Abstract
The nucleolar localization elements (NoLEs) of U17 small nucleolar RNA (snoRNA), which is essential for rRNA processing and belongs to the box H/ACA snoRNA family, were analyzed by fluorescence microscopy. Injection of mutant U17 transcripts into Xenopus laevis oocyte nuclei revealed that deletion of stems 1, 2, and 4 of U17 snoRNA reduced but did not prevent nucleolar localization. The deletion of stem 3 had no adverse effect. Therefore, the hairpins of the hairpin–hinge–hairpin–tail structure formed by these stems are not absolutely critical for nucleolar localization of U17, nor are sequences within stems 1, 3, and 4, which may tether U17 to the rRNA precursor by base pairing. In contrast, box H and box ACA are major NoLEs; their combined substitution or deletion abolished nucleolar localization of U17 snoRNA. Mutation of just box H or just the box ACA region alone did not fully abolish the nucleolar localization of U17. This indicates that the NoLEs of the box H/ACA snoRNA family function differently from the bipartite NoLEs (conserved boxes C and D) of box C/D snoRNAs, where mutation of either box alone prevents nucleolar localization.
INTRODUCTION
The processing and modification of the ribosomal RNA precursor (pre-rRNA) in the nucleoli of eukaryotic cells is accomplished by a large number of small nucleolar RNAs (snoRNAs) complexed with proteins in ribonucleoprotein particles (snoRNPs). The transport of snoRNAs from their nucleoplasmic sites of transcription to their site of function, the nucleolus, is a prerequisite for ribosome biosynthesis. Although cues that direct some snoRNAs to the nucleolus are beginning to be elucidated, nothing is known about the signals that localize snoRNAs of the box H/ACA family to nucleoli, a question that is addressed in the present report.
The first of the three families of snoRNAs is characterized by two phylogenetically conserved sequences, boxes C and D. Only a few snoRNAs of the box C/D family are essential for cell growth because of their participation in rRNA processing (reviewed by Gerbi, 1995; Maxwell and Fournier, 1995; Sollner-Webb et al., 1995; Venema and Tollervey, 1995). Most box C/D snoRNAs are nonessential and are used as guide RNAs to direct 2′-O-ribose methylation in rRNA (Cavailléet al., 1996; Kiss-Lászlóet al., 1996, 1998; Maden, 1996; Maden and Hughes, 1997; Nicoloso et al., 1996; Tollervey, 1996; Tycowski et al., 1996; Smith and Steitz, 1997; Lowe and Eddy, 1999) and snRNA (Tycowski et al., 1998). Within this family, boxes C and D are the cis-acting nucleolar localization elements (NoLEs), which direct box C/D snoRNA molecules from the nucleoplasm to the nucleolus (Lange et al., 1998b,c; Samarsky et al., 1998); controversy exists about the importance of box C′ as a NoLE in U3 snoRNA (Narayanan et al., 1999). Box D is also important for 5′-cap hypermethylation and nuclear retention of U3 box C/D snoRNA (Terns et al., 1995). Furthermore, boxes C and D are required for the splicing of intronic box C/D snoRNAs, such as U14, from the host RNA (Watkins et al., 1996; Xia et al., 1997), an event that occurs in the nucleoplasm (Samarsky et al., 1998).
A second (minor) family of snoRNAs is composed of only two species: 7-2/MRP snoRNA and the RNA component of RNase P. In 7-2/MRP snoRNA, which is essential for 5.8S rRNA processing (Schmitt and Clayton, 1993; Chu et al., 1994; Lygerou et al., 1996), nucleotides 23–62, which contain the To antigen binding site, are required for nucleolar localization (Jacobson et al., 1995). The nucleolar localization of the ribonucleoprotein enzyme RNase P, which catalyzes the 5′ processing of pre-tRNA, is also mediated at least in part by the nucleolar To antigen binding site and RNase P-associated proteins (Jacobson et al., 1997; Bertrand et al., 1998; Jarrous et al., 1999).
The third family of snoRNAs is characterized by two evolutionarily conserved sequences: box H (ANANNA) and box ACA. Both sequences are required for accumulation and stability of box H/ACA snoRNAs in yeast cells (Balakin et al., 1996; Bousquet-Antonelli et al., 1997; Ganot et al., 1997b). Some snoRNAs of the box H/ACA family are essential for rRNA processing (snR10 [Tollervey, 1987], snR30 [Morrissey and Tollervey, 1993], and E1 = U17, E2, and E3 [Mishra and Elicieri, 1997]), but the majority function as guide RNAs for pseudouridine modifications in rRNA (Ganot et al., 1997a; Ni et al., 1997; Smith and Steitz, 1997). All box H/ACA snoRNAs possess a characteristic hairpin–hinge–hairpin–tail secondary structure with the single-stranded hinge region containing box H and the single-stranded tail containing box ACA (Balakin et al., 1996; Ganot et al., 1997b). To target pseudouridylation, a bulge structure within one or both hairpins base pairs with the rRNA on either side of the substrate uridine, forming a modification pocket. Either box H or the box ACA motif is located 14–16 nucleotides (nt) downstream of this pocket (Ganot et al., 1997a; Ni et al., 1997; Smith and Steitz, 1997). Several proteins have been reported to associate with box H/ACA snoRNAs in yeast: Gar1p (Girard et al., 1992; Balakin et al., 1996; Bousquet-Antonelli et al., 1997; Ganot et al., 1997b), the putative rRNA pseudouridine synthase Cbf5p (Nap57/dyskerin) (Jiang et al., 1993; Meier and Blobel, 1994; Cadwell et al., 1997; Lafontaine et al., 1998), Nhp2p, and Nop10p (Kolodrubetz and Bergum, 1991; Henras et al., 1998). All of these proteins with the exception of Gar1 are required for accumulation and stability of box H/ACA snoRNAs in yeast (Bousquet-Antonelli et al., 1997; Henras et al., 1998, and references therein). To date, however, it is unknown if any of these proteins contribute to box H/ACA snoRNA transport to the nucleolus nor have the structural requirements within box H/ACA snoRNAs been studied that are essential for nucleolar localization.
In the present report we have employed an assay previously used to analyze NoLEs of box C/D snoRNA (Lange et al., 1998a–c) to study the localization of a box H/ACA snoRNA. U17 snoRNA is one of the most abundant box H/ACA snoRNAs (Pelczar and Filipowicz, 1998) and is essential for the cleavage of pre-rRNA within the 5′ external transcribed spacer, resulting in the formation of 18S rRNA (Enright et al., 1996; Mishra and Elicieri, 1997). U17 snoRNA is of intronic origin and has been characterized from various vertebrate organisms (Kiss and Filipowicz, 1993; Nag et al., 1993; Rimoldi et al., 1993; Ruff et al., 1993; Cecconi et al., 1996) including Xenopus laevis (Cecconi et al., 1994, 1995; Selvamurugan et al., 1997). The secondary structure of U17 is similar to that of guide RNAs with hairpin structures flanking the single-stranded box H region within the molecule and single-stranded box ACA at the 3′ end (Cecconi et al., 1994, 1996; Selvamurugan et al., 1997) (Figure 1, top).
Figure 1.
