Abstract
Nuclear domains, called cleavage bodies, are enriched in the RNA 3′-processing factors CstF 64 kDa and and CPSF 100 kDa. Cleavage bodies have been found either overlapping with or adjacent to coiled bodies. To determine whether the spatial relationship between cleavage bodies and coiled bodies was influenced by the cell cycle, we performed cell synchronization studies. We found that in G1 phase cleavage bodies and coiled bodies were predominantly coincident, whereas in S phase they were mostly adjacent to each other. In G2 cleavage bodies were often less defined or absent, suggesting that they disassemble at this point in the cell cycle. A small number of genetic loci have been reported to be juxtaposed to coiled bodies, including the genes for U1 and U2 small nuclear RNA as well as the two major histone gene clusters. Here we show that cleavage bodies do not overlap with small nuclear RNA genes but do colocalize with the histone genes next to coiled bodies. These findings demonstrate that the association of cleavage bodies and coiled bodies is both dynamic and tightly regulated and suggest that the interaction between these nuclear neighbors is related to the cell cycle–dependent expression of histone genes.
INTRODUCTION
The cell nucleus contains various distinct structural and functional domains, each with its own morphology and protein composition (de Jong et al., 1996; Lamond and Earnshaw, 1998). One well-studied nuclear domain is the coiled body, which owes its name to its appearance as a ball of coiled threads in the electron microscope (Monneron and Bernhard, 1969). Coiled bodies are small spherical structures, one to five per nucleus, with a diameter of 0.2–1.0 μm. They are evolutionary conserved from plants to mammals, indicating that they have a crucial role in the nucleus (for reviews, see Lamond and Carmo-Fonseca, 1993; Gall et al., 1995). Over the years, many nuclear factors have been found concentrated in coiled bodies. Among these are nucleolar constituents, such as fibrillarin (Raška et al., 1990) and Nopp140 (Meier and Blobel, 1992), U3 small nucleolar RNA (Jiménez-García et al., 1994), and nucleoplasmic factors, such as the general transcription factors TFIIH and TFIIF (Grande et al., 1997; Jordan et al., 1997) and the RNA-processing factors U1, U2, U4/U6, and U7 small nuclear ribonuclear protein (Carmo-Fonseca et al., 1992; Wu and Gall, 1993; Frey and Matera, 1995). The protein p80-coilin is especially enriched inside coiled bodies (Andrade et al., 1991; Raška et al., 1991) and is a hallmark for this nuclear domain. It has become clear that coiled bodies are dynamic structures. Disruption of cellular processes, e.g., by heat shock or drug treatment, rapidly alters the distribution and number of coiled bodies (Carmo-Fonseca et al., 1992). Coiled bodies also undergo changes during the cell cycle. During mitosis they disassemble, and only small remnants are left in the mitotic cell (Carmo-Fonseca et al., 1993). Coilin enters the nucleus in telophase, but coiled bodies are not formed until later in G1 (Carmo-Fonseca et al., 1993; Ferreira et al., 1994). Thus several lines of investigation indicate that there is a direct relationship among the transcriptional, metabolic, and developmental states of the cell and the number, morphology, and distribution of coiled bodies.
The emerging view is that many nuclear factors are not only concentrated in domains, but that these domains can be spatially and functionally associated with each other and with specific genomic elements (for review, see Schul et al., 1998a). Several genes and domains enriched in specific proteins have now been found juxtaposed to coiled bodies. One of the first examples of such a juxtaposition are the domains enriched in the CstF 64-kDa and CPSF 100-kDa proteins (Schul et al., 1996). CstF and CPSF are essential 3′-processing factors for virtually all mRNAs, together with factors such as CFI, CFII, and poly(A) polymerase (for review, see Wahle and Kühn, 1997). Immunolocalization studies showed that the CstF 64-kDa and CPSF 100-kDa subunits distributed diffusely throughout the nucleus and concentrated in a few small foci. These foci were found to associate with coiled bodies (Schul et al., 1996). Because these domains could be observed as morphologically distinct structures in the electron microscope and were enriched in RNA 3′ cleavage factors, we called them “cleavage bodies.” Cleavage bodies were mainly found either adjacent to coiled bodies or overlapping with them. The degree of overlap may be correlated with the transcriptional activity of the locus. For example, treatment of cells with transcription inhibitors resulted in a complete colocalization of coiled bodies and cleavage bodies (Schul et al., 1996). Additionally, ∼20% of the cleavage bodies were found to contain newly synthesized RNA. Coiled bodies, however, are known to be devoid of newly synthesized RNA (Fakan and Bernhard, 1971; Moreno Diaz de la Espina et al., 1982; Callan and Gall, 1991; Schul et al., 1996; Jordan et al., 1997; Schul et al., 1998b). Thus only cleavage bodies that do not overlap with a coiled body, i.e., the cleavage bodies adjacent to coiled bodies, contain newly synthesized RNA. We hypothesized that specific genes, located at the periphery of coiled bodies, might recruit the necessary cleavage factors and cause the formation of cleavage bodies next to coiled bodies (Schul et al., 1996).
