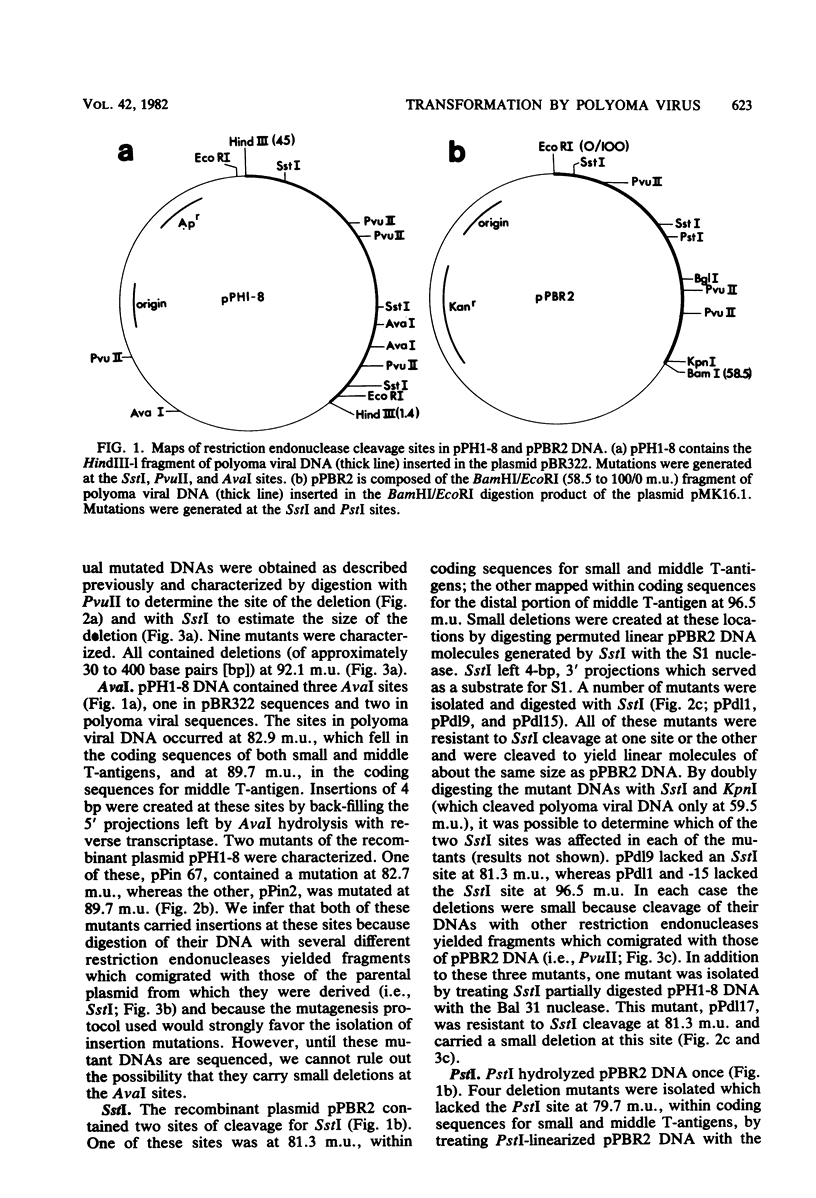
Abstract
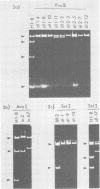
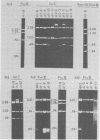
To determine whether small or middle T-antigen (or both) of polyoma virus is required for transformation, we constructed mutants of recombinant plasmids which bear the viral oncogene and measured the capacity of these mutants to transform rat cells in culture. Insertion and deletion mutations in sequences encoding small and middle T-antigens (79.7, 81.3, and 82.9 map units) rendered the DNA incapable of causing transformation by the focus assay. Similar mutations in sequences that encoded middle but not small T-antigen (89.7, 92.1, and 96.5 map units) generally abolished the transforming activity of the DNA. However, two mutants (pPdl1-4 and PPd12-7) that carried deletions at 92.1 map units retained the capacity to transform cells; pPdl1-4 did so at frequencies equal to those of the parental plasmid, whereas pPdl2-7 transformed at 10% the frequency of its antecedent. From these studies we conclude that small T-antigen alone is insufficient to cause transformation and that middle T-antigen is required for transformation, either in combination with small T-antigen or by itself.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrand J. R., Keller W., Roberts R. J. Extent of terminal repetition in adenovirus 2 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):401–407. doi: 10.1101/sqb.1974.039.01.052. [DOI] [PubMed] [Google Scholar]
- Bacheler L. T. Virus-secific transcription in 3T3 cells transformed by the ts-a mutant of polyoma virus. J Virol. 1977 Apr;22(1):54–64. doi: 10.1128/jvi.22.1.54-64.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig M. M., Thomas T., Folk W. R. Viable deletion mutant in the medium and large T-antigen-coding sequences of the polyoma virus genome. J Virol. 1980 Mar;33(3):1215–1220. doi: 10.1128/jvi.33.3.1215-1220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael G. G., Benjamin T. L. Identification of DNA sequence changes leading to loss of transforming ability in polyoma virus. J Biol Chem. 1980 Jan 10;255(1):230–235. [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- FRIED M. ISOLATION OF TEMPERATURE-SENSITIVE MUTANTS OF POLYOMA VIRUS. Virology. 1965 Apr;25:669–671. doi: 10.1016/0042-6822(65)90098-x. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Benjamin T. L. Comparisons of two early gene functions essential for transformation in polyoma virus and SV-40. Virology. 1979 Jul 15;96(1):205–228. doi: 10.1016/0042-6822(79)90185-5. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Frisque R. J., Sambrook J. Origin-defective mutants of SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):293–300. doi: 10.1101/sqb.1980.044.01.033. [DOI] [PubMed] [Google Scholar]