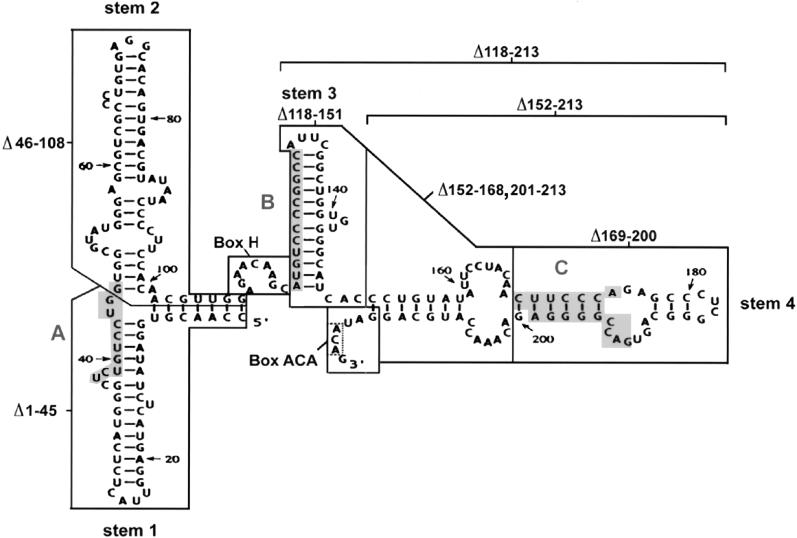
Structure and mutations of U17 snoRNA. The sequence and structure of Xenopus laevis U17 snoRNA (copy f), found in the sixth intron of the gene for ribosomal protein S7 (formerly S8), is from Cecconi et al. (1994), with sequence corrections at positions 33 (G instead of A), 91 and 161 (additional U). Shaded areas A and B in stems 1 and 3 of U17 are complementary to sequences in 18S rRNA (Cecconi et al., 1994). Shaded area C in stem 4 is complementary to the external transcribed spacer of pre-rRNA (Cecconi et al., 1994). The regions of U17 that were mutated in the present study are enclosed by lines. Box ACA, consisting of three nucleotides (enclosed by a dotted line), lies within the single-stranded 3′ end of the molecule. The sequences of the mutants of U17 designed for the present study are listed in the lower portion; nucleotides that are the same as in wild-type U14 snoRNA are shown by dots in the sequence alignment, and deletions are indicated by dashes. The double mutants Δbox H/Δbox ACA+, Δbox H/Δbox ACA, sub. box H/sub. box ACA+, Δ46–108/Δbox H, and Δ46–108/Δ152–213 are not shown, because they are simply the sum of the individual single mutations.
The present study shows that box H and the box ACA region but not the hairpins of the hairpin–hinge–hairpin–tail secondary structure are essential NoLEs of U17; the combined substitution or deletion of those two single-stranded areas but not of either box alone abolished nucleolar localization. These two elements presumably act by binding protein(s) that either transport the snoRNA from the nucleoplasm to the nucleolus and/or anchor it within the nucleolus, whereas direct U17 snoRNA–rRNA interactions do not appear to be critical for nucleolar localization. The integrity of the hairpin–hinge–hairpin–tail structure contributes to nucleolar localization, probably by assisting the major NoLEs, box H, and box ACA. This is the first identification of nucleolar localization sequences for a member of the box H/ACA snoRNA family.
MATERIALS AND METHODS
Plasmid Constructs
U17 templates for in vitro transcription reactions were constructed by PCR using the primers listed below. Plasmid pU17f′ containing U17 snoRNA copy f from intron 6 of the X. laevis gene for ribosomal protein S7 (formerly S8; Cecconi et al., 1994) served as the template for the PCR reactions.
U17 5′-End Primers (T7 promotor shown in italics).
Wild type, 5′-TAA TAC GAC TCA CTA TAG GGC CAA CGT GGA TAT CTC ATG-3′;
Δ1–45, 5′-TAA TAC GAC TCA CTA TAG GGG TGG CGT ATG GGA GCG-3′;
Δ46–108, 5′-TAA TAC GAC TCA CTA TAG GGC CAA CGT GGA TAT CTC ATG AGG TTA CTC TCA TGG GCT CTG TCC TGA GAA CAA GCA TGT CC-3′;
Δ46–108/Δbox H, 5′-TAA TAC GAC TCA CTA TAG GGC CAA CGT GGA TAT CTC ATG AGG TTA CTC TCA TGG GCT CTG TCC TGA TGT CCC CGG CCA TTC-3′.
U17 3′-End Primers (substitutions shown by lowercase letters).
Wild type, 5′-CTG TAT CCT GCA TGG TTT-3′;
sub. box H, 5′-CTG TAT CCT GCA TGG TTT GTC TCC CCG GTC ACT GCC CGA GGG CTC TGG GAA GTT GTA GGA ATA TAC AGG GTG ATG CCC ACA CCA GCC GAA TGG CCG GGG ACA Tca cca cca cCC AAC GTT GTG GAA GG-3′;
Δbox H, 5′-CTG TAT CCT GCA TGG TTT GTC TCC CCG GTC ACT GCC CGA GGG CTC TGG GAA GTT GTA GGA ATA TAC AGG GTG ATG CCC ACA CCA GCC GAA TGG CCG GGG ACA TCC AAC GTT GTG GAA GG-3′;
Δ118–213, 5′-CTG TAT CGC TTG TTC TCC AAC GTT-3′;
Δ118–151, 5′-CTG TAT CCT GCA TGG TTT GTC TCC CCG GTC ACT GCC CGA GGG CTC TGG GAA GTT GTA GGA ATA TAC AGG GCT TGT TCT CCA ACG TTG-3′;
Δ152–213, 5′-CTG TAT CGT GAT GCC CAC ACC AGC CG-3′;
Δ169–200, 5′-CTG TAT CCT GCA TGG TTT GTT TGT AGG AAT ATA CAG GGT-3′;
Δ152–168,201–213, 5′-CTG TAT CCT CCC CGG TCA CTG CCC GAG GGC TCT GGG AAG GTG ATG CCC ACA CCA GCC-3′;
sub. box ACA, 5′-Caa gAT CCT GCA TGG TTT G-3′;
sub. box ACA+, 5′-gaa gga aCT GCA TGG TTT GTC TCC C-3′;
Δbox ACA+, 5′-CCT GCA TGG TTT GTC T-3′;
Δbox ACA, 5′-CAT CCT GCA TGG TTT GTC-3′.
For PCR mutagenesis of a given U17 mutant, one of the mutant primers listed above was used in combination with the wild-type primer at the other end.
For the double mutation “Δbox H/Δbox ACA+,” the PCR construct “Δbox H” served as the template, and the wild-type 5′ primer and Δbox ACA+ 3′ primer were used. For the double mutation “sub. box H/sub. box ACA+,” the PCR construct “sub. box H” served as the template, and the wild-type 5′ primer and sub. box ACA+ 3′ primer were used. For the double mutation “Δ46–108/Δ152–213,” the PCR construct “Δ46–108” served as the template, and the Δ46–108 5′ primer and the Δ152–213 3′ primer were used. The wild-type as well as all the mutant PCR products were cloned into pCR3.1 (Invitrogen, Carlsbad, CA), and their sequences were confirmed, with the exception of constructs “Δ169–200” and “Δ152–168,201–213,” which, however, were created by using the sequenced wild-type clone and wild-type 5′ primer as well as the appropriate 3′ primers listed above. The “Δbox H/Δbox ACA” mutation was created by using the sequenced clone of the Δbox H/Δbox ACA+ mutation as a template, and the wild-type 5′ primer and the Δbox ACA 3′ primer were used.