There have also been reports of other nuclear domains that are closely associated with coiled bodies. Liu and Dreyfuss (1996) identified domains enriched in the protein SMN that often occur adjacent to coiled bodies, called “gemini of coiled bodies” or “gems.” Yannoni and White (1997) found domains in the nuclei of Drosophila neurons that were enriched in the protein ELAV, referred to as “ELAV dots” and the “ELAV web,” which were often associated with coiled bodies. It is unknown whether there is any relationship between nuclear domains, cleavage bodies, and the coiled body–associated genes.
There is, however, increasing evidence that the spatial association of different nuclear domains and genomic elements is a fundamental organizational principle of the cell nucleus. The grouping of specific genes and domains enriched in transcription and processing factors, as found in the nucleolus, also occurs at other sites in the nucleus (discussed by Schul et al., 1998a). It is becoming increasingly clear that the spatial relationship between different domains and genes is a common organizational principle, probably to allow an efficient and controlled synthesis and processing of a range of gene transcripts. In keeping with this idea, several genes have been found to preferentially associate with coiled bodies: the histone gene clusters (Gall et al., 1981; Callan et al., 1991; Frey and Matera, 1995), the U1 and U2 small nuclear RNA (snRNA) genes (Frey and Matera, 1995; Smith et al., 1995), and the U3 small nucleolar RNA genes (Gao et al., 1997) are all frequently located adjacent to coiled bodies. It remains unclear, however, whether there is any relationship between these genes and cleavage bodies.
To study the spatial relationship among coiled bodies, cleavage bodies, and several coiled body–associated genes, we performed immunofluorescent and in situ hybridization double-labeling experiments on cells in different stages of the cell cycle. Confocal laser scanning microscopy, in combination with image restoration to correct for diffraction-induced distortions, allowed a detailed analysis of the small nuclear domains. We found that the overlap between coiled bodies and cleavage bodies changes during the cell cycle. These changes can be correlated with both the spatial juxtaposition and transcription activity of the cell cycle–regulated histone gene cluster on human chromosome 6p21.
MATERIALS AND METHODS
Cell Culture and Synchronization
T24 cells (from human bladder carcinoma) were grown on circular glass coverslips at 37°C under a 10% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (Life Technologies, Gaithersburg, MD) supplemented with 1% glutamine (Life Technologies), 10% fetal calf serum (Life Technologies), and antibiotics (100 IU/ml penicillin and 100 mg/ml streptomycin; Life Technologies).
Cells were synchronized in G1 and S phase by mitotic shake-off, essentially as described by Zwanenburg (1983). Exponentially growing cells were washed twice with fresh medium to remove free-floating material. Tapping the culture flask dislodged mitotic cells, which were subsequently harvested by removing the medium. The mitotic cells were allowed to reattach to coverslips and were cultured for 4–10 h, allowing them to enter G1 and S phase.
Cells were synchronized in S and G2 phase by a hydroxyurea block. Cells on coverslips were cultured in the presence of 2 mM hydroxyurea for 20 h. The medium was then replaced by fresh medium to release the block. Cells were cultured for another 4–10 h to enter S and G2 phase.
Immunofluorescence Labeling
All steps were performed at room temperature unless stated otherwise. Coverslips with attached cells were rinsed once in PBS and incubated with 2% paraformaldehyde in PBS for 15 min. After fixation cells were rinsed twice with PBS and permeabilized with 0.5% Triton X-100 (Sigma, St. Louis, MO) in PBS for 5 min. Cells were subsequently rinsed twice in PBS, incubated in PBS containing 100 mM glycine (Sigma) for 10 min, and incubated for 10 min in PBS containing 0.5% BSA (Sigma) and 0.05% gelatin from cold water fish skin (Sigma) (PBG).
For immunolabelling polyclonal antibody 204/5 from rabbit against p80-coilin (a gift from Dr. A.I. Lamond, Department of Biochemistry, University of Dundee, Dundee, United Kingdom) (Bohmann et al., 1995), a monoclonal antibody from mouse against CstF 64 kDa (gift from Drs. Y. Takagaki and J.L. Manley, Department of Biological Sciences, Columbia University, New York, NY) (Takagaki et al., 1990), and anti-5-bromodeoxyuridine (BrdU) polyclonal antibody from rat (Seralab, Crawley Down, United Kingdom) were used.
Fixed cells were incubated overnight at 4°C or for 2 h at room temperature with primary antibodies diluted in PBG. Subsequently, cells were washed four times for 5 min each in PBG and incubated with secondary antibodies diluted in PBG for 1.5 h. Secondary antibodies were donkey anti-mouse immunoglobulin G (IgG) coupled to DTAF (4,6-dichlorotriazinyl amino fluorescein) or Cy3 (Jackson ImmunoResearch, West Grove, PA), donkey anti-rabbit IgG coupled to FITC or Cy3 (Jackson), and donkey anti-rat IgG coupled to Cy3 (Jackson).
