck) for 2 min (54). For immunogold labeling, mycelia were freeze-substituted with 0.1% (wt/vol) uranyl acetate and 0.01% (wt/vol) osmium tetroxide in acetone. SPC-enriched fractions were high-pressure frozen (53) and freeze-substituted with 0.1% (wt/vol) uranyl acetate and 0.01% (wt/vol) osmium tetroxide in acetone. Thereafter, samples were low-temperature embedded in Lowicryl HM20 (36).
Immunogold labeling.
Lowicryl HM20 sections were incubated overnight with anti-SPC18 (1:30) or anti-recSPC18 (1:60) antibody. The antigen-antibody complexes were visualized by incubation with secondary goat-anti-rabbit antibodies (1:10) conjugated with 10 nm of gold particles (Aurion, Wageningen, The Netherlands) for 1 h at room temperature. Following immunolabeling, sections were contrasted with 4% (wt/vol) aqueous uranyl acetate for 2 min.
Transmission electron microscopy.
Sections were viewed with a TECNAI 10 model (FEI Company, Eindhoven, The Netherlands) transmission electron microscope at an acceleration voltage of 100 kV.
RESULTS
SPCs of R. solani.
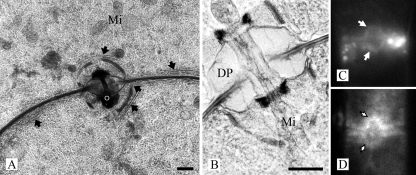
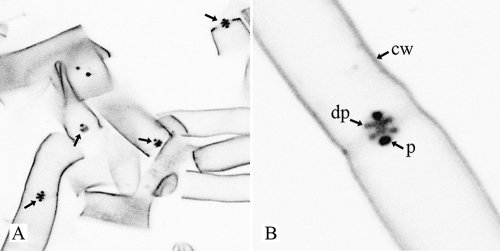
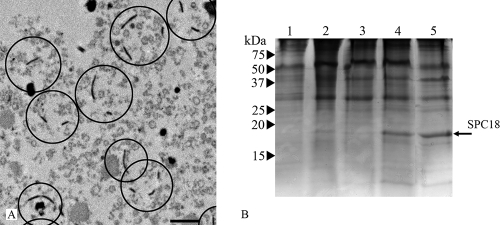
The dolipore septa of R. solani are associated with perforate SPCs of about 1.6 to 2.0 μm in diameter (Fig. 1A and B). The dolipore channel may be plugged with electron-dense occluding material (Fig. 1A), thereby blocking protoplasmic streaming. When they are open, the SPC and the dolipore channel allow passage of mitochondria (Fig. 1B). ER parallel to the septum is connected to the SPC base (Fig. 1A). In addition, the ER-specific fluorochromes ER-Tracker and BODIPY-BFA stained both the ER and the SPCs (Fig. 1C and D). The hyphal fragments obtained after the French-press treatment, which is the first step of the SPC enrichment procedure, were analyzed by staining with Alexa 488 conjugated to WGA, which labels the cell walls, the dolipore, and the SPCs (52). It was shown that three treatments of 500 lb/in2 each were sufficient to break the mycelia into small hyphal fragments and to release the mycelial content. Furthermore, fluorescence microscopy showed that cell walls, dolipore, and plug material were highly fluorescent, whereas the septal plate showed hardly any fluorescence (Fig. 2). SPCs from R. solani were enriched after the combined use of the French press, isopycnic centrifugation, and Triton X-100 (Fig. 3A) (53).
FIG. 1.
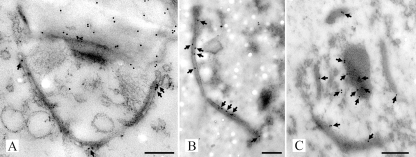
SPCs in R. solani. (A and B) Transmission electron micrographs of a high-pressure frozen, freeze-substituted, and Lowicryl HM20-embedded R. solani hyphae showing dolipore septa associated with perforate SPCs. ER (arrows) is connected with the SPC. The dolipore channel is plugged with electron-dense occluding material (o) (panel A) or can be open, thereby allowing passage of mitochondria (Mi) (panel B). DP, dolipore swelling. The dolipore channel is surrounded by a ring-like structure of electron-dense material (panel B). Fluorescence micrographs show hyphae stained with ER-Tracker (C) and BODIPY-BFA (D). In both cases fluorescence was localized to the SPCs. Bars, 500 nm (A) and 200 nm (B).
