Abstract
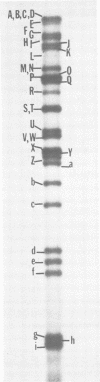
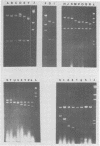
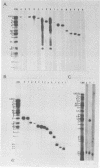
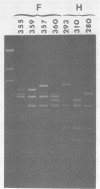
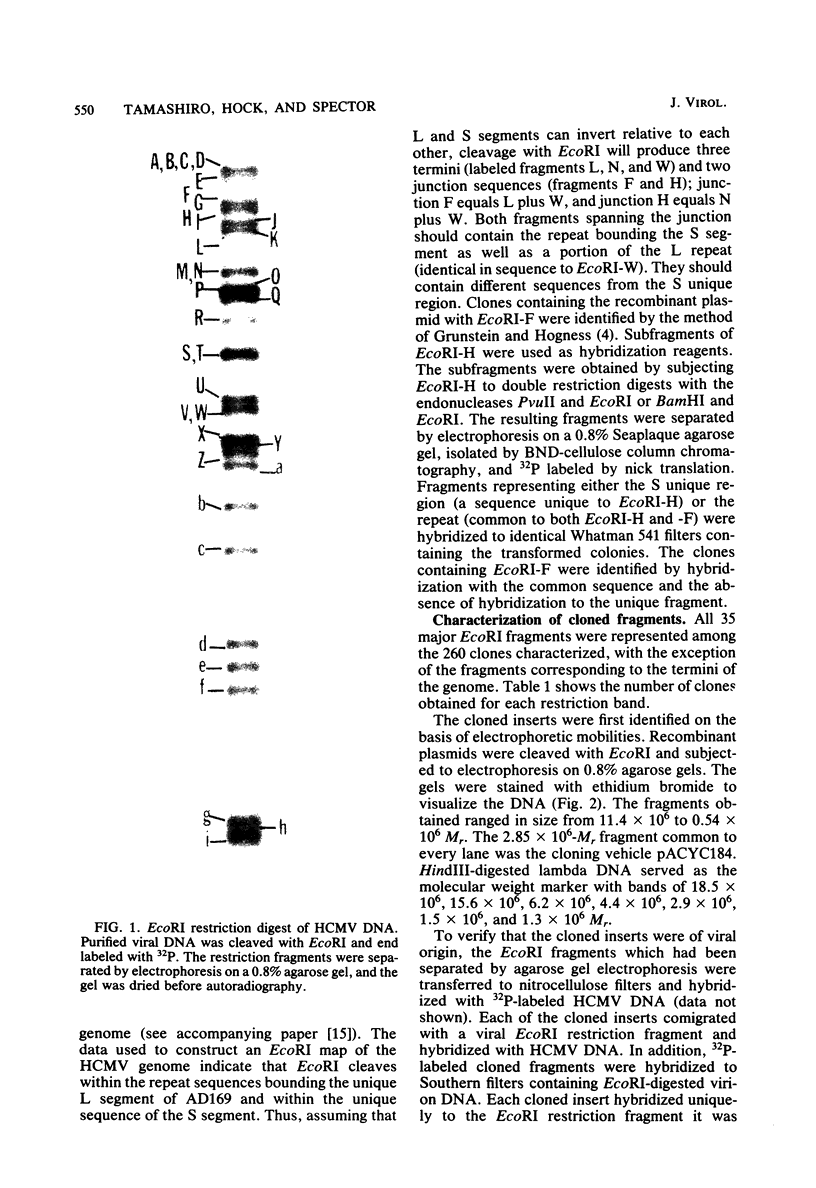
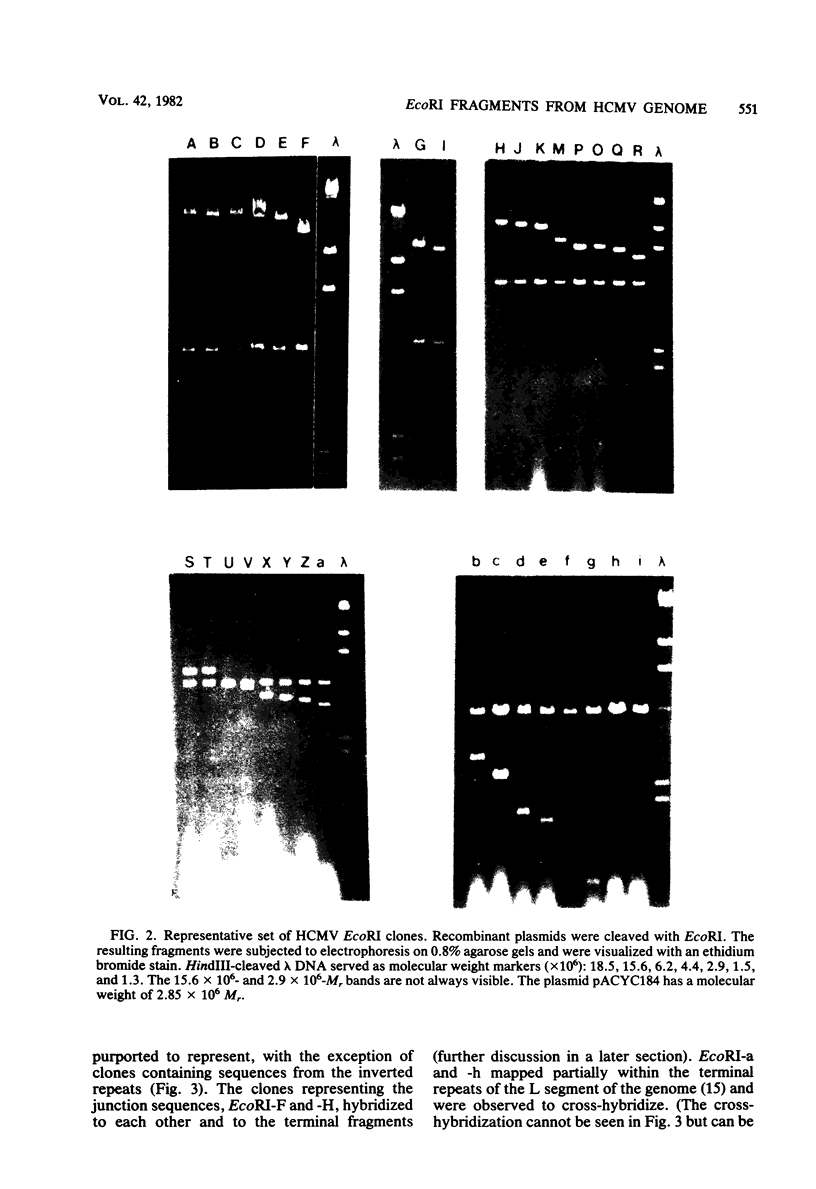
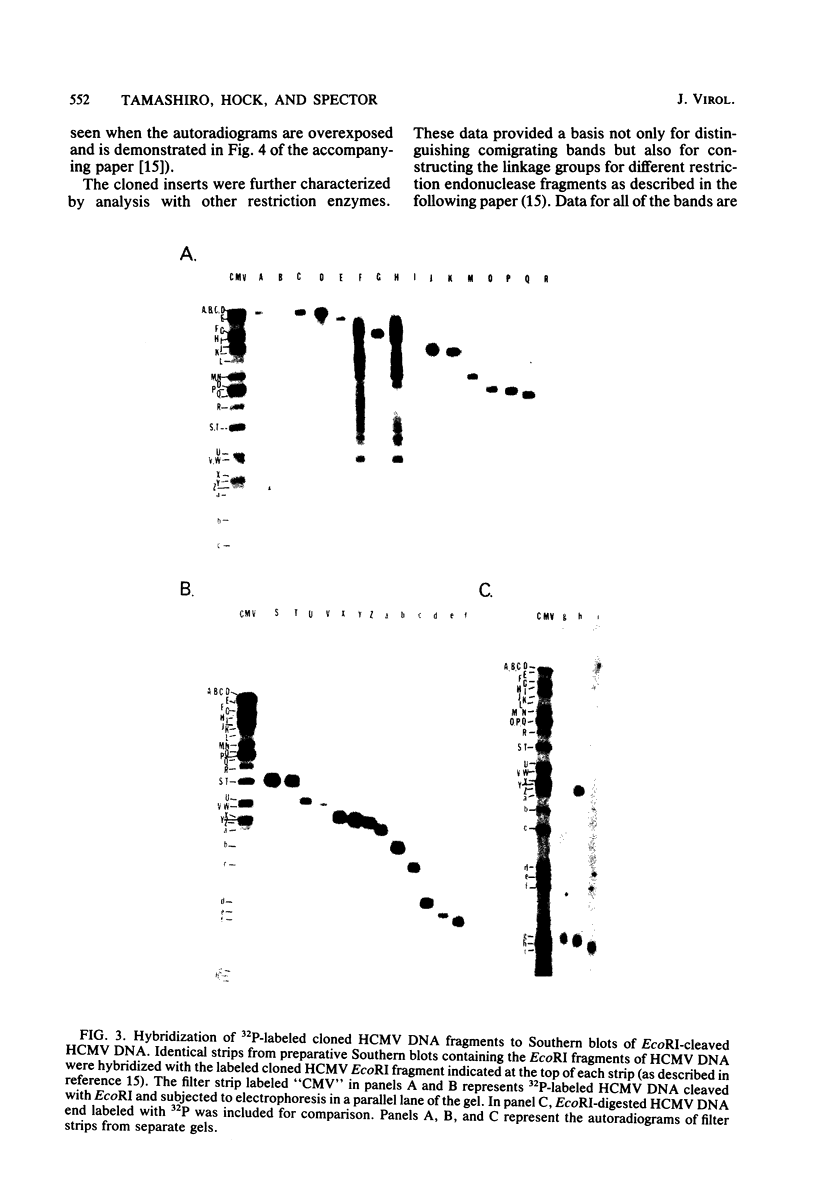
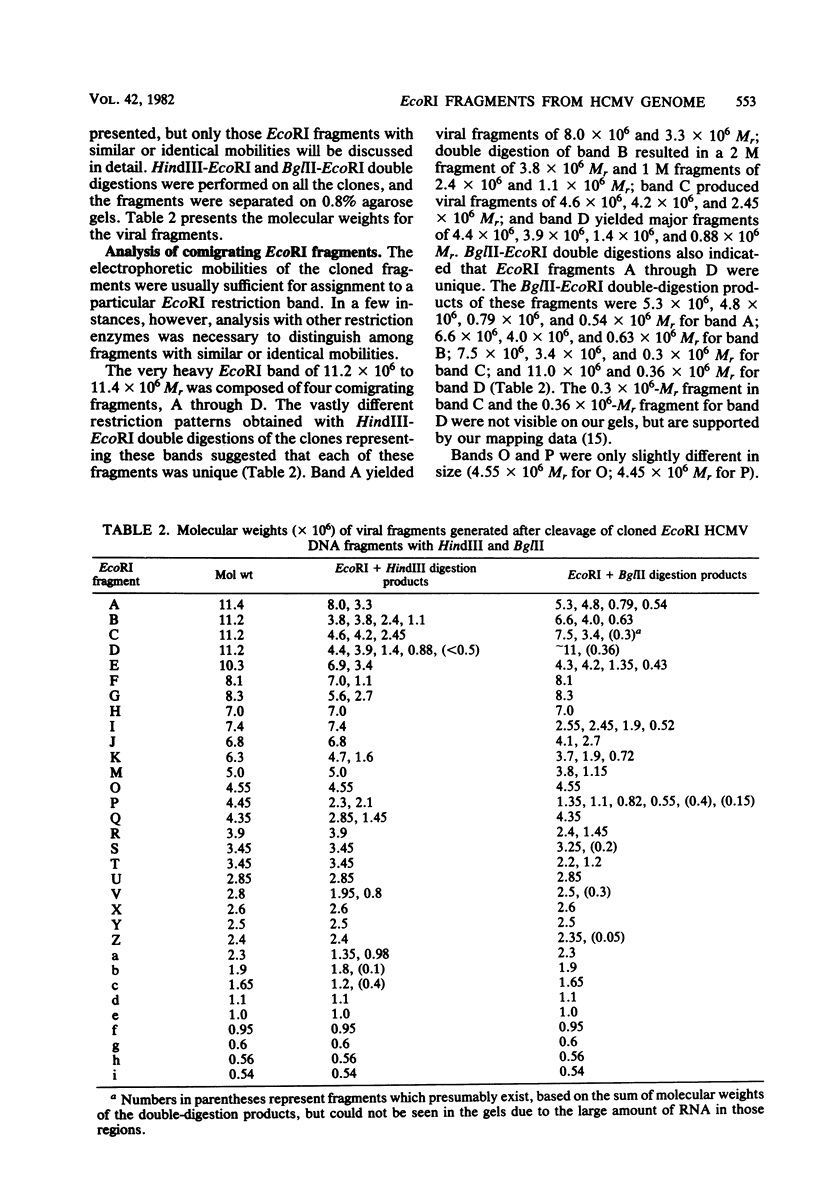
The DNA genome of human cytomegalovirus (HCMV) strain AD169 is 158 x 10(6) Mr. Cleavage of the HCMV DNA with the restriction endonuclease EcoRI yields 35 major fragments ranging in size from 0.54 x 10(6) Mr. We have constructed a cloned library of the EcoRI fragments of this strain of HCMV, using the plasmid pACYC184 and the recipient bacterium Escherichia coli strain HB101 RecA-. The viral origin of the cloned inserts was determined by hybridization to viral DNA. The fragments were