After labeling, cells were washed two times for 5 min each in PBG and two times for 5 min each in PBS followed by incubation in PBS containing 0.4 μg/ml Hoechst 33258 (Sigma) for 5 min. All coverslips were mounted in Vectashield (Vector Laboratories, Burlingame, CA).
Fluorescent In Situ Hybridization in Combination with Immunofluorescence Labeling
When immunofluorescence labeling was combined with in situ hybridization, the following adaptations and additions to the above protocol were implemented. PBG was substituted with PBS containing 0.1 mg/ml nuclease-free acetylated BSA (Sigma) and 0.1 μg/ml herring sperm DNA (PBH). After primary and secondary antibody labeling, the cells were fixed for 5 min with 2% formaldehyde in PBS, washed twice in PBS, incubated 10 min in 100 mM glycine in PBS, and washed in PBS.
Cells were dehydrated by subsequent incubations in 70, 90, and 100% ice-cold ethanol for 4 min per incubation and air dried. Genomic DNA was denatured by incubating the coverslips in 2× SSC containing 70% formamide, pH 7.2, at 80°C for 5 min. Immediately after, the cells were treated with the 70, 90, and 100% ice-cold ethanol for 4 min each and air dried. The cells were incubated overnight in probe solution at 37°C.
The probes were produced from human genomic clones of the RNU1 locus at 1p36, of the RNU2 locus at 17q21 (gifts of Dr. A.M. Weiner, Department of Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, CT) (Frey and Matera, 1995), of the histone gene cluster locus at 6p21, or of the dihydrofolate reductase gene locus at 5q12–13 (clone CHB203; American Type Culture Collection, Manassas, VA). The probes were labeled by nick translation using digoxigenin-labeled dUTP essentially as described by Rigby et al. (1977) and Langer et al. (1981). The probe was heat denatured in 70% deionized formamide together with COT-1 DNA (Boehringer Mannheim, Indianapolis, IN) at 80°C for 10 min. The final probe solution contained 2× SSC, 50% formamide, 10% dextran sulfate, COT-1 and herring sperm DNA, and the labeled probe.
After incubation with probe solution, the coverslips were washed three times for 5 min each in 2× SSC containing 50% formamide, pH 7.2, at 39°C and three times for 5 min each in 1× SSC at room temperature. The cells were washed twice in PBS and incubated for 30 min in PBH. Subsequently, the coverslips were incubated for 60 min in PBH containing FITC-conjugated anti-digoxigenin antibody (Sigma). The cells were then washed four times in PBS. The cells were stained with Hoechst and embedded and mounted as described above.
Confocal Laser Scanning Microscopy and Image Analysis
Images of double-labeled cells were produced on a Leica (Nussloch, Germany) confocal laser scanning microscope with a 100×, 1.35 oil immersion lens. A dual-wavelength laser was used to excite green (DTAF or FITC) and red (Cy3) fluorochromes simultaneously at 488 and 514 nm, respectively. The fluorescence signals from the two fluorochromes were recorded simultaneously. Optical cross-talk was quantified and subtracted as described previously (Manders et al., 1992). Image analysis was performed using Scil-Image software, developed at the University of Amsterdam (Van Balen et al., 1994). For detailed analysis of the spatial relationship between two labeling patterns, the images were subjected to a restoration procedure to correct for diffraction-induced distortions using a measured point spread function (Van der Voort and Straster, 1995).
BrdU Labeling
To detect cells in S phase, BrdU was added to the culture medium of cells at a final concentration of 10 μM. The cells were incubated for 10 min at 37°C and subsequently washed twice with culture medium for 1 min at 37°C. The cells were fixed and permeabilized as described above. Before immunolabelling, DNA was denatured in 2 N HCl at 37°C for 30 min, after which the coverslips were washed twice with PBS.
Flow Cytometry
The degree of synchronization was also analyzed by flow cytometry. Cells were fixed and stained essentially as described by Krishan (1975). Approximately 106 cells were harvested and concentrated by centrifugation at 1200 rpm for 10 min and subsequently resuspended in 2 ml of PBS. Slowly, while gently mixing, 6 ml of 96% ethanol were added. The fixed cells were stored at 4°C. The suspension of fixed cells was centrifuged for 5 min at 1000 rpm, and the pellet was resuspended in 0.25 ml of PBS. Then 0.25 ml of 4 mg/ml RNase (Sigma) in PBS and 0.5 ml 1 mg/ml propidium iodine plus 0.2 μg/ml saponin (Sigma) in PBS were added, and the mixture was incubated at 37°C for 15 min. The suspension was passed through a 0.7-mm needle twice to reduce aggregation of cells before flow cytometric measurements. Measurements were performed using a Becton Dickinson (Mountain View, CA) FACStar plus.
