Abstract
The ubiquitin-dependent proteolysis of mitotic cyclin B, which is catalyzed by the anaphase-promoting complex/cyclosome (APC/C) and ubiquitin-conjugating enzyme H10 (UbcH10), begins around the time of the metaphase–anaphase transition and continues through G1 phase of the next cell cycle. We have used cell-free systems from mammalian somatic cells collected at different cell cycle stages (G0, G1, S, G2, and M) to investigate the regulated degradation of four targets of the mitotic destruction machinery: cyclins A and B, geminin H (an inhibitor of S phase identified in Xenopus), and Cut2p (an inhibitor of anaphase onset identified in fission yeast). All four are degraded by G1 extracts but not by extracts of S phase cells. Maintenance of destruction during G1 requires the activity of a PP2A-like phosphatase. Destruction of each target is dependent on the presence of an N-terminal destruction box motif, is accelerated by additional wild-type UbcH10 and is blocked by dominant negative UbcH10. Destruction of each is terminated by a dominant activity that appears in nuclei near the start of S phase. Previous work indicates that the APC/C–dependent destruction of anaphase inhibitors is activated after chromosome alignment at the metaphase plate. In support of this, we show that addition of dominant negative UbcH10 to G1 extracts blocks destruction of the yeast anaphase inhibitor Cut2p in vitro, and injection of dominant negative UbcH10 blocks anaphase onset in vivo. Finally, we report that injection of dominant negative Ubc3/Cdc34, whose role in G1–S control is well established and has been implicated in kinetochore function during mitosis in yeast, dramatically interferes with congression of chromosomes to the metaphase plate. These results demonstrate that the regulated ubiquitination and destruction of critical mitotic proteins is highly conserved from yeast to humans.
INTRODUCTION
Progress through mitosis is driven largely by posttranslationally regulated changes in existing proteins. At the G2–M border, preformed complexes of the kinase cdc2 and its positive regulatory subunit cyclin B are activated by the removal of inhibitory phosphorylations from cdc2 (for review, see Morgan, 1997). Activated cyclin B/cdc2 then leads to phosphorylation of numerous target proteins, either directly or indirectly, resulting in dramatic changes in the organization or activity of those proteins. The result is formation of the mitotic spindle, chromosome condensation and congression to the metaphase plate and, in animal cells, breakdown of the nuclear envelope (McIntosh and Koonce, 1989; Osmani et al., 1991; Nigg et al., 1996). Sister chromatids are linked by cohesion proteins from the time of DNA replication, and cohesion is maintained during chromosome condensation and alignment at the metaphase plate (for review, see Biggins and Murray, 1998; Yanagida, 1998). During the process of congression, incompletely aligned chromosomes activate checkpoint control pathways that prevent the onset of chromosome separation until congression is complete (for review, see Wells, 1996; Rieder and Salmon, 1998). Anaphase onset, sister chromatid separation and segregation to the daughter cells, and the formation of postmitotic G1 nuclei requires the selective, ubiquitin-dependent proteolysis of several regulatory proteins. Anaphase onset is driven by the destruction of the anaphase-inhibitory proteins Pds1p in budding yeast (Cohen-Fix et al., 1996; Yamamoto et al., 1996a,b; Guacci et al., 1997; Michaelis et al., 1997; Ciosk et al., 1998) and Cut2p in fission yeast (Funabiki et al., 1996a,b, 1997), which results in loss of chromosome cohesion. The budding yeast protein Ase1p, which is involved in spindle morphology and elongation during anaphase and telophase, is also degraded when cells exit mitosis (Juang et al., 1997). The degradation of mitotic cyclins, which are essential positive regulatory subunits for cdc2, results in the release of inactive cdc2 (for review, see King et al., 1996; Townsley and Ruderman, 1998). After inactivation of cdc2, mitotic phosphorylations are removed, and the dephosphorylated proteins return to their interphase states, leading to chromosome decondensation, breakdown of the mitotic spindle, formation of the interphase array of microtubules and, in animal cells, reformation of the nuclear envelope.
Ubiquitin-dependent proteolysis occurs through the sequential function of four enzymatic activities (for review, see Hochstrasser, 1996; Ciechanover, 1998). Ubiquitin is first activated by formation of a thioester with the ubiquitin-activating enzyme E1. Ubiquitin is then transferred to one of several E2s, also known as ubiquitin-conjugating enzymes (Ubcs). In most cases, a third activity termed an E3 or ubiquitin ligase catalyzes transfer of ubiquitin from the Ubc to the target protein. Polyubiquitinated proteins are then recognized and degraded by the 26S proteasome. Specialized ubiquitin carrier proteins (E2s/Ubcs) and ubiquitin ligases (E3s) are responsible for the highly selective and regulated ubiquitination of target proteins. One of the best understood examples comes from studies with cyclin B. Ubiquitin-dependent proteolysis of cyclin B begins around the time of the anaphase onset; the exact timing may vary among different organisms and types of cell cycles (reviewed by King et al., 1996; Townsley and Ruderman, 1998). A large multisubunit E3 known as the anaphase-promoting complex or cyclosome (APC/C) (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995; Tugendreich et al., 1995) catalyzes the transfer of ubiquitin from a specialized E2, which, in humans, is called UbcH10 (Hershko et al., 1994; Aristarkhov et al., 1996; Yu et al., 1996; Osaka et al., 1997; Townsley et al., 1997). Ubiquitination of cyclin B depends on the presence of a small N-terminal motif known as the destruction box (D box) (Glotzer et al., 1991). Other APC/C targets also contain D boxes that are essential for their mitotic destruction. Well-characterized examples include cyclin A (Luca et al., 1991), the budding yeast anaphase inhibitor Pds1p (Cohen-Fix et al., 1996), its fission yeast homologue Cut2p (Funabiki et al., 1996b), the budding yeast spindle-associated protein Ase1p (Juang et al., 1997), and the Xenopus protein geminin, an inhibitor of DNA replication (McGarry and Kirschner, 1998). Prc1, a human protein resembling Ase1, has also been described recently by Jiang et al. (1998).