For the stability assays described below, we used U14 snoRNA transcripts from the murine hsp70 intron 5 (Liu and Maxwell, 1990; Leverette et al., 1992) as an internal control. Wild-type U14 template was derived by PCR from plasmid pSP64T7 using the following primers:
U14 5′-End Primer.
5′-TAA TAC GAC TCA CTA TAG GGT TCG CTG TGA TGA TGG ATT CCA AAA-3′.
U14 3′-End Primer.
5′-TTC GCT CAG ACA TC-3′.
U2 snRNA was used as a control in stability as well as localization assays. Plasmid pXlU2 that contains the X. laevis U2 snRNA gene (Mattaj and Zeller, 1983) served as the template for PCR to add the T7 promotor sequence by using the following primers:
U2 5′-End Primer.
5′-TAA TAC GAC TCA CTA TAG GGA TCG CTT CTC GGC CTT TTG GC-3′.
U2 3′-End Primer.
5′-AAG TGC ACC GGT CCT GGA GG-3′.
In Vitro Transcription and Labeling of RNA
All transcripts were obtained using a T7 megascript in vitro transcription kit (Ambion, Austin, TX) according to the method of Lange et al. (1998b) with an incubation time of 4 h at 37°C. In contrast to some non-intronic snoRNAs that normally contain a monomethyl G cap, which is subsequently converted to a trimethyl G cap (Terns and Dahlberg, 1994; Terns et al., 1995), U17 snoRNA is processed from the intron of another gene and lacks a 5′ cap. However, because we observed a higher degradation of in vitro-transcribed U17 with an unprotected 5′ end after injection into oocytes (our unpublished results), stability of the transcripts was improved by capping the 5′ ends with the m7G(5′)ppp(5′)G cap analogue (Ambion). Previously it was shown for intronic as well as non-intronic snoRNAs that the presence or absence of a cap did not affect nucleolar localization of a given snoRNA in Xenopus oocyte nucleoli (Lange et al., 1998a–c). After the 5′ addition of a monomethyl G, all mutated U17 snoRNAs were sufficiently stable to be within the range of concentrations that would be detectable by fluorescence microscopy if they had localized to nucleoli (see Figures 2 and 5). The transcripts were purified (Lange et al., 1998b), and their integrity was confirmed by 8% polyacrylamide, 8 M urea gel electrophoresis. The amount of fluorescent transcript was determined by spectrophotometry at 260 nm. In addition, wild-type and mutated snoRNA transcripts were run on the same gel, and their fluorescence intensity as well as their staining by methylene blue were compared and adjusted accordingly for injection of equivalent amounts.
Figure 2.
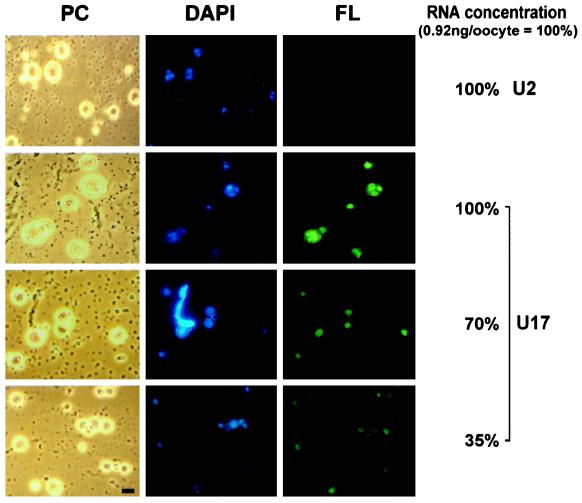
Nucleolar localization of wild-type U17 snoRNA injected into X. laevis oocytes. Fluorescein-labeled wild-type U17 snoRNA or spliceosomal U2 snRNA as a control was injected into the nuclei of X. laevis oocytes. After 1.5 h, nucleoli were prepared and analyzed by phase-contrast (PC) or fluorescence (FL) microscopy. The nucleolar rDNA is stained (DAPI, blue). Injection at an amount of 0.9 ng per oocyte (100%) resulted in a strong nucleolar labeling by U17 snoRNA but not by U2 snRNA (FL, green). After dilution of U17, even 35% of this amount yields detectable nucleolar labeling 1.5 h after injection. Oocyte nucleoli vary in size (Wu and Gall, 1997) and can fuse into multinucleolar clusters (Shah et al., 1996). A lampbrush chromosome is visible by DAPI staining (see PC and DAPI panels for 70% of U17 injected). Bar, 10 μm.
Figure 5.
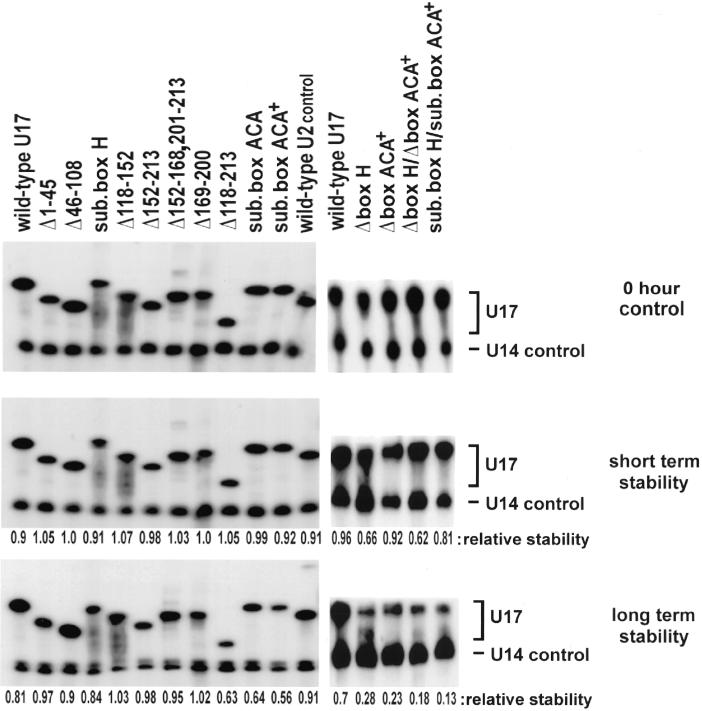
Stability of wild-type and mutated U17 snoRNA. 32P-labeled U17 snoRNA (mutants or wild-type) were injected into oocyte nuclei, and the RNAs were isolated and analyzed by gel 8% polyacrylamide, 8 M urea gel electrophoresis. The upper panel shows controls (sample recovery immediately after injection, 0 h), the middle panel shows short-term stability at 1.5 h (the time when localization assays were carried out), and the lower panel shows long-term stability (left gel at 15 h, right gel at 16 h) of U17 snoRNA. 32P-labeled U14 snoRNA (lower band) was coinjected as an internal control to normalize for any differences in injection or recovery of the samples. The relative stability was calculated as [U17/U14 after incubation]/[U17/U14 at 0 h]. The ratio of U17/U14 shows that all the mutants are stable at the 1.5-h time point used for analysis of nucleolar localization (middle panel).