RESULTS
Synchronization of Cells
The observed spatial associations between coiled bodies and cleavage bodies fall into three categories: bodies completely overlap; they only partially overlap; or they are adjacent to each other. Each of these conditions is detectable in an asynchronous cell population; however, it remains unclear what causes these differences (Schul et al., 1996). To investigate whether the cell cycle has any influence on this spatial relationship, we synchronized T24 cells by a hydroxyurea block or by mitotic shake-off.
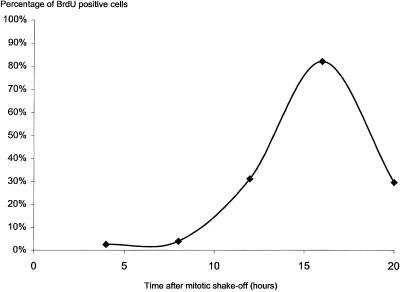
To obtain cells in G1 and S phase we used mitotic shake-off. This yields mitotic cells that synchronously enter G1 phase and proceed to S phase in several hours. The time at which these cells enter S phase was determined by detecting DNA synthesis using BrdU labeling at several time points after mitotic shake-off. Figure 1 shows that the percentage of cells that contain BrdU labeling starts to increase ∼10 h after mitotic shake-off, marking the beginning of S phase.
Figure 1.
Graph displaying the percentage of cells in S phase, indicated by BrdU labeling, at different time points after synchronization by mitotic shake-off.
To obtain cells in S and G2 phase we used a hydroxyurea block. Hydroxyurea is able to block the cell cycle at the beginning of S phase, and a subsequent release from this block yields a cell population that is enriched in S phase cells (Fox et al., 1987; Tobey et al., 1988). The synchronization of cells and their progression through the cell cycle were monitored by flow cytometry. Approximately two-thirds of the cells in an asynchronous culture were in G1 phase, whereas approximately one-third of the population was in S or G2 phase (Figure 2A). After the hydroxyurea block most cells were found to accumulate in G1 phase (Figure 2B), and 4 h after release from the block the majority of cells had progressed to S phase (Figure 2C). Approximately 8 h after release from the block, the population was enriched in cells in late S and G2 phase (Figure 2D).
Figure 2.
Histograms of flow cytometry measurements on hydroxyurea-synchronized cells. (A) Unsynchronized cells contain cells in G1, S, and G2 phase. (B) Cells treated with hydroxyurea are mainly found in G1 phase. (C) Approximately 4 h after release from the hydroxyurea block the cells have progressed to S phase. (D) After 8 h the population contains few cells in G1 phase and is enriched in cells in late S and G2 phase. These data show the degree of synchronization of the cells used in the immunofluorescent labeling experiments.
Because it is known that not all cells recover from a synchronization procedure with the same efficiency, and some cells advance faster through the cell cycle than others, there cannot be an absolute synchronous progression through the cell cycle of all cells (Grdina et al., 1984). Only a substantial enrichment of cells in a certain stage of the cell cycle can practically be achieved. Additionally, the longer after the release from the synchronization block, the more asynchronous a cell population will become. We therefore only used hydroxyurea-synchronized cells to study middle to late S phase and G2 phase and mitotic shake-off to study G1 and early S phase.
Spatial Association between Coiled Bodies and Cleavage Bodies Changes during the Cell Cycle
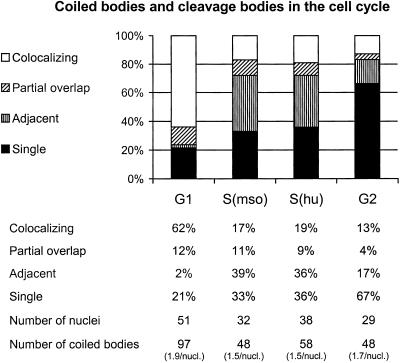
Double labeling of synchronized cells with antibodies against p80-coilin and CstF 64 kDa allowed the analysis of the spatial association between coiled bodies and cleavage bodies in the cell cycle. Cells in middle and late G1 phase, synchronized by mitotic shake-off, mainly contained coiled bodies and cleavage bodies that completely overlapped (Figures 3, first column, and 4A). When we allowed this population to proceed to S phase, the percentage of colocalizing bodies was greatly reduced, and most coiled bodies and cleavage bodies were either adjacent or only partially overlapping (Figures 3, second column, and 4B). Similarly, cells synchronized in S phase by hydroxyurea treatment showed the same percentages of adjacent and partially overlapping bodies (Figure 3, third column). A cell population in late S and G2 phase, 8 h after release from the hydroxyurea block, still contained coiled bodies but had fewer cleavage bodies. The cleavage bodies that we did observe were often less bright and less defined in shape (Figure 4C). As a result, the percentage of solo coiled bodies, i.e., coiled bodies unassociated with a cleavage body, was significantly higher in these cells in late S and G2 phase (Figure 3, fourth column). The number of coiled bodies per cell stayed approximately the same in the cell cycle (Figure 3).
Figure 3.