FIG. 2.
Confocal microscopy image (inverted) shows R. solani hyphal fragments stained with WGA conjugated to Alexa 488. Cell walls (cw), dolipore swellings (dp), and plugs (p) are labeled, whereas the septal plate hardly shows any fluorescence. Plugs are more intensely labeled than dolipore swellings and cell walls. (A) Overview; (B) detail image.
FIG. 3.
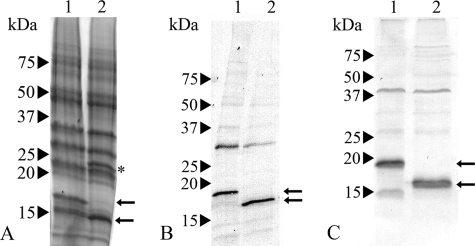
SPC enrichment. (A) Transmission electron micrograph of chemically fixed SPC-enriched fraction with isolated SPCs encircled. Bar, 1 μm. (B) SDS-PAGE analysis of the subcellular fractions of R. solani obtained after isopycnic centrifugation. Lanes 1, fraction on top of 30% sucrose layer; 2, fraction on top of 50% sucrose layer; 3, fraction 50% sucrose layer; 4, fraction on top of 70% sucrose layer (SPC-containing fraction); 5, SPC-enriched fraction (SPC-containing fraction treated with 2% Triton X-100). The arrow indicates the position of SPC18 protein. Numbers at left are molecular masses.
Molecular cloning and sequence analysis of SPC18.
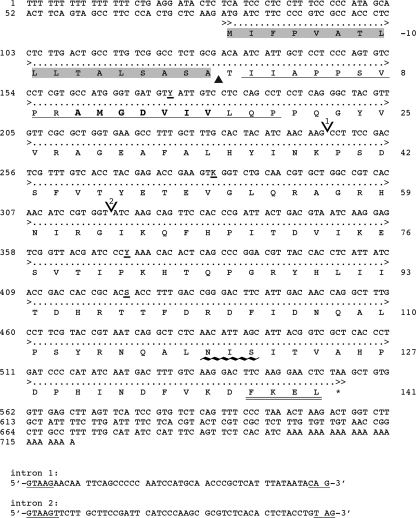
A protein with an apparent molecular mass of about 18 kDa was enriched in the SPC-enriched fraction (Fig. 3) and was named septal pore cap 18 (SPC18) protein. The N-terminal amino acid sequence of the SPC18 protein was determined to be RIIAPPSVPRAMGDVIVLQP, and, after we performed RT-PCR and 5′ RACE PCR, we obtained the complete cDNA sequence (Fig. 4). The open reading frame consisted of 477 bp encoding a protein of 158 amino acids, with a predicted molecular mass of 17,664 Da. Homologous genes or proteins were not found in the sequence databases or in any of the sequenced fungal genomes. A comparison of the cDNA and the genomic sequence revealed two short introns of 51 bp and 52 bp (Fig. 4), with 5′ and 3′ splice sites typical of filamentous fungi (2, 25). Southern analysis showed one band after hybridization (result not shown). The genomic sequence, however, showed nucleotide variation at three sites, suggesting different alleles of SPC18 in the R. solani genome. The deduced SPC18 protein sequence encompasses a putative hydrophobic N-terminal signal sequence with a predicted signal sequence cleavage site between positions 17 and 18 of the protein (position 0 of the mature protein [Fig. 4]) that corresponded with the determined N terminus of mature SPC18. The cleaved form of SPC18 has a calculated molecular mass of 15,963 Da. The differences in apparent molecular weight as determined by SDS-PAGE are due to glycosylation of the protein. Residues 137 to 139 (N-I-S) of the protein represent a potential N-glycosylation site (residues 119 to 121 of the mature protein [Fig. 4]). Indeed, enzymatic deglycosylation of SPC18 resulted in a reduction of the molecular mass from about 18 kDa to about 16 kDa (Fig. 5A).
FIG. 4.