In all cases examined so far, the APC/C is the regulated component of the system; it becomes activated after completion of chromosome congression, remains active through the completion of mitosis, and, in somatic cells of both fungi and animals, remains active toward cyclin B until the end of G1 (reviewed by King et al., 1996; Osmani and Ye, 1997; Townsley and Ruderman, 1998). Activation of the APC/C depends on a pathway involving phosphorylation of unknown components and on association with noncatalytic activators of the cdc20 family, different members apparently providing additional temporal and substrate level discrimination among different mitotic targets (reviewed by Peters, 1998). In contrast to the considerable progress in identifying components of the APC/C activation pathway, much less is known about the mechanism of APC/C inactivation and the timing of its inactivation toward different substrates. Many subunits of the active APC/C are highly phosphorylated, but dephosphorylation in vitro does not result in APC/C inactivation (Fang et al., 1998; Kotani et al., 1998). In budding yeast, the appearance of active G1 cyclin/cdk complexes is required to turn off APC/C activity, but the targets are not known (Amon et al., 1994). In Drosophila, the appearance of cyclin E/cdk2 activity seems required for APC/C inactivation, but again the mechanism is not known (Knoblich et al., 1994).
We have used cell-free systems from rat and human somatic tissue culture cells to study regulatory mechanisms underlying APC/C activity during different phases of the cell cycle. These in vitro systems reproduce the regulated ubiquitin-dependent proteolysis of several cell cycle regulators, including cyclins A and B, geminin H, and Cut2p, that occurs in vivo. For all four targets, destruction seems to be regulated coordinately: in each case, destruction is blocked by the addition of dominant negative UbcH10; destruction requires the N-terminal D box-containing domain; destruction continues through G1 and depends on the activity of a PP2A-like phosphatase and is terminated by a dominant activity that appears in S phase nuclei. When dominant negative UbcH10 is injected into cells at prophase, chromosome congression to the metaphase plate proceeds on schedule, but anaphase onset is substantially delayed, providing further support that inhibitors of anaphase onset play an important role in the M–G1 transition in mammalian cells.
MATERIALS AND METHODS
Cell Culture and Cell Synchronization
Fisher rat fibroblasts (FR3T3, a gift from J. Rommelaere, INSERM, Heidelberg, Germany) and HeLa cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum plus 100 μg/ml streptomycin and 100 U/ml penicillin (Life Technologies, Frederick, MD) at 37°C under 5% CO2 and 95% air. FR3T3 cells were arrested in G0 by serum starvation for 72 h. Cell cycle reentry was induced by addition of 10% serum. Cells were taken at 0, 3, 16, and 24 h after addition of serum, and cell cycle progression was monitored by fluorescence-activated cell sorting (FACS) analyses and bromodeoxyuridine incorporation as described previously (Bastians et al., 1998). Cells arrested in mitosis were obtained by culturing in the presence of 0.3 μM nocodazole for 14–16 h. HeLa cells were synchronized in G1 phase by a nocodazole arrest and release protocol. Briefly, cells were arrested in mitosis by culturing in the presence of 0.3 μM nocodazole (Sigma, St. Louis, MO) for 14 h. Mitotic cells were shaken off and allowed to enter G1 phase by growth in fresh medium for an additional 3–5 h. Cells synchronized in S phase were obtained by arresting cells in the presence of 2 mM hydroxyurea (Sigma). Alternatively, cells were arrested at G1–S by a double thymidine block and released into S phase for 2 h. Cells in G2 were obtained by incubation with 0.3 μM nocodazole. Mitotic cells were removed by shake off, and residual cells were harvested. These cells had accumulated in G2 as judged by FACS analysis. Samples of cells were subjected to FACS analyses following standard protocols (Adams et al., 1997). PtK1 (rat kangaroo kidney) cells used for microinjection studies were cultured in minimal essential medium (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum, 20 mM HEPES buffer, 1× nonessential amino acids, 1 mM sodium pyruvate, 60 μg/ml penicillin, and 100 μg/ml streptomycin.