Cells synchronized by mitotic shake-off (mso) and hydroxyurea (hu) treatment revealed that the spatial association between coiled bodies and cleavage bodies is cell cycle dependent. Coiled bodies were scored as colocalizing, partially overlapping, adjacent, or unassociated with a cleavage body (cleavage bodies were almost never seen unassociated with a coiled body). G1 cells were analyzed 4 h after mso; S phase cells were analyzed 10 h after mso or 4 h after release from hu block; and G2 cells were analyzed 8 h after release from hu block. These show that the nuclear bodies were mainly colocalizing in G1 and adjacent in S phase.
Figure 4.
Confocal optical sections of double-labeled cells. (A) Cells synchronized in G1 mainly contained cleavage bodies (green) that overlapped with coiled bodies (red; also see enlarged area). (B) Cells synchronized in S phase contained many cleavage bodies (green) adjacent to coiled bodies (red; also see enlarged area). (C) Cells synchronized in late S phase and G2 phase contained irregularly shaped cleavage bodies (green) adjacent to coiled bodies (red; also see enlarged area). (D) In situ hybridization showed that histone gene loci (red) are preferentially found adjacent to coiled bodies (green) in human T24 cells. (E) Histone gene loci (red) were regularly found colocalizing with cleavage bodies (green), specifically in S phase. (F) This series of optical sections show that the histone gene loci (red) are inside the cleavage bodies (green). (G) The U2 snRNA gene loci (red) were also often found adjacent to coiled bodies, but they were only found adjacent to cleavage bodies (green) or at a small distance from a cleavage body (H). Bar: A–E and G, 2 μm; F, 1 μm.
Summing up, we have found that cleavage bodies preferentially overlap with coiled bodies during G1 phase, reside adjacent to coiled bodies during S phase, and seem to disintegrate during late S and G2 phase (also see Figure 4).
Cleavage Bodies Coincide with Histone Genes but Not with snRNA Genes Adjacent to Coiled Bodies
Several human genes have been shown to preferentially associate with coiled bodies (Frey and Matera, 1995; Smith et al., 1995; Gao et al., 1997). Given the peculiar cell cycle–dependent spatial relationship between coiled bodies and cleavage bodies, we investigated whether cleavage bodies had any relationship with the U1 snRNA, U2 snRNA, and histone genes.
Because the degree of association between coiled bodies and the aforementioned genes can vary greatly between cell types (Frey and Matera, 1995), we first determined to which extent the genes and coiled bodies are associated in T24 cells. We used fluorescent in situ hybridization, with probes against the U1 gene cluster on chromosome 1p36, the U2 gene cluster on chromosome 17q21, and the major histone gene cluster on chromosome 6p21, in combination with immunofluorescent labeling with an antibody against p80-coilin to compare the distribution of the genes and coiled bodies. We found that 38% of the T24 cells contained coiled bodies located next to a histone gene cluster (Figure 4D), 56% next to a U2 snRNA gene cluster, and 25% next to a U1 snRNA gene cluster. These associations persisted throughout the cell cycle and did not appear to be specific for one particular stage. As a control, the dihydrofolate reductase gene at chromosome 5q12–13 was compared with coiled bodies and was not found associated with this nuclear domain in any of the 34 nuclei analyzed. In agreement with reports that coiled bodies do not contain DNA (Thiry, 1994, 1995), we never found any of the genes colocalizing with coiled bodies. Now that we had established that the snRNA and histone genes were often associated with coiled bodies in T24 cells, they seemed good candidates to be involved with cleavage bodies adjacent to coiled bodies.
The distribution of cleavage bodies was compared with the location of the genes using an antibody against CstF 64 kDa in combination with fluorescent in situ hybridization. Careful analysis of series of confocal sections of these double-labeled cells from a random culture revealed a frequent overlap of cleavage bodies with the histone genes but not with the snRNA genes. Approximately 37% of the nuclei displayed a partial or complete overlap between a histone gene cluster and a cleavage body (Figure 4, E and F). In cells synchronized in G1 by mitotic shake-off we did not observe this colocalization, which seemed to occur specifically in S phase cells. Conversely, no overlap between cleavage bodies and the U1 and U2 snRNA genes was detected in any of the 50 cells that were analyzed for each double labeling. We did sometimes observe cleavage bodies adjacent to the snRNA genes or at a small distance (<1.0 μm) (Figure 4, G and H), as can be expected for two domains that are both associated with coiled bodies. These findings reveal a specific relationship between cleavage bodies and the histone gene cluster adjacent to coiled bodies.