Nucleotide sequence and deduced amino sequence of the SPC18 gene from R. solani. Numbers at the left mark the position of the nucleotide residues. Numbers at the right mark the position of amino residues (mature protein). The single underline represents amino acid residues determined by Edman degradation. Amino acid residues of the signal peptide are shaded; the amino acid residues that are underlined with ∼∼∼ represent the N-glycosylation site; and the FKEL N terminus is doubly underlined. The position shown by the ▴ is the cleavage site of the putative signal peptide; the residues in boldface represent the position of the degenerate primer; the positions showed by V (1 and 2) are the two intron sites; and the asterisk (*) represents the stop codon. The intron sequences are shown separately, and the typical fungal 5′ and 3′ intron splice sequences are underlined. The GenBank accession number is EF636668.
FIG. 5.
SDS-PAGE analysis of EndoH-treated SPC-enriched fraction (A) and Western blot analysis of SPC18 protein as detected by anti-SPC18 (B) and anti-recSPC18 (C) antibodies. Lanes 1, SPC-enriched fraction; 2, EndoH-treated SPC-enriched fraction. The upper and lower arrows indicate the position of SPC18 before and after EndoH treatment, respectively. * indicates the EndoH protein. Numbers at left are molecular masses.
Localization of SPC18.
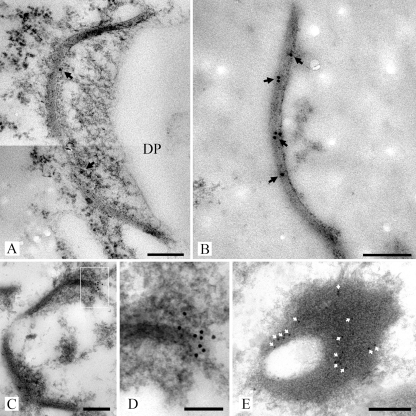
The polyclonal antibodies that were raised against mature SPC18 (anti-SPC18) and against the recSPC18 (anti-recSPC18) recognized SPC18 before and after enzymatic deglycosylation (Fig. 5B and C). Anti-SPC18 antibodies localized SPC18 at the electron-translucent portion of the dolipore swelling, the SPCs, and the plug material (Fig. 6). The dolipore swelling was more densely labeled with gold particles than the SPC (Fig. 6A). The label intensity at the SPCs was denser in sections of SPC-enriched fractions (Fig. 6B) than in hyphal sections (Fig. 6A). Furthermore, label preferentially localized at the plug periphery (Fig. 6C). Because anti-SPC18 antibodies were raised against the mature SPC18 that was glycosylated, sugar residues may have induced an antigen response. Therefore, sections were treated with a periodate or were labeled with an antiserum directed against beta-1,6-glucan to block these sites for subsequent labeling with anti-SPC18. Both treatments did not inhibit anti-SPC18 antibodies from labeling the electron-translucent portion of the dolipore swelling (results not shown). Still, recognition of sugar residues by anti-SPC18 antibodies cannot be completely ruled out. Therefore, antibodies were generated against the nonglycosylated recSPC18 protein. The anti-recSPC18 antibody labeling is specific to SPC18, as was shown with preabsorption experiments. Anti-recSPC18 antibodies were incubated with the recSPC18 protein that was bound to nitrocellulose membrane. Binding of the antibodies to SPC18 was confirmed by secondary antibody labeling using goat anti-rabbit peroxidase (result not shown). Antibodies that were not preincubated with recSPC18 protein revealed about 80% SPCs (of 10 SPCs) with 3 to 9 gold labels, whereas antibodies preincubated with 300 μg of recSPC18 protein showed significantly reduced labeling, namely only 5 labeled SPCs out of 14, with 1 to 2 gold labels each (Fig. 7). These anti-recSPC18 antibodies localized SPC18 at the SPCs and the plug material but not at the dolipore swelling (Fig. 8). The label intensity at the SPCs was denser in sections of SPC-enriched fractions than in sectioned hyphal tissue, as was also shown with anti-SPC18 antibodies. Label preferentially localized at the basal parts of the SPCs (Fig. 8C and D) and at the periphery of the plug (Fig. 8E).
FIG. 6.
Transmission electron micrographs of high-pressure frozen, freeze-substituted, and Lowicryl HM20-embedded R. solani hyphae (A) and SPC-enriched fractions (B and C) labeled with anti-SPC18 antibodies. Antigen-antibody complexes were visualized with secondary goat-anti-rabbit antibodies conjugated with 10 nm gold. Gold particles (arrows) were located at the dolipore swelling and SPC in hyphal cells (A). Furthermore, sections of the SPC-enriched fractions showed gold label present at the SPC (B and C) and at the periphery of plug material that was connected to the SPC (C). Bars, 250 nm.