- Grady L. J., North A. B., Campbell W. P. Polyoma genome transcription in transformed mouse cells growing in culture and as tumors in syngeneic mice. Int J Cancer. 1977 Feb 15;19(2):236–239. doi: 10.1002/ijc.2910190213. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Ostrander D. A., Hodnett J. L., Legerski R. J., Robberson D. L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975 Sep;2(9):1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori J., Carmichael G. G., Benjamin T. L. DNA sequence alterations in Hr-t deletion mutants of polyoma virus. Cell. 1979 Mar;16(3):505–513. doi: 10.1016/0092-8674(79)90025-4. [DOI] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Hourihan S. L., Rowe W. P., Martin M. A. Biological activity of polyoma viral DNA in mice and hamsters. J Virol. 1979 Mar;29(3):990–996. doi: 10.1128/jvi.29.3.990-996.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Dulbecco R. Characterization of polyoma virus T antigen. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1259–1263. doi: 10.1073/pnas.74.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C., Figurski D., Meyer R., Remaut E., Helinski D. R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Kamen R., Lindstrom D. M., Shure H., Old R. W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- Lania L., Griffiths M., Cooke B., Ito Y., Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (Hr-t) mutant. Cell. 1979 Nov;18(3):793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxman M. N., Takemoto K. K., Eckhart W. Polyoma T antigen synthesis by temperature-sensitive mutants of polyoma virus. Virology. 1972 Sep;49(3):675–682. doi: 10.1016/0042-6822(72)90524-7. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin D., Cuzin F. Polyoma virus T antigen. I. Synthesis of modified heat-labile T angiten in cells transformed with the ts-a mutant. J Virol. 1975 Feb;15(2):393–397. doi: 10.1128/jvi.15.2.393-397.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R., Benjamin T. L. Cellular alterations dependent upon the polyoma virus Hr-t function: separation of mitogenic from transforming capacities. Cell. 1978 Jul;14(3):587–599. doi: 10.1016/0092-8674(78)90244-1. [DOI] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J., Schaffhausen B., Benjamin T. Tumor antigens induced by nontransforming mutants of polyoma virus. Cell. 1978 Oct;15(2):485–496. doi: 10.1016/0092-8674(78)90018-1. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Villa-Komaroff L., Efstratiadis A., Broome S., Lomedico P., Tizard R., Naber S. P., Chick W. L., Gilbert W. A bacterial clone synthesizing proinsulin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3727–3731. doi: 10.1073/pnas.75.8.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]