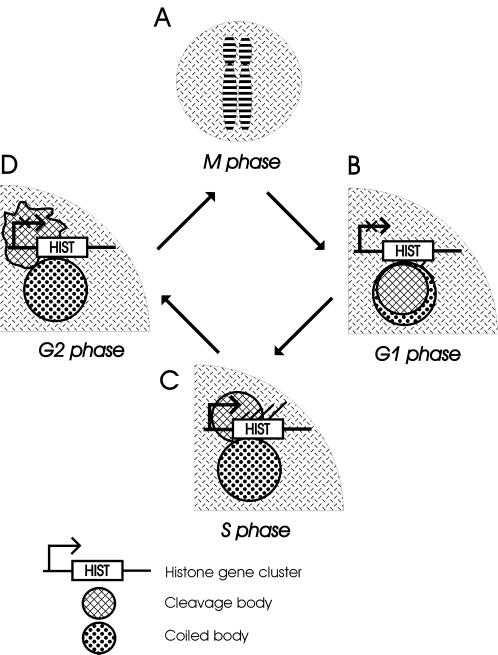
Taken together, the following picture of bodies and genes emerges (Figure 5). In G1 phase, cleavage bodies colocalize with coiled bodies and are often located next to histone gene clusters (Figure 5B). In S phase, cleavage bodies are preferentially located adjacent to coiled bodies (Figure 5C) and often overlap with a histone gene cluster but not with other coiled body–associated genes. Afterward, in G2 phase, the cleavage bodies appear to disintegrate (Figure 5D). It is well documented that the expression of the histone genes increases at the beginning of the S phase of the cell cycle. Our findings suggest a link between cleavage bodies and the cell cycle–regulated expression of histone genes.
Figure 5.
Overview of the various observations in the different stages of the cell cycle. (A) Cleavage bodies and coiled bodies are not observed during M phase. (B) In G1 phase the factors CstF 64 kDa and p80-coilin reenter the nucleus and concentrate together in overlapping cleavage bodies and coiled bodies. Histone gene clusters, inactive in G1 phase, are preferentially found adjacent to coiled bodies. (C) In S phase the cleavage bodies are mostly found adjacent to coiled bodies. They overlap with the histone gene clusters, which have become active in S phase. Newly synthesized RNA is found inside cleavage bodies adjacent to coiled bodies. (D) In G2 phase the histone genes have become inactive again. Cleavage bodies seem to desintegrate. Upon entry into M phase the cleavage bodies and coiled bodies have disappeared.
DISCUSSION
Cleavage bodies, enriched in the RNA 3′-processing factors CstF 64 kDa and CPSF 100 kDa, have a complex spatial relationship with coiled bodies. This relationship varies from close juxtaposition to complete colocalization (Schul et al., 1996). To investigate whether this is a cell cycle–related phenomenon, we performed double-labeling experiments on cells that were synchronized in different stages of the cell cycle. This analysis revealed that the cleavage bodies mainly colocalize with coiled bodies during G1 phase and are preferentially found adjacent to coiled bodies during S phase (Figure 5). Coiled bodies are known to disassemble during mitosis, and only small remnants can sometimes be seen in mitotic cells (Carmo-Fonseca et al., 1993). Cleavage bodies are also no longer observed during mitosis (Schul et al., 1996). The coiled body protein p80-coilin and the cleavage factors CstF and CPSF enter the nucleus in late telophase when the nuclear membrane has just reformed (Ferreira et al., 1994; Schul et al., 1996). However, coiled bodies are not immediately formed in early G1 phase and only appear after a lag period (Andrade et al., 1993; Carmo-Fonseca et al., 1993). Our data show that CstF 64 kDa already accumulates in these newly formed coiled bodies in G1 phase. At this point in the cell cycle coiled bodies and cleavage bodies probably constitute the same nuclear domain.
Cleavage bodies clearly formed separate domains that are adjacent to coiled bodies when cells entered S phase. Previously we have shown that cleavage bodies that are next to coiled bodies regularly contain newly synthesized RNA, suggesting a colocalization with active genes (Schul et al., 1996). We hypothesized that the shift of CstF 64 kDa and CPSF 100 kDa from the coiled body to a neighboring location could be caused by one or more active genes that recruit the cleavage factors from the coiled body (Schul et al., 1996). A good indication that gene activity is required for the juxtaposition of cleavage bodies and coiled bodies came from treating cells with transcription inhibitors. Without transcription, coiled bodies and cleavage bodies were always found to colocalize (Schul et al., 1996). Because cleavage bodies seem to be specifically associated with gene activity during S phase, it seems likely that the putative gene or genes inside cleavage bodies are only active in this stage of the cell cycle.
Histone genes are primarily active during S phase to supply the replicating DNA with new histones (Heintz, 1991; Stein et al., 1994). In humans the major histone genes are clustered on chromosomes 1q21 and 6p21 (Tripputi et al., 1986). The largest cluster, on chromosome 6, also includes the genes for the various histone H1 isotypes (Albig et al., 1993). Frey and Matera (1995) have recently shown that the two human histone gene clusters are frequently positioned next to coiled bodies in HeLa-ATCC and HEp-2 cells. In our experiments we found a similar spatial association in T24 cells. When the location of the histone gene cluster was compared with cleavage bodies, we found that they were mostly next to each other in G1 phase and overlapping during S phase. This cell cycle–dependent overlap might suggest a functional relationship between the cleavage bodies and the histone genes.