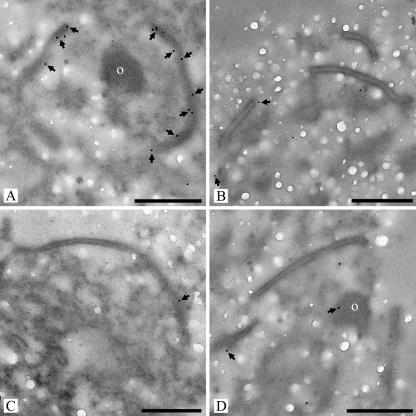
FIG. 7.
Transmission electron micrographs of high-pressure frozen, freeze-substituted, and Lowicryl HM20 embedded R. solani septal pore cap-enriched fractions labeled with anti-recSPC18 antibodies (A) or anti-recSPC18 antibodies preincubated with recSPC18 protein (B-D). After preincubation with 12 μg (B), 60 μg (C), and 300 μg (D) recSPC18 protein, the labeling was heavily diminished compared with the nonpreincubated antibody labeling (A). o, electron-dense occluding material (A and D). Bars, 500 nm.
FIG. 8.
Transmission electron micrographs of high-pressure frozen, freeze-substituted, and Lowicryl HM20-embedded R. solani hyphae (A) and SPC-enriched fractions (B to E) labeled with anti-recSPC18 antibodies. Antigen-antibody complexes were visualized with secondary goat-anti-rabbit antibodies conjugated with 10 nm gold. Gold label (arrows) was present at the SPC (A to D). Gold label was preferentially localized at the base of the SPC (C and D). Panel D shows an enlargement of the box in panel C. Furthermore, label (white arrows) was preferentially localized at the periphery of the plug, as shown in this detailed image of a plug (E). Bars, 200 nm (A to C and E) and 100 nm (D).
DISCUSSION
The divergence of the Dikarya lineages (Ascomycota and Basidiomycota) from the nonseptate basal lineages of the Fungi (Glomeromycota, Chytridiomycota, and Zygomycota) coincided with the occurrence of regularly septate hyphae. It appears that following these evolutionary innovations, two different strategies evolved to plug the septal pores during stress or hyphal damage. Filamentous Ascomycota (i.e., Pezizomycotina) seal their septal pores with Woronin bodies that are associated with the septum (24, 32, 56). In contrast, SPCs seem to fulfill this role in higher Basidiomycota, viz., Agaricomycotina (1, 31, 38, 50). Several main morphotypes of the SPC exist, namely, vesicular-tubular, imperforate, and perforate, and it seems that they have arisen from the ER or that SPCs and ER both share a common ancestor (51). About 50 years after the first transmission electron microscopic observation of SPCs in Trametes versicolor (cited as Polystictus versicolor [17]), we enriched and biochemically analyzed perforate SPCs from the Agaricomycotina group for the first time, using R. solani as the model species. Here, we describe a novel SPC protein that resides in the SPCs and in the septal pore-plugging material in the hyphae of R. solani. Therefore, the newly identified SPC18 protein may have a role in dolipore plugging in basidiomycetous fungi.
SPC18 has an apparent molecular mass of 18 kDa and was isolated from the SPC-enriched fraction of R. solani. This protein has a hydrophobic signal sequence and is N glycosylated. These features are characteristic for proteins that are targeted and imported into the ER (6, 20, 28). The putative ER nature of SPC18, the connection of the ER at the base of SPCs, as well as the clear staining of SPCs with ER-specific fluorochromes (12) strongly support the possibility that SPCs are part or are even specialized subdomains of the ER, as has been speculated before (9, 38, 55). Southern analysis showed the presence of a single SPC18 gene in the genome, but allelic variations may occur, as was shown by the nucleotide variation of the genomic sequence. The cause of this variation is unknown, but it may be caused by the multinucleate nature of the R. solani strain that was used. However, the observed sequence variation did not alter the amino acid sequence. It is rather surprising that SPC18 was not found in the genomes of other basidiomycetous fungi that have dolipore septa associated with SPCs, like Coprinopsis cinerea (33) and Phanerochaete chrysosporium (cited as Chrysosporium xerophilum [7]). This suggests that SPC18 is a species-specific component of the SPC of R. solani and, possibly, of its close relatives. Subsequent characterizations of other SPC proteins and/or SPCs of other basidiomycetous fungi could reveal a common SPC component that also would provide evidence for the evolutionary history of these organelles.