Oocyte Microinjections and Fractionation
A portion of the ovary was surgically removed from female X. laevis (Nasco, Fort Atkinson, WI) following National Institutes of Health– and Institutional Animal Care and Use Committee–approved procedures and transferred to OR2 saline buffer (Wallace et al., 1973). Single oocytes were obtained by digesting the connective tissue with collagenase type I and II (3000 U/ml each; Sigma, St. Louis, MO) in OR2 for 2 h at room temperature. Stage V oocyte nuclei were injected 16–40 h after isolation (Nanoject; Drummond, Broomall, PA). For fluorescence analysis of snoRNA nucleolar localization as well as for stability assays, oocyte nuclei were injected with in vitro-transcribed RNA in H2O (0.1 μg/μl stock solution; 9.2 nl injected = 0.92 ng/oocyte). To distinguish elements needed for nucleolar localization from those used for intronic processing of U17, the mature form of Xenopus U17 snoRNA was injected into oocyte nuclei. After subsequent incubation for 1.5 or 15–16 h at 20°C, oocytes were transferred to an isolation buffer containing 83 mM KCl, 17 mM NaCl, 1 mM MgCl2, 6.5 mM Na2HPO4, and 3.5 mM KH2PO4, pH 7.5, and the nuclear envelopes were manually removed.
Nucleolar Localization Assay
Following a method for preparation of lampbrush chromosomes (Gall et al., 1991), the nuclear contents of one oocyte were dispersed in a chamber on a slide containing a solution of 20.75 mM KCl, 4.25 mM NaCl, 0.5 mM MgCl2, 10 μM CaCl2, 0.1% paraformaldehyde, 6.5 mM Na2HPO4, and 3.5 mM KH2PO4, pH 7.2. The slides were centrifuged at 4000 × g for 40 min at 4°C, incubated in 2% paraformaldehyde in PBS (137 mM NaCl, 3 mM KCl, 6.4 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.0) for 1 h and washed once in 100% ethanol and 0.3 M ammonium acetate. DNA in the nucleoli was stained by adding 20 ng/ml 4’-6-diamidino-2-phenylindole (DAPI) in PBS for 5 min. For fluorescence analysis, a Zeiss (Thornwood, NY) Axiophot Epifluorescence microscope equipped with a 100× Neofluar Ph 3 objective and a 100-W mercury lamp and Zeiss filter sets 48-7709 (for fluorescein) and 48-7702 (for DAPI) were used. Nucleolar preparations were embedded in PBS under a coverslip, and pictures were taken with constant exposures for each filter set using Ektachrome 400x professional film (Eastman Kodak, Rochester, NY).
snoRNA Stability Assay
To determine the stability of the various U17 snoRNA constructs after injection into oocyte nuclei, wild-type U14 snoRNA transcripts were coinjected and served as an internal control to normalize for any differences in injection or recovery of the samples. One and one-half hours after injection of the oocytes with [α-32P]UTP-labeled wild-type U14 and U17 mutants, the RNA of five oocytes per sample was recovered and analyzed as described previously (Lange et al., 1998a,b). For any given mutation, the ratio of U17/U14 between 1.5 h (or 15–16 h for long term stability) and 0 h (sample recovery immediately after injection) determines the relative stability of a U17 mutant compared with wild-type U17.
RESULTS
Detection of U17 snoRNA Localization to Nucleoli
Nucleolar localization of U17 snoRNA was monitored by direct visualization of nucleolar preparations after injection of fluorescein-labeled in vitro transcripts of U17 into Xenopus oocyte nuclei. We previously used this technique to analyze the NoLEs of box C/D snoRNAs (Lange et al., 1998a–c). One and one-half hours after injection with labeled U17 transcripts, oocyte nuclei were manually dissected, and the nuclear envelope was removed. Subsequently, the nuclear contents including chromosomes, nucleoli, and coiled bodies (snurposomes) were centrifuged onto a microscope slide. This technique is valuable because it permits a direct qualitative assessment of nucleolar localization of the labeled RNA. As shown in Figure 2, strong fluorescent signals depicting nucleolar localization of wild-type U17 snoRNA were seen 1.5 h after injection of 0.9 ng of transcript per oocyte nucleus. In favorable preparations, signals were found in ring-like structures within the nucleoli (e.g., Figure 2). These structures appear to correspond to the dense fibrillar component of nucleoli, which surrounds the rDNA-containing fibrillar center (Shah et al., 1996). This supposition was supported by DAPI staining of DNA, located in the center of the labeled areas.
The nucleolar localization of fluorescent U17 snoRNA was specific, because injection of the same concentration (0.9 ng/oocyte) of U2, a small nuclear RNA that is part of the splicing machinery, did not give nucleolar signals (Figure 2). Additional controls demonstrated that the fluorescent signals we observed were not due to degradation of fluorescent snoRNA and subsequent reuse of the label by other nuclear components. For example, as published previously (Lange et al., 1998b,c), injection of a 200-fold molar excess of fluorescein-UTP alone did not label the nucleoli. Moreover, stability assays of 32P-labeled wild-type and mutant U17 snoRNA transcripts (summarized below) demonstrated the short-term stability of all mutants at 1.5 h after injection into oocyte nuclei (the time at which the localization assays were carried out). To determine the amount of fluorescent U17 transcripts necessary for reliable detection, a dilution series was carried out: it revealed that 35% of the original amount of wild-type U17 snoRNA (0.9 ng/oocyte) can still be detected (Figure 2). This also indicates that should a mutant U17 snoRNA be three times less stable than the wild-type U17 at 1.5 h after injection, it would still be sufficient in amount to be detected in this localization assay.
Small nucleoli can be distinguished from coiled bodies (snurposomes) by the presence of DAPI staining, because only nucleoli contain DNA (Wu and Gall, 1997). Fluorescent U17 snoRNA was not observed to localize to coiled bodies at the time point (1.5 h) when the assay was carried out. Similarly, another box H/ACA snoRNA, U65, also failed to localize to coiled bodies 15–240 min after oocyte injection, although members of the box C/D snoRNA family seem to traffic through coiled bodies (Narayanan et al., 1999). In addition to not staining coiled bodies, U17 does not stain lampbrush chromosomes (Figure 2). In summary, these results indicate that nucleolar localization of U17 snoRNA is specific and therefore mediated by defined intrinsic features within the molecule, such as unique structures and/or defined sequences.
Are the Hairpin Structures Essential for Nucleolar Localization of U17 snoRNA?
To define the elements of U17 snoRNA necessary for nucleolar localization, the localization of mutant transcripts was compared with that of wild-type U17. Figure 1 summarizes the various U17 mutants designed for the present study as well as the sequence of mature wild-type U17 snoRNA of Xenopus U17 snoRNA (Cecconi et al., 1994).