The U1 and U2 snRNA gene clusters, like the histone genes, have frequently been observed adjacent to coiled bodies (Frey and Matera, 1995; Smith et al., 1995). We observed this same spatial association in T24 cells. The U1 and U2 snRNA genes and the cleavage bodies were also often located close together. This is expected because all three are associated with coiled bodies. However, the snRNA genes and cleavage bodies never overlapped with each other. This indicates that the colocalization between the histone genes and cleavage bodies is not just a fortuitous overlap. It reveals a specific association between the cleavage bodies and the histone gene locus during the precise period that the histone genes are known to be highly transcribed. It is tempting to speculate that when cells enter S phase and histone synthesis peaks, the cleavage factors are recruited to the histone genes to fulfill a function there. It should be noted, however, that most histone mRNAs are not polyadenylated and therefore may not require the factors CstF and CPSF for 3′ end formation. There have been reports of histone mRNAs that do have a poly(A) tail, but their expression is not cell cycle dependent, and most of their genes are not located in the major histone gene cluster. However, the possibility that coiled bodies and cleavage bodies do have some role in histone mRNA synthesis, as suggested by our data, cannot be ruled out. Further studies will have to clarify this issue.
Earlier indications toward the involvement of coiled bodies in the production of histone mRNA have mainly come from studies on lampbrush chromosomes in the germinal vesicles of amphibian oocytes by Gall et al., (1981) and Callan et al. (1991). They showed that the histone gene loci on the lampbrush chromosomes of the newt Notophthalmus viridescens and the frog Xenopus laevis are associated with distinct round structures, 5–10 μm in diameter, known as spheres or sphere organelles but now thought to be coiled bodies (Gall et al., 1981; Callan et al., 1991). Importantly, histone transcripts were found to be produced from the chromatin loops immediately adjacent to the attached spheres. Although the spheres themselves only slowly incorporated tritiated uridine (Callan and Gall, 1991), histone transcripts were found closely associated with the periphery of the spheres. This intimate spatial organization of histone genes and distinct nuclear structures strongly suggests a functional relationship between the two. Moreover, it demonstrates that the association between coiled bodies and histone genes is evolutionary conserved from amphibians to humans, indicating a fundamental role of coiled bodies in histone gene expression (Frey and Matera, 1995). It should be noted that coiled bodies are enriched in U7 snRNA, which is essential for histone mRNA 3′ processing.The process of histone mRNA 3′ processing is not well understood, and little is known about the protein factors involved. A protein that binds to a conserved stem–loop structure at the 3′ ends of histone transcripts, called hairpin-binding factor (HBF) or stem–loop-binding protein 1 (SLBP1), and a less-characterized heat-labile factor have so far been identified (Gick et al., 1987; Melin et al., 1992; Wang et al., 1996). Recently, stem–loop-binding protein 1 has also been found inside coiled bodies (Abbott et al., 1999). Our results indicate an association among coiled bodies, cleavage bodies, and histone genes, but a rationale for this association remains unclear.
The spatial association of genes and specific nuclear domains, enriched in factors necessary for expression of these genes, seems to be a recurring principle of nuclear organization (for discussion, see Schul et al., 1998a). The most well-known example is the nucleolus where genes coding for rRNA are organized in and around specific nuclear compartments that are enriched in essential transcription and processing factors (Hernandez-Verdun, 1986). Similarly, Xing et al. (1993, 1995) have shown for several highly expressed genes that they are associated with the periphery of domains enriched in RNA polymerase II, polyadenylation, and splicing factors, known as nuclear speckles. For coiled bodies we have recently shown that they contain elevated levels of the transcription factors PTF and TBP, which are both essential for the transcription of the neighboring U1 and U2 snRNA genes (Schul et al., 1998b). These findings suggest that such nuclear domains are involved in facilitating and/or regulating gene expression by controlling the supply of factors and enzymes to specific genes. The results we present here on RNA 3′-processing factors being redistributed from coiled bodies to adjacent genes in conjunction with gene expression agree with this theory. It is still unclear, however, what the molecular basis is of the association between genes and the various nuclear domains (Misteli and Spector, 1998).
Our data on the behavior of cleavage bodies, coiled bodies, and specific genes during different stages of the cell cycle have revealed a dynamic spatial and functional relationship between these components. Gene expression and cell proliferation appear to play an important role in this relationship. In addition, these findings provide indications that the general RNA 3′-processing factors CstF 64 kDa and CPSF 100 kDa may be involved in the regulation or facilitation of histone gene expression.
ACKNOWLEDGMENTS
We thank Carel van Oven for assistance with the flow cytometry measurements, Dr. R. Dirks for help and advice with in situ hybridization, Dr. A.M. Weiner for supplying the genomic clones, and Drs. Y. Takagaki, J.L. Manley, and A.I. Lamond for supplying the antibodies.