After we performed immunogold labeling studies, we observed that SPC18 was located at both the SPC and the plugging material. The putative presence of glycoproteins in both the SPCs and the plug material was shown by labeling of SPCs (52) and plugs (Fig. 2) with WGA, a lectin that detects N-acetylglucosamine groups (40). In addition, a previous study by Müller et al. (36) already showed the presence of sugar moieties in the plug of Trichosporon gamsii (cited as T. sporotrichoides) by 1,6-β-glucan labeling. Next to the labeling of the SPC and plugging material, the anti-SPC18 antibodies also showed labeling of the dolipore swelling, which was not observed with the anti-recSPC18 antibodies. This unspecific labeling may be the result of antibodies that were raised against the glycosylated SPC18 isolated from the protein pool extracted from the SPC-enriched fraction. Therefore, the antibodies raised against the recSPC18 are superior to the anti-SPC18 antibodies because there is no cross-reactivity with sugar residues or with other fungal proteins or components. Furthermore, we showed that the anti-recSPC18 antibodies specifically recognized the SPC18 protein on nitrocellulose membrane, as well as on sectioned material. After the preabsorption with recSPC18 protein was performed, we observed much less labeled SPCs. Those that were labeled showed a significant reduction of gold label. We found only one or two gold labels on those SPCs that were labeled, compared to between three and nine gold labels on the nonpreabsorbed control of labeled SPCs. These results clearly indicate that the anti-recSPC18 antibodies specifically bind to the SPC18 protein.
In the labeling studies, we also found that SPCs in the SPC-enriched fraction labeled more intensely than when intact hyphae were sectioned and labeled. This difference could be explained by the fact that the SPC-enriched fractions were treated with Triton X-100, which might increase the accessibility of protein epitopes in the SPCs to the antibodies. Furthermore, it should be noted that the plugging material was not always labeled and that labeling was observed mainly at the periphery of the plug. In contrast, sections of R. solani hyphae hardly showed plugged dolipores and these plugs were not labeled with antibodies (result not shown) or WGA label (52). Possibly, the treatment to enrich the SPCs may have generated freshly formed dense plugs, and SPC18 that localized in these plugs came from the SPCs that act as repositories of SPC18. However, in time, pore plugs lose their reversible character and become more permanent (11). During this process, SPC18 can be modified in such a way that the antibodies cannot detect it anymore. Finally, the base of the SPC, which is the part that is connected with ER, was also found to be preferentially labeled. This difference in label intensity suggests that SPC18 proteins are extremely packed in the SPC or exist in different conformations or both. Alternatively, other unidentified proteins may reside in the central parts of the SPC and plug. In addition, the buoyant density of the isolated SPCs lies between 1.24 g/cm3 (50% sucrose at 4°C [14]) and 1.36 g/cm3 (70% sucrose at 4°C [14]) and is higher than the 1.1-g/cm3 density of ER (8). The higher density can be explained by these packed proteins stored in the SPCs (27).
We hypothesize that the SPC18 protein is targeted to ER where glycosylation takes place. As the ER is continuous with the base of the SPC, SPC18 may then be transported from the ER lumen to the SPC (37). Furthermore, modifications of the N-glycan group on SPC18 may occur in the ER or possibly also in the SPC that define the destiny of the protein. The transport route of SPC18 from ER to SPC to the cytoplasm at the dolipore is different from the secretory pathway where proteins are destined for the Golgi compartment, endosomes, lysosomes, the plasma membrane, or the extracellular environment. It more closely resembles the retrotranslocation route from ER to cytoplasm of misfolded proteins (34) or an unknown mechanism for the export of small glycoproteins (46). SPC18 is then deposited in the SPCs until it is instantly released in case it is needed to plug the dolipore. The filaments between the inside of the SPC and the plugging material (38, 39, 41, 53) may transport or guide SPC18 from the SPC to the orifice of the dolipore. During this transport, the SPC matrix may become more electron translucent at sites where filaments are attached at the inside of the SPC (36, 38). Furthermore, other posttranslational modifications of the SPC18 protein may be of importance and need further investigation. Juvvadi et al. (26) showed that protein kinase C phosphorylation of the HEX-1 protein is needed for multimerization of the protein and proper localization in Woronin bodies in Ascomycota. As ATPase activity was found at the plug site (42), phosphorylation of the SPC18 protein or other unidentified proteins that are part of the SPC-dolipore-plugging complex might also play a role in the dolipore-plugging process of basidiomycetous fungi.