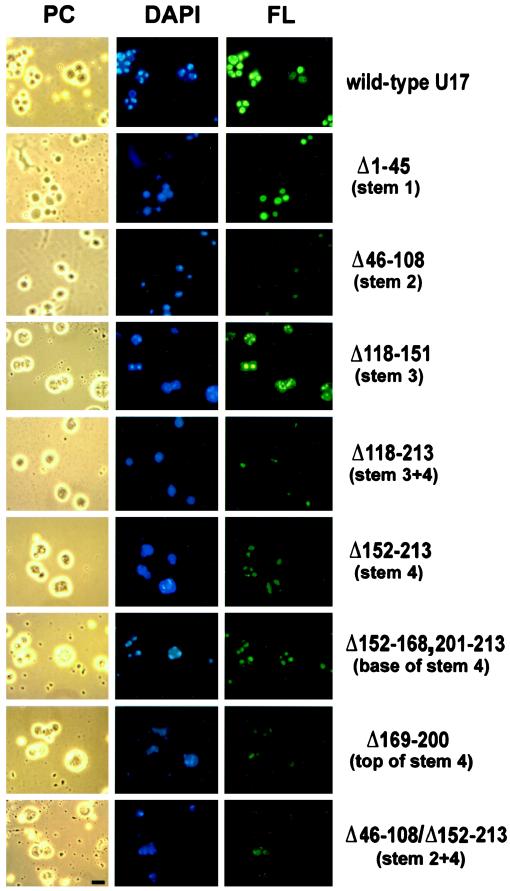
As described in INTRODUCTION, box H/ACA snoRNAs have a characteristic secondary structure (hairpin–hinge–hairpin–tail). We tested whether the hairpins flanking box H are important for nucleolar localization by deleting the entire stem 2 or stem 3 (Figure 1). Also, mutants with a deletion of stem 4 alone or in combination with stem 3 were tested, because the two hairpins may form a functional unit by separating box H from the ACA region (Figure 1). The analysis revealed that deletion of stem 3 did not affect nucleolar localization (Figure 3; Δ118–151). The deletion of stem 2 (Δ46–108), stem 4 (Δ152–213), or the stem3/stem4 structure (Δ118–213) resulted in reduced nucleolar localization but did not abolish it. This indicates that the hairpins of the hairpin–hinge–hairpin–tail structure of U17 snoRNA are helpful but not critical for nucleolar localization. This conclusion is supported by the observation that even after the combined deletion of both stems 2 and 4, which individually contribute somewhat to nucleolar localization, U17 localization was still detectable (Figure 3, Δ46–108/Δ152–213).
Figure 3.
Nucleolar localization of U17 snoRNA after deletion of sequences with stem structures and rRNA binding sites. Fluorescein-labeled U17 snoRNA was injected into the nuclei of X. laevis oocytes at an amount of 0.9 ng per oocyte. After 1.5 h, nucleolar preparations were analyzed by phase-contrast (PC) or fluorescence (FL) microscopy; the nucleolar rDNA was stained (DAPI, blue). U17 snoRNA carrying a deletion of stem 3 (Δ118–151) localized as well to nucleoli as the wild-type molecule (FL, green). U17 deleted in stem 1 (Δ1–45) localized strongly to nucleoli, and deletions of stem 2 (Δ46–108), stem 4 (Δ152–213), or a combination of both (Δ46–108/Δ152–213) showed significantly less but not abolished localization. Dissection of stem 4 (Δ169–200 or Δ152–168,201–213) did not reveal any major site of importance for nucleolar localization. The deletion of the entire structure of stems 3 and 4 between the single-stranded regions of conserved boxes H and ACA (Δ118–213) reduced but did not completely abolish nucleolar localization. Bar, 10 μm.
Another important question is whether elements of U17 snoRNA that may tether it to pre-rRNA by base pairing (see Figure 1, shaded areas A, B, and C in stems 1, 3, and 4) also have a role in localization. Deletion of stem 1 (containing area A) led to a somewhat reduced signal but did not abolish nucleolar localization of U17 (Figure 3, Δ1–45). As noted above, deletion of stem 3 (containing area B) did not affect U17 localization (Δ118–151), and deletion of stem 4 (Δ152–213) (containing area C) only reduced but did not abolish nucleolar localization. Also, the deletion of just the top part of stem 4 (Δ169–200) with the complementary sequence to the ETS of pre-rRNA did not exert any stronger effect on nucleolar localization of U17 than the deletion of the bottom half of stem 4 (Δ152–168,201–213). Even deletion of the entire stem3/stem4 structure (Δ118–213) did not appreciably perturb nucleolar localization. Therefore, we conclude that direct snoRNA–rRNA interactions are not critical for the localization of U17 snoRNA to nucleoli.
Role of Evolutionarily Conserved Box H and Box ACA for Nucleolar Localization of U17 snoRNA
Recently, we defined the NoLEs of box C/D snoRNAs, showing that the conserved boxes C and D are the cis-acting and primary NoLEs for this family of snoRNAs (Lange et al., 1998b,c). By analogy, it could be hypothesized for snoRNAs of the box H/ACA family that the evolutionary conservation of specific sequences might reflect their function in nucleolar localization. To address this question, we designed several mutations in conserved regions of U17 snoRNA, namely boxes H and ACA, to be tested in the nucleolar localization assay.
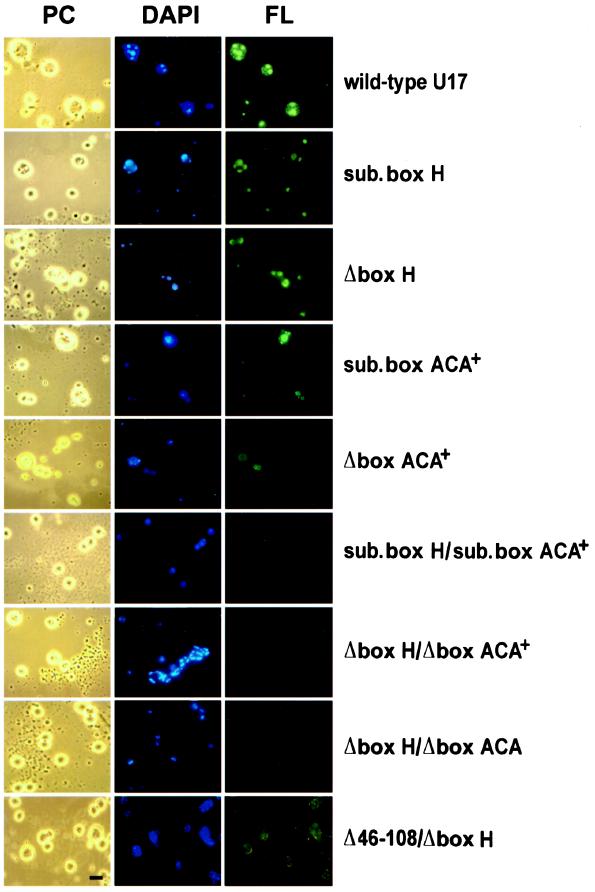
As can be seen in Figure 4, neither the substitution (sub. box H) nor the deletion of the box H region alone (Δbox H) significantly hindered the localization of mutant U17 transcripts to nucleoli. Mutations of the single-stranded 3′-tail region carrying the conserved box ACA generally reduced but did not abolish U17 localization to nucleoli (Figure 4, sub. box ACA+ and Δbox ACA+). In contrast to the mutations of box H, however, box ACA mutations showed some variability in signals. Figure 4 shows an example in which some nucleoli after injection of the mutant with a substituted box ACA region (sub. box ACA+) are stained weakly and one nucleolus is stained strongly. Similarly, the mutant with a deleted ACA region (Δbox ACA+) showed some stained and some unstained nucleoli. Variable results were also observed for U17 snoRNA with a substitution of the three nucleotides constituting box ACA (our unpublished results).
Figure 4.
Role of evolutionarily conserved box H and box ACA in nucleolar localization of U17 snoRNA. U17 snoRNA carrying a substitution (sub. box H) or deletion (Δbox H) of just conserved box H alone retained the ability to localize to nucleoli, resulting in moderate to strong nucleolar labeling. Nucleolar localization signals with mutants substituted in box ACA, deleted in the ACA region (Δbox ACA+), or substituted in the ACA region (sub. box ACA+) were highly variable. The double mutants of box H and the box ACA region, being either depleted (Δbox H/Δbox ACA+) or substituted (sub. box H/sub. box ACA+) in both sequences, were not capable of localizing to nucleoli. Nucleolar localization was also abolished when the deletion of box H was coupled with the deletion of just box ACA itself (Δbox H/Δbox ACA). A combination of the deletion of stem 2 and box H (Δ46–108/Δbox H) did not enhance the effect of a stem 2 deletion alone (see Figure 3), and localization was still apparent. Bar, 10 μm.