REFERENCES
- Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albig W, Drabent B, Kunz J, Kalff-Suske M, Grzeschik KH, Doenecke D. All known human H1 histone genes except the H1(0) gene are clustered on chromosome 6. Genomics. 1993;16:649–654. doi: 10.1006/geno.1993.1243. [DOI] [PubMed] [Google Scholar]
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Chan EKL, Raška I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80-coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan HG, Gall JG. Association of RNA with the B and C snurposomes of Xenopus oocytes nuclei. Chromosoma. 1991;101:69–82. doi: 10.1007/BF00357056. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis—evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong L, Grande MA, Mattern KA, Schul W, van Driel R. Nuclear domains involved in RNA synthesis, RNA processing and replication. Crit Rev Eukaryot Gene Expr. 1996;6:215–246. doi: 10.1615/critreveukargeneexpr.v6.i2-3.60. [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W. Localization of rapidly and slowly labeled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res. 1971;67:129–141. doi: 10.1016/0014-4827(71)90628-8. [DOI] [PubMed] [Google Scholar]
- Ferreira J, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MH, Read RA, Bedford JS. Comparison of synchronized chinese hamster ovary cells obtained by mitotic shake-off, hydroxyurea, aphidicolin, or methotrexate. Cytometry. 1987;8:315–320. doi: 10.1002/cyto.990080312. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17pq11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O, Krämer A, Vasserot A, Birnstiel ML. Heat-labile regulatory factor is requried for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- Grdina DJ, Meistrich ML, Meyn RE, Johnson TS, White RA. Cell synchrony techniques. I. A comparison of methods. Cell Tissue Kinet. 1984;17:223–236. doi: 10.1111/j.1365-2184.1984.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Structural organization of the nucleolus in mammalian cells. Methods Achiev Exp Pathol. 1986;12:26–62. [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez MdeL, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleogenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Cunha C, Carmo-Fonseca M. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol Biol Cell. 1997;8:1207–1217. doi: 10.1091/mbc.8.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodine staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Langer PR, Waldrop AA, Ward DC. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci USA. 1981;78:6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Manders EMM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Melin L, Soldati D, Mital R, Streit A, Schümperli D. Biochemical demonstration of complex formation of histone premRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus is some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Moreno Diaz de la Espina S, Risueno M, Medina F. Ultrastructural, cytochemical and autoradiographic characterization of coiled bodies in the plant cell nucleus. Biol Cell. 1982;44:229–238. [Google Scholar]
- Raška I, Andrade LEC, Ochs RL, Chan EKL, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Raška I, Ochs RL, Andrade LEC, Chan EKL, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Rigby PWJ, Dieckmann M, Rhodes C, Berg P. Labeling DNA to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbors: The spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998a;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schul W, Groenhout B, Koberna K, Takagaki Y, Jenny A, Manders EMM, Raška I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell. 1998b;9:1025–1036. doi: 10.1091/mbc.9.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, van Wijnen AJ, Lian JB. Histone gene transcription: a model for responsiveness to an integrated series of regulatory signals mediating cell cycle control and proliferation/differentiation interrelationships. J Cell Biochem. 1994;54:393–404. doi: 10.1002/jcb.240540406. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian premRNAs. Genes & Dev. 1990;3:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Thiry M. Cytochemical and immunocytochemical study of coiled bodies in different cultured cell lines. Chromosoma. 1994;103:268–276. doi: 10.1007/BF00352251. [DOI] [PubMed] [Google Scholar]
- Thiry M. Nucleic acid compartmentalization within the cell nucleus by in situ transferase-immunogold techniques. Microsc Res Techn. 1995;31:4–21. doi: 10.1002/jemt.1070310103. [DOI] [PubMed] [Google Scholar]
- Tobey RA, Valdez JG, Crissman HA. Synchronization of human diploid fibroblasts at multiple stages of the cell cycle. Exp Cell Res. 1988;179:400–416. doi: 10.1016/0014-4827(88)90279-0. [DOI] [PubMed] [Google Scholar]
- Tripputi P, Emanuel BS, Croce CM, Green LG, Stein GS, Stein JL. Human histone genes map to multiple chromosomes. Proc Natl Acad Sci USA. 1986;83:3185–3188. doi: 10.1073/pnas.83.10.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Balen R, ten Kate T, Koelma D, Mosterd B, Smeulders AWM. Scilimage: a multi-layered environment for use and development of image processing software. In: Christensen HI, Crowley LL, editors. Experimental Environments for Computer Vision and Image Processing. Singapore: World Scientific Press; 1994. pp. 107–126. [Google Scholar]
- Van der Voort HTM, Straster K. Restoration of confocal images for quantitative image analysis. J Microsc. 1995;178:165–181. [Google Scholar]
- Wahle E, Kühn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic premRNA. Prog Nucl Acid Res Mol Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone premRNA processing. Genes & Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wu CHH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Carol VJ, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific premRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoni YM, White K. Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma. 1997;105:332–341. doi: 10.1007/BF02529748. [DOI] [PubMed] [Google Scholar]
- Zwanenburg TSB. Standardized shake-off to synchronize cultured CHO cells. Mutat Res. 1983;120:151–159. doi: 10.1016/0165-7992(83)90157-4. [DOI] [PubMed] [Google Scholar]