Acknowledgments
We thank H. Yiggitop, A. Overbeek, and L. de Kok for technical assistance, E. Kuramea (CBS, The Netherlands) and J. Leunissen (Wageningen UR, The Netherlands) for help with bioinformatics tools, and L. Lugones (Utrecht University, The Netherlands) for Southern analysis.
The Odo van Vloten Foundation financially supported this work.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Aylmore, R. C., G. E. Wakley, and N. K. Todd. 1984. Septal sealing in the basidiomycete Coriolus versicolor. J. Gen. Microbiol. 1302975-2982. [Google Scholar]
- 2.Balance, D. J. 1991. Transformation systems for filamentous fungi and an overview of fungal gene structure. p. 1-29. In S. A. Leong, and R. M. Berka (ed.), Molecular industrial mycology, systems and applications for filamentous fungi. Marcel Dekker Inc., New York, NY.
- 3.Bauer, R., D. Begerow, J. P. Sampaio, M. Weiss, and F. Oberwinkler. 2006. The simple-septate basidiomycetes: a synopsis. Mycol. Prog. 541-66. [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 5.Berbee, M. L., and J. W. Taylor. 2001. Fungal molecular evolution: gene trees and geologic time. p. 229-245. In D. J. McLaughlin, E. G. McLaughlin, and P. A. Lemke (ed.), The mycota VII, systematics and evolution, part B. Springer-Verlag, Berlin, Germany.
- 6.Blobel, G., and B. Dobberstein. 1975. Transfer of proteins across membranes. I. The presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boekhout, T., C. A. N. Van Oorschot, J. A. Stalpers, W. H. Batenburg-Van der Vegte, and A. C. M. Weijman. 1989. The taxonomic position of Chrysosporium xerophilum and septal morphology in Chrysosporium, Sporotrichum and Disporotrichum. Stud. Mycol. 3129-39. [Google Scholar]
- 8.Borgeson, C. E., and B. J. Bowman. 1983. Isolation and characterization of the Neurospora crassa endoplasmic reticulum. J. Bacteriol. 156362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracker, C. E., and E. E. Butler. 1963. The ultrastructure and development of septa in hyphae of Rhizoctonia solani. Mycologia 5535-58. [Google Scholar]
- 10.Bracker, C. E., and E. E. Butler. 1964. Function of the septal pore apparatus in Rhizoctonia solani during protoplasmic streaming. J. Cell Biol. 21152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracker, C. E. 1967. Ultrastructure of fungi. Annu. Rev. Phytopathol. 5343-374. [Google Scholar]
- 12.Cole, L., D. Davies, G. J. Hyde, and A. E. Ashford. 2000. ER-tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and Golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. J. Microsc. 197239-249. [DOI] [PubMed] [Google Scholar]
- 13.Curach, N. C., V. S. J. Te'o, M. D. Gibbs, P. L. Bergquist, and K. M. H. Nevalainen. 2004. Isolation, characterization and expression of the hex1 gene from Trichoderma reesei. Gene 331133-140. [DOI] [PubMed] [Google Scholar]
- 14.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research. Oxford University Press, New York, NY.
- 15.De E. Castro, C. J. A. Sigrist, A. Gattiker, V. Bulliard, P. S. Langendijk-Genevaux, E. Gasteiger, A. Bairoch, and N. Hulo. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34W362-W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server. p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 17.Girbardt, M. 1958. Über die Substruktur von Polystictus versicolor L. Arch. Mikrobiol. 28255-269. (In German.) [PubMed] [Google Scholar]
- 18.Girbardt, M. 1961. Licht- und Elektronenmikroskopische Untersuchungen an Polystictus versicolor. Arch. Mikrobiol. 39351-359. (In German.) [PubMed] [Google Scholar]
- 19.Gull, K. 1978. Form and function of septa in filamentous fungi. p. 78-93. In J. E. Smith (ed.), The filamentous fungi: developmental mycology. Edward Arnold, London, United Kingdom.