Interestingly, and in contrast to all other mutations tested in the present report, U17 molecules that carried a combined substitution (sub. box H/sub. box ACA+) or combined deletion (Δbox H/Δbox ACA+) of box H and the box ACA region were fully incapable of localization to nucleoli (Figure 4). The lack of nucleolar localization was observed when the deletion of box H was coupled with a 5-nt deletion of the box ACA region or a 3-nt deletion of just box ACA itself (Δbox H/Δbox ACA) (Figure 4). This clearly shows that although box H as well as box ACA can function by themselves to mediate U17 snoRNA localization to nucleoli, nucleolar localization is entirely blocked when both are destroyed. This conclusion is supported by a control showing that the combination of a box H and stem 2 deletion does not enhance the effect of the stem 2 deletion alone; the double deletion of box H and stem 2 (Figure 4, Δ46–108/Δbox H) still localizes to nucleoli, because box ACA remains intact.
It was important to ascertain the stability of each mutant U17 snoRNA, to guard against the possibility that failure of some mutants to localize to nucleoli was simply due to their degradation. Stability assays using 32P-labeled transcripts demonstrated that all transcripts were sufficiently stable 1.5 h after injection into oocyte nuclei (the time when localization assays were carried out) (Figure 5, middle panel): all mutants were as stable as wild-type U17 snoRNA, except for mutants with a box H deletion or double deletion of box H and the ACA region where two-thirds of the mutant transcripts remained 1.5 h after injection into oocyte nuclei compared with wild-type U17 snoRNA. Even for these latter two mutants, however, the amount of transcript remaining is two times more than needed for reliable detection in the localization assay (Figure 2). The double mutant Δbox H/Δbox ACA was as stable as Δbox H/Δbox ACA+ (our unpublished results). These results clearly show that the failure of U17 molecules containing the box H/ACA double deletion to localize to nucleoli was not due to major degradation of the transcripts but rather to the loss of both NoLEs. Similarly, the failure of U17 carrying the box H/ACA double substitution (sub. box H/sub. box ACA+) to localize was not simply due to degradation, because it was almost as stable as the wild-type U17 snoRNA. Also, note that the transcripts carrying just a box H deletion and showing a slightly decreased stability 1.5 h after injection still localized to nucleoli (Figure 4).
Although well beyond the time frame of the localization assay, we were also interested to see whether any of the U17 mutants would show significant instability after longer incubation periods. This approach could reveal which elements in U17 are likely to be stabilizing elements. By assaying the long-term stability 15–16 h after injection of transcripts into oocyte nuclei, we found that deletion of either box H or the box ACA region individually (Figure 5, lower panel, Δbox H or Δbox ACA+; Δbox ACA [our unpublished results]) or in combination (Figure 5, lower panel, Δbox H/Δbox ACA+; Δbox H/Δbox ACA [our unpublished results]) significantly reduced the relative stability of the molecules compared with wild-type U17. Also, double substitutions of both box H and the ACA region, unlike the substitution in only one of these regions, resulted in minimal stability of the molecule (sub. box H/sub. box ACA+). The substitution of the 3 nts of box ACA (sub. box ACA) or substitution of the entire box ACA region (sub. box ACA+) decreased the stability of U17 somewhat. The only other mutant molecule with some reduced stability was the one lacking stems 3 and 4, in which almost one-half of the molecule was depleted (Figure 5, Δ118–213, lower panel). U17 carrying a box H substitution did not show any instability even 15–16 h after oocyte injection, although in yeast the substitution of box H completely abolished accumulation of different box H/ACA snoRNAs (Ganot et al., 1997b; Bortolin et al., 1999).
Our results show that both the deletion and substitution of the box ACA sequence destabilize U17 snoRNA. This destabilization is further increased when the box ACA+ substitution (or deletion) is coupled with a substitution (or deletion) of box H, indicating a stabilizing role as well for conserved box H, as also evidenced by deletion of box H alone.
In summary, evolutionarily conserved box H as well as box ACA at the 3′-tail of U17 snoRNA function as major NoLEs, whereas direct U17 snoRNA–rRNA interactions do not appear to be critically important for nucleolar localization. The integrity of the hairpin–hinge–hairpin–tail secondary structure contributes to nucleolar localization, probably by assisting the binding of proteins to the major NoLEs, which may either transport the snoRNA from the nucleoplasm to the nucleolus and/or anchor it within the nucleolus. This is the first identification of nucleolar localization sequences for a member of the Box H/ACA snoRNA.
DISCUSSION
In the present study the localization of U17 snoRNA was analyzed by microscopy of nucleolar preparations after injection of fluorescein-labeled in vitro transcripts into X. laevis oocyte nuclei. We found that U17, known to be essential for pre-rRNA processing and 18S rRNA production (Enright et al., 1996; Mishra and Elicieri 1997), preferentially localizes to the dense fibrillar component of nucleoli. Various controls confirmed that the nucleolar localization of U17 snoRNA was specific and, therefore, most likely mediated by defined intrinsic features. The results presented here discern which areas of the U17 snoRNA molecule are important for its nucleolar localization. The hairpins of the characteristic hairpin–hinge–hairpin–tail secondary structure are not essential, because their deletion resulted in a reduction of the localization signal but did not abolish it. This is in contrast to the critical role in pseudouridylation played by the hairpin structures (Bortolin et al., 1999). The stems can be regarded as accessory localization elements, which themselves are not absolutely essential but probably support box H and box ACA to function as NoLEs, as discussed below. We previously identified such accessory elements that faciliate the nucleolar localization of box C/D snoRNAs with a complex secondary structure such as U3 or U8 (Lange et al., 1998a,c).
The mutational analysis of the stem structures of U17 snoRNA also addressed whether elements of U17 snoRNA that potentially tether it to pre-rRNA may also have a role in localization. This question arises from the idea that nucleolar localization of a snoRNA could occur passively by diffusion of the snoRNA through the nucleoplasm into the nucleolus, where it may become trapped by base pairing with pre-rRNA. Two regions in stem 1 and 3 of U17 snoRNA (Figure 1, shaded areas A and B) are complementary to sequences in 18S rRNA (Rimoldi et al., 1993; Cecconi et al., 1994) and a sequence in stem 4 (Figure 1, shaded area C) is complementary to the external transcribed spacer of pre-rRNA (Cecconi et al., 1994). Furthermore, psoralen cross-linking has supported the notion that U17 stem 1 base pairs with 18S rRNA (Rimoldi et al., 1993). However, in our study the deletion of stem 3 had no adverse effect on nucleolar localization, and the deletion of stems 1 and 4 only resulted in a reduced signal but did not abolish nucleolar localization of U17. This is not surprising, because the nucleolar localization of members of the other major family of snoRNAs (box C/D snoRNAs) was shown to be independent from their interaction with pre-rRNA: for example, boxes A and A′ that contain regions of complementarity to 18S rRNA and are crucial for rRNA processing (Borovjagin and Gerbi, unpublished data) are not essential for nucleolar localization of X. laevis U3 snoRNA (Lange et al., 1998c; Narayanan et al., 1999). Similarly, the 5′ region of U8 snoRNA needed for rRNA processing and hypothesized to bind to the 5′ end of 28S rRNA (Peculis and Steitz, 1993, 1994; Peculis, 1997) is not essential for nucleolar localization (Lange et al., 1998a). Moreover, the middle part of U14 snoRNA that contains regions of complementarity to 18S rRNA, crucial for rRNA processing and 18S rRNA methylation, is dispensible for U14 snoRNA nucleolar localization (Lange et al., 1998b, and references therein; Samarsky et al., 1998). Taken together with the present results, we conclude that direct snoRNA–rRNA interactions do not critically regulate the nucleolar localization of snoRNAs of the box H/ACA or box C/D families.