characterized further by digestion with other restriction enzymes. Several clones were obtained which contained sequences spanning the junction between the long (L) and short (S) components of the viral DNA sequences. These clones differed in molecular weight by multiples of 0.3 x 10(6) to 0.4 x 10(6) Mr. The variability found in the clones was also reflected in the genome. Each clone containing a junction sequence hybridized to a series of bands on Southern filters of EcoRI-digested HCMV DNA. This "ladder effect" provided evidence for a region of heterogeneity within the L-S junction.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarchi J. M., Blankenship M. L., Brown G. D., Kaplan A. S. Size and complexity of human cytomegalovirus DNA. Virology. 1978 Sep;89(2):643–646. doi: 10.1016/0042-6822(78)90209-x. [DOI] [PubMed] [Google Scholar]
- Geelen J. L., Walig C., Wertheim P., van der Noordaa J. Human cytomegalovirus DNA. I. Molecular weight and infectivity. J Virol. 1978 Jun;26(3):813–816. doi: 10.1128/jvi.26.3.813-816.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin C., Lussier G. Concentration of human cytomegalovirus from large volumes of tissue culture fluids. J Gen Virol. 1979 Jan;42(1):193–197. doi: 10.1099/0022-1317-42-1-193. [DOI] [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C., Figurski D., Meyer R., Remaut E., Helinski D. R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- Kilpatrick B. A., Huang E. S. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J Virol. 1977 Oct;24(1):261–276. doi: 10.1128/jvi.24.1.261-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakeman A. D., Osborn J. E. Size of infectious DNA from human and murine cytomegaloviruses. J Virol. 1979 Apr;30(1):414–416. doi: 10.1128/jvi.30.1.414-416.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spector D. H., Hock L., Tamashiro J. C. Cleavage maps for human cytomegalovirus DNA strain AD169 for restriction endonucleases EcoRI, BglII, and HindIII. J Virol. 1982 May;42(2):558–582. doi: 10.1128/jvi.42.2.558-582.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Summers W. C. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J Virol. 1978 Aug;27(2):374–387. doi: 10.1128/jvi.27.2.374-387.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth B. B., French L. Plaque assay of cytomegalovirus strains of human origin. Proc Soc Exp Biol Med. 1970 Nov;135(2):253–258. doi: 10.3181/00379727-135-35031. [DOI] [PubMed] [Google Scholar]