- 20.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 2912364-2369. [DOI] [PubMed] [Google Scholar]
- 21.Hibbett, D. S., and R. G. Thorn. 2001. Homobasidiomycetes. p. 121-170. In D. J. McLaughlin, E. G. McLaghlin, and P. A. Lemke (ed.), The mycota VII, systematics and evolution, part B. Springer-Verlag, Berlin, Germany.
- 22.Hibbett, D. S., M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk, R. Lücking, H. Thorsten Lumbsch, F. Lutzoni, P. B. Matheny, D. J. McLaughlin, M. J. Powell, S. Redhead, C. L. Schoch, J. W. Spatafora, J. A. Stalpers, R. Vilgalys, M. C. Aime, A. Aptroot, R. Bauer, D. Begerow, G. L. Benny, L. A. Castlebury, P. W. Crous, Y. C. Dai, W. Gams, D. M. Geiser, G. W. Griffith, C. Gueidan, D. L. Hawksworth, G. Hestmark, K. Hosaka, R. A. Humber, K. D. Hyde, J. E. Ironside, U. Kõljalg, C. P. Kurtzman, K. H. Larsson, R. Lichtwardt, J. Longcore, J. Miadlikowska, A. Miller, J. M. Moncalvo, S. Mozley-Standridge, F. Oberwinkler, E. Parmasto, V. Reeb, J. D. Rogers, C. Roux, L. Ryvarden, J. P. Sampaio, A. Schüssler, J. Sugiyama, R. G. Thorn, L. Tibell, W. A. Untereiner, C. Walker, Z. Wang, A. Weir, M. Weiss, M. M. White, K. Winka, Y. J. Yao, and N. Zhang. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111509-547. [DOI] [PubMed] [Google Scholar]
- 23.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G. H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schussler, J. E. Longcore, K. O'Donnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443818-822. [DOI] [PubMed] [Google Scholar]
- 24.Jedd, G., and N.-H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2226-231. [DOI] [PubMed] [Google Scholar]
- 25.Johansson, T., and P. O. Nyman. 1996. A cluster of gene encoding major isoenzymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene 17031-38. [DOI] [PubMed] [Google Scholar]
- 26.Juvvadi, P. R., J. Maruyama, and K. Kitamoto. 2007. Phosphorylation of the Aspergillus oryzae Woronin body protein, AoHex1, by protein kinase C: evidence for its role in the multimerization and proper localization of the Woronin body protein. Biochem. J. 405533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, M. S., J. Au-Young, and E. Cabib. 1985. Modification of yeast plasma membrane density by concanavalin A attachment. J. Biol. Chem. 26012680-12684. [PubMed] [Google Scholar]
- 28.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70755-775. [DOI] [PubMed] [Google Scholar]
- 29.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157105-132. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structure proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 31.Markham, P. 1994. Occlusions of septal pores in filamentous fungi. Mycol. Res. 981089-1106. [Google Scholar]
- 32.Markham, P., and A. J. Collinge. 1987. Woronin bodies of filamentous fungi. FEMS Microbiol. Lett. 461-11. [Google Scholar]
- 33.McLaughlin, D. J. 1974. Ultrastructural localization of carbohydrate in the hymenium and subhymenium of Coprinus. Protoplasma 82341-364. [DOI] [PubMed] [Google Scholar]
- 34.Molinari, M. 2007. N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3313-320. [DOI] [PubMed] [Google Scholar]
- 35.Moore, R. T., and J. H. McAlear. 1962. Fine structures of mycota. Observations on septa of ascomycetes and basidiomycetes. Am. J. Bot. 4986-94. [Google Scholar]
- 36.Müller, W. H., R. C. Montijn, B. M. Humbel, A. C. Van Aelst, E. J. M. C. Boon, T. P. Van der Krift, and T. Boekhout. 1998. Structural differences between two types of basidiomycete septal pore caps. Microbiology 1441721-1730. [DOI] [PubMed] [Google Scholar]
- 37.Müller, W. H., J. A. Stalpers, A. C. Van Aelst, T. P. Van de Krift, and T. Boekhout. 1998. Field emission gun-scanning electron microscopy of septal pore caps of selected species in the Rhizoctonia s.l. complex. Mycologia 90170-179. [Google Scholar]
- 38.Müller, W. H., A. J. Koster, B. M. Humbel, U. Ziese, A. J. Verkleij, A. C. Van Aelst, T. Van der Krift, R. C. Montijn, and T. Boekhout. 2000. Automated electron tomography of the septal pore cap in Rhizoctonia solani. J. Struct. Biol. 13110-18. [DOI] [PubMed] [Google Scholar]
- 39.Müller, W. H., J. A. Stalpers, A. C. Van Aelst, M. D. M. De Jong, T. P. Van de Krift, and T. Boekhout. 2000. The taxonomic position of Asterodon, Asterostroma and Coltricia inferred from the septal pore cap ultrastructure. Mycol. Res. 1041485-1491. [Google Scholar]
- 40.Nagata, Y., and M. M. Burger. 1974. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J. Biol. Chem. 2493116-3122. [PubMed] [Google Scholar]
- 41.Orlovich, D. A., and A. E. Ashford. 1994. Structure and development of the dolipore septum in Pisolithus tinctorius. Protoplasma 17866-80. [Google Scholar]
- 42.Schramm, K.-H. 1971. Enzymmuster bei Pilzen. Ph.D. thesis, Freie Universität Berlin, Berlin, Germany.