Conserved Box H and Box ACA Are Major NoLEs
The characteristic and name-giving feature of box H/ACA snoRNAs is the presence of the conserved box H (ANANNA) within the hinge region and box ACA within the 3′-tail (Balakin et al., 1996; Ganot et al., 1997b). It can be hypothesized that the conservation of specific elements in box H/ACA snoRNAs might at least partly reflect their functional importance for nucleolar localization, as previously demonstrated for the box C/D snoRNA family (Lange et al., 1998b,c; Samarsky et al., 1998). Our data support this notion. The experiments presented here revealed that both box H and box ACA play an essential role in the nucleolar localization of U17 snoRNA. Only U17 molecules that carried a combined substitution or deletion of box H and box ACA were entirely defective in nucleolar localization. This indicates that box H and box ACA are each individually able to support nucleolar localization somewhat, even when one of the two regions is depleted or substituted. This conclusion is supported by the observation that the combination of a box H deletion with another mutation that weakened nucleolar localization, such as the deletion of stem 2, did not show any additional deleterious effect, because box ACA was still intact.
By analogy to box C/D snoRNAs, evolutionarily conserved box H and box ACA might be general NoLEs for the entire family of box H/ACA snoRNAs. It is intriguing to think that the NoLEs may be recognized by proteins specific for the box C/D or box H/ACA family, respectively. These proteins might either transport the snoRNA from the nucleoplasm to the nucleolus and/or anchor it within the nucleolus. Such proteins have not been identified yet, but candidate proteins that are known to interact with box H/ACA snoRNAs will be discussed below.
We observed some differences between the NoLEs of the two families of snoRNAs. In contrast to boxes H and ACA, boxes C and D in C/D snoRNAs seem to act in concert, because neither sequence by itself in the absence of the other box is sufficient for nucleolar localization: mutation of either box C or box D alone obliterates nucleolar localization of U8 or U14 box C/D snoRNAs (Lange et al., 1998b; Narayanan et al., 1999) as well as of U3 snoRNA (Lange et al., 1998c), although Narayanan et al. (1999) claim that for U3 snoRNA box C′ rather than box C functions as a NoLE. It could be that binding of putative localization proteins to the box C/D motif requires the presence of both box C and box D; when either one is missing, then protein binding would not occur, and nucleolar localization would be abolished. However, in the present situation, nucleolar localization is only obliterated when both box H and box ACA are absent. This suggests that these regions are redundant with one another and/or additive in their roles as NoLEs. For example, two copies of the putative nucleolar localization protein(s) might bind to U17 snoRNA, with one copy binding to box H and the second copy binding to box ACA. Efficient nucleolar localization would require both copies, but less efficient localization would be possible with just one copy. As an alternative model, there could be just one putative localization protein with two binding sites: one for box H and the other for box ACA. This protein might still bind to U17 snoRNA when just one binding site is present, and hence nucleolar localization would still be seen.
The premise that proteins may bind the NoLEs is supported by the observation that the box H and box ACA regions not only act as NoLEs but also are important for intronic processing of U17 and for stability of the molecule. From studies in yeast it has been suggested that the conserved ACA region protects Box H/ACA snoRNAs from processing exonucleases, whereas box H was proposed to contribute to 5′-end formation and maintenance of box H/ACA snoRNAs (Balakin et al., 1996; Ganot et al., 1997a; Henras et al., 1998; Bortolin et al., 1999). In the present case, proteins could bind to these regions and help define the boundaries of the U17 snoRNA during intronic processing and subsequently act to localize U17 to the nucleolus. However, intronic processing and nucleolar localization are not obligatorily coupled, because we injected the mature form of U17 snoRNA, which nonetheless was properly localized to nucleoli. Once, in the nucleolus, protein(s) bound to the NoLEs would confer long-term stability. Studies in yeast have shown that box H and box ACA are needed for cellular accumulation of box H/ACA snoRNAs (Balakin et al., 1996; Ganot et al., 1997b; Bortolin et al., 1999). Human telomerase RNA, a small percentage of which is found in nucleoli, also contains boxes H and ACA that are essential for its cellular accumulation (Mitchell et al., 1999). However, accumulation of a given RNA in those experiments depends on a multitude of factors, including synthesis, processing, and/or stability. The present study is the first to examine stability of a box H/ACA snoRNA in a manner distinguishable from processing. Our data indicate that both box H and box ACA confer long-term stability to mature U17 snoRNA in Xenopus oocytes. Thus, box H and box ACA serve a dual function as elements for nucleolar localization and long-term stability.
Candidate Proteins That May Interact with the NoLEs of Box H/ACA snoRNA
For both U17 box H/ACA snoRNA as well as the box C/D snoRNA family, the NoLEs are phylogenetically highly conserved sequences (Lange et al., 1998a–c; Samarsky et al., 1998; this study). Similarly, for the two species of the third and minor family of snoRNA (7-2/MRP snoRNA and the RNA component of RNase P), a preserved motif, the To antigen binding site, appears to mediate nucleolar localization (Jacobson et al., 1995, 1997). This suggests that snoRNA family-specific proteins bind to the NoLEs, thereby mediating nucleolar localization.