- 43.Schuren, F. H. J., M. C. Harmsen, and J. G. H. Wessels. 1993. A homologous gene-reporter system for the basidiomycete Schizophyllum commune based on internally deleted homologous genes. Mol. Gen. Genet. 23891-96. [DOI] [PubMed] [Google Scholar]
- 44.Soundararajan, S., G. Jedd, X. Li, M. Ramos-Pamplona, N.-H. Chua, and N. I. Naqvi. 2004. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 161564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spurr, A. R. 1969. A low viscosity resin embedding medium for electron microscopy. J. Ultrastruct. Res. 2631-43. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, T., and W. J. Lennarz. 2000. In yeast the export of small glycopeptides from the endoplasmic reticulum into the cytosol is not affected by the structure of their oligosaccharide chains. Glycobiology 1051-58. [DOI] [PubMed] [Google Scholar]
- 47.Swann, E. C., and J. W. Taylor. 1995. Phylogenetic perspectives on basidiomycete systematics: evidence from the 18S rRNA gene. Can. J. Bot. 73S862-S868. [Google Scholar]
- 48.Swann, E. C., E. M. Frieders, and D. J. McLauglin. 2001. Urediniomycetes. p. 37-56. In D. J. McLaughlin, E. G. McLaghlin, and P. A. Lemke (ed.), The mycota VII, systematics and evolution, part B. Springer-Verlag, Berlin, Germany.
- 49.Tenney, K., I. Hunt, J. Sweigard, J. I. Pounder, C. McClain, E. J. Bowman, and B. J. Bowman. 2000. hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31205-217. [DOI] [PubMed] [Google Scholar]
- 50.Thielke, C. 1972. Die dolipore der Basidiomyceten. Arch. Mikrobiol. 8231-37. (In German.) [Google Scholar]
- 51.van Driel, K. G. A. 2007. Septal pore caps in Basidiomycetes, composition and ultrastructure. Ph.D. thesis, Utrecht University, Utrecht, The Netherlands.
- 52.van Driel, K. G. A., T. Boekhout, H. A. B. Wösten, A. J. Verkleij, and W. H. Müller. 2007. Laser microdissection of fungal septa as visualised by scanning electron microscopy. Fungal Genet. Biol. 44466-473. [DOI] [PubMed] [Google Scholar]
- 53.van Driel, K. G. A., A. F. van Peer, H. A. B. Wösten, A. J. Verkleij, T. Boekhout, and W. H. Müller. 2007. Enrichment of perforate septal pore caps from the basidiomycetous fungus Rhizoctonia solani by combined use of French press, isopycnic centrifugation, and Triton X-100. J. Microbiol. Methods 71298-304. [DOI] [PubMed] [Google Scholar]
- 54.Venable, J. H., and R. Coggeshall. 1965. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 25407-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilsenach, R., and M. Kessel. 1965. On the function and structure of the septal pore of Polyporus rugulosus. J. Gen. Microbiol. 40397-400. [DOI] [PubMed] [Google Scholar]
- 56.Woronin, M. 1864. Zur Entwicklungsgeschichte des Ascobolus pulcherrimus Cr. und Pezizen. Abh. Senkenb. Naturforsch. 5333-344. (In German.) [Google Scholar]