There are several candidate proteins that have been described to bind various box H/ACA snoRNAs. In addition, one protein (Ssb1p), seems to be specific for just snR10 and snR11 snoRNP (Clark et al., 1990). We hypothesize that a protein important for nucleolar localization of box H/ACA snoRNAs is more likely to be among those that are common to the entire family, rather than a protein specific to just one or a few snoRNPs. So far, four proteins common to the box H/ACA family have been identified: Gar1p (Girard et al., 1992; Balakin et al., 1996; Bousquet-Antonelli et al., 1997; Ganot et al., 1997b), Cbf5p (Nap57/dyskerin) (Jiang et al., 1993; Meier and Blobel, 1994), Nhp2p, and Nop10p (Kolodrubetz and Burgum, 1991; Henras et al., 1998), which are predicted to be present in two copies each per snoRNA (Watkins et al., 1998). All of the proteins mentioned above are required for 18S rRNA production or pseudouridylation (Girard et al., 1992; Bousquet-Antonelli et al., 1997; Cadwell et al., 1997; Henras et al., 1998; Lafontaine et al., 1998). Cbf5p is the candidate enzyme for ribosomal pseudouridylation (Koonin, 1996; Lafontaine et al., 1998; Watkins et al., 1998). Because Cbf5p lacks an apparent RNA binding motif, it is unlikely to bind directly to a snoRNA sequence element and probably is held in the snoRNP by interaction with other proteins of the complex (Watkins et al., 1998; Bortolin et al., 1999). Nhp2p would be a more likely candidate to bind directly to box H/ACA NoLEs because it contains an RNA binding motif, which happens to be similar to that in some ribosomal proteins (Koonin et al., 1994; Henras et al., 1998; Watkins et al., 1998). Interestingly, as Henras et al. (1998) pointed out, the ribosomal binding site for one of these proteins, L32, closely resembles the box H sequence of H/ACA snoRNA. It was recently suggested, however, that Nop10p, rather than Nhp2p, might contact one of the conserved boxes of H/ACA snoRNAs, because snR30 box H/ACA snoRNA remains detectable in Nhp2p-depleted cells but not in Nop10p-depleted cells (Henras et al., 1998). The fourth protein, Gar1p, has the potential to bind pre-rRNA but seems to bind to snoRNPs through interaction with Cbf5p, rather than by direct interaction with the snoRNA (Henras et al., 1998, and references therein). In fact, box H/ACA snoRNAs remain stable in yeast cells depleted of Gar1p but not in cells lacking Cbf5p, Nhp2p, or Nop10p (Girard et al., 1992; Bousquet-Antonelli et al., 1997). Those three proteins might also be responsible for the stability of U17 snoRNA in Xenopus oocytes by directly or indirectly binding to box H and box ACA. Direct binding of one or two different proteins, such as Nop10p and Nhp2p, to both major NoLEs, box H and box ACA together or individually, might initiate the localization of U17 and other box H/ACA snoRNAs to nucleoli. It cannot be excluded that a snoRNP complex providing strong interaction of all assembled factors has to be fully formed to either transport the snoRNA from the nucleoplasm to the nucleolus and/or anchor it within the nucleolus.
The present report provides the foundation for further studies to define the exact mechanism of nucleolar localization of box H/ACA snoRNAs.
ACKNOWLEDGMENTS
We thank E. Stuart Maxwell for the gift of pSP64T7 with the wild-type gene for U14 snoRNA and Annette W. Coleman for use of her fluorescence microscope. We are grateful to Anja-Katrin Bielinsky and Melanie T. North for helpful discussions as well as to Lola M. Brito for proofreading the manuscript. This work was funded in part by National Institutes of Health grant GM20261 to S.A.G.
REFERENCES
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different Box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes & Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of rRNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Henry Y, Gelugne JP, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNA. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Crosio C, Mariottini P, Cesareni G, Giorgi M, Brenner S, Amaldi F. A functional role for some Fugu introns larger than the typical short ones: the example of the gene coding for ribosomal protein S7 and snoRNA U17. Nucleic Acids Res. 1996;24:3167–3172. doi: 10.1093/nar/24.16.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Mariottini P, Loreni F, Pierandrei-Amaldi P, Campioni N, Amaldi F. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a protein S8 gene. Nucleic Acids Res. 1994;22:732–741. doi: 10.1093/nar/22.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MW, Yip ML, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990;111:741–751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B. 5′ ETS rRNA processing facilitated by four small RNAs: U14, E3, U17 and U3. RNA. 1996;2:1094–1099. , 1318. [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Callan HG, Wu Z, Murphy C. Lampbrush chromosomes. In: Kay BK, Peng HB, editors. Methods in Cell Biology. Vol. 36. New York: Academic Press; 1991. pp. 149–166. [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in pre-rRNA is guided by small nucleolar RNAs. Cell. 1997a;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. The family of Box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 1997b;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Gerbi SA. Small nucleolar RNA. Biochem Cell Biol. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- Girard JP, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao L-G, Taneja K, Singer RH, Wang Y-L, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao L-G, Wang Y-L, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999;146:559–571. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon HJ, Fouquet C, Carbon J. An essential yeast protein, Cbf5p, binds in vitro to centromeres and microtubules. Mol Cell Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of pre-rRNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNA essential for site-specific methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D, Burgum A. Sequence and genetic analysis of NHP2: a moderately abundant high mobility group-like nuclear protein with an essential function in Saccharomyces cerevisiae. Yeast. 1991;7:79–90. doi: 10.1002/yea.320070202. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Borovjagin AV, Gerbi SA. Nucleolar localization elements (NoLEs) in U8 snoRNA differ from sequences required for rRNA processing. RNA. 1998a;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Borovjagin A, Maxwell ES, Gerbi SA. Conserved Boxes C and D are essential nucleolar localization elements of U8 and U14 snoRNAs. EMBO J. 1998b;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Ezrokhi M, Borovjagin AV, Rivera-León R, North MT, Gerbi SA. Nucleolar Localization Elements of Xenopus laevis U3 snoRNA. Mol Biol Cell. 1998c;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverette RD, Andrews MT, Maxwell ES. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- Liu J, Maxwell ES. Mouse U14 snRNA in an intron of the cognate hsc70 heat shock gene. Nucleic Acids Res. 1990;18:6565–6571. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic precursor rRNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Maden BEH. Click here for methylation. Nature. 1996;383:675–676. doi: 10.1038/383675a0. [DOI] [PubMed] [Google Scholar]
- Maden BEH, Hughes JMX. Eukaryotic rRNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Zeller R. Xenopus laevis U2 snRNA genes: tandemly repeated transcription units sharing 5′ and 3′ flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2:1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–933. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Elicieri GL. Three small nucleolar RNAs that are involved in rRNA precursor processing. Proc Natl Acad Sci USA. 1997;94:4972–4977. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag MK, Thai TT, Ruff EA, Selvamurugan N, Kunnimalaiyaan M, Eliceiri GL. Genes for E1, E2, and E3 small nucleolar RNAs. Proc Natl Acad Sci USA. 1993;90:9001–9005. doi: 10.1073/pnas.90.19.9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nicoloso M, Qu LH, Michot B, Bachellerie JP. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- Peculis BA. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol. 1997;17:3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes & Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- Pelczar P, Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol Cell Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi OJ, Raghu B, Nag MK, Eliceiri GL. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol Cell Biol. 1993;13:4382–4390. doi: 10.1128/mcb.13.7.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EA, Rimoldi OJ, Raghu B, Eliceiri GL. Three small nucleolar RNAs of unique nucleotide sequences. Proc Natl Acad Sci USA. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamurugan N, Joost OH, Haas ES, Brown JW, Galvin NJ, Elicieri GL. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SB, Terry CD, Wells DA, DiMario PJ. Structural changes in oocyte nucleoli of Xenopus laevis during oogenesis and meiotic maturation. Chromosoma. 1996;105:111–121. doi: 10.1007/BF02509521. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B, Tycowski KT, Steitz JA. rRNA processing in eukaryotes. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Synthesis. Boca Raton, FL: CRC Press; 1995. pp. 469–490. [Google Scholar]
- Terns MP, Dahlberg JE. Retention and 5′ cap trimethylation of U3 RNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Terns MP, Grimm C, Lund E, Dahlberg JE. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. Small nucleolar RNAs guide rRNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Smith CM, Shu M-D, Steitz JA. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, You Z-H, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega NW. Protein incorporation by isolated amphibian oocytes. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauerl G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Leverette RD, Ling X, Andrews MT, Maxwell ES. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide Boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Gall JG. “Micronucleoli” in the Xenopus germinal vesicle. Chromosoma. 1997;105:438–443. doi: 10.1007/BF02510480. [DOI] [PubMed] [Google Scholar]
- Xia L, Watkins NJ, Maxwell ES. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]