Cell Extracts
Cells were harvested by trypsin-EDTA treatment and washed twice in ice-cold PBS and once in low-salt buffer (50 mM HEPES-NaOH, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT plus protease inhibitors [complete protease inhibitor tablets; Boehringer Mannheim, Mannheim, Germany]). To prepare whole-cell extracts (Brandeis and Hunt, 1996; Arvand et al., 1998; Bastians et al., 1998), the supernatant was removed, and cells were resuspended in the residual buffer, incubated on ice for 20 min, and lysed by sonication at 4°C. Cellular debris was removed by centrifugation at 14,000 × g for 20 min. To prepare cytoplasmic and nuclear extracts, cells were harvested and washed as described above. Cells were resuspended in 0.5× vol of low-salt buffer, incubated on ice for 20 min, and lysed by douncing using a loose fitting pestle. The lysate was centrifuged at 2000 × g for 5 min to collect intact nuclei. The cytoplasmic supernatant was obtained by recentrifugation at 14,000 × g for 20 min. The nuclear pellet was resuspended in 0.5× vol of low-salt buffer and lysed by sonication, and soluble nuclear extracts were obtained by centrifugation of the nuclear lysate at 14,000 × g for 20 min. All extracts were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Plasmids
The following plasmids were used for in vitro transcription and translation: pGEM-human cyclin B (Pines and Hunter, 1989), pET5-human cyclin B Δ1-86, pGEM-human cyclin A (Pines and Hunter, 1990), pcDNA3-human cyclin A Δ1-70, pCS2-Xenopus geminin H (McGarry and Kirschner, 1998), pcDNA3-Xenopus geminin H Δ1-61, pGEM-Schizosaccharomyces pombe cut2 after subcloning from pGEX-cut2 and pGEM-S. pombe cut2 Δ1-73 after subcloning from pGEX-cut2 Δ1-73 (Funabiki et al., 1996b), pET11-Saccharomyces cerevisiae ase1 (Juang et al., 1997), pCS2-S. cerevisiae pds1(Cohen-Fix et al., 1996), pT7–7-p27Kip1 (Pagano et al., 1995), and pcDNA3-p27Kip1Δ186-198.
In Vitro Transcription and Translation
A coupled transcription and translation system using reticulocyte lysate (TNT; Promega, Madison, WI) was used with [35S]methionine (1175 Ci/mmol) to prepared radiolabeled in vitro translation products.
Recombinant Proteins
For bacterial expression of human Ubc proteins, the coding sequences of human UbcH10, wild-type and dominant negative variants (Townsley et al., 1997), were amplified by PCR and cloned into pET29a to create UbcH10 sequences containing a C-terminal hexahistidine tag. Plasmids encoding hexahistidine-tagged wild-type or dominant negative human Ubc3 were described by Pagano et al. (1995). Recombinant proteins were expressed in Escherichia coli BL21(DE3) and purified under native conditions using Ni-NTA spin columns (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Eluted proteins were subjected to gel filtration on NAP-5 columns (Pharmacia, Uppsala, Sweden), eluted in 50 mM HEPES-NaOH, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, and 1 mM DTT, and concentrated using Microcon-10 microconcentrators (Amicon, Beverly, MA). Purified recombinant mutant UbcH5b protein (Jensen et al., 1995) was generously provided by Dr. Vincent Chau (ProScript, Cambridge, MA) and was concentrated as described above.
In Vitro Degradation Assays
Assays were performed in a final volume of 10 μl. Five microliters of cell extracts were thawed on ice and supplemented with an ATP-regenerating system (1.5 mM ATP, 40 mM phosphocreatine, 80 μg/ml creatine kinase) and, when indicated, 2.5 μM purified recombinant wild-type or dominant negative human UbcH10 or Ubc3 protein (Arvand et al., 1998; Bastians et al., 1998). One to 2 μl of radiolabeled substrate proteins were mixed with the cell extracts and incubated at 30°C. Two-microliter samples were taken at the indicated time points, and proteins were resolved on 12.5% SDS-PAGE followed by autoradiography. To deplete ATP, cell extracts were preincubated with 10 mM glucose and 1 μg/μl hexokinase (Sigma) for 30 min at 30°C. To inhibit polyubiquitination or degradation by the 26S proteasome, cell extracts were incubated with 1 μg/μl methyl ubiquitin (Boston Biochem, Boston, MA) or with 100 μM lactacystin (Boston Biochem), ALLN (Calbiochem, San Diego, CA) or MG132 (Calbiochem), respectively. The following concentrations of kinase and protein phosphatase inhibitors were used: 20 nM staurosporine (Sigma), 5 mM 6-dimethylaminopurine (6-DMAP; Sigma), 50 μM olomoucine (Alexis Biochemicals, San Diego, CA), protein phosphatase inhibitor mix (20 mM sodium-p-nitrophenylphosphate, 25 mM sodium glycerophosphate, 50 mM sodium fluoride, 5 mM sodium molybdate, 0.2 mM sodium orthovanadate, 5 mM l-phenylalanine, 150 μM 1,10-phenanthroline, 5 mM EDTA), 1 μM okadaic acid (OA; Calbiochem), 1 mM sodium orthovanadate (Sigma), and 200 nM heat-stable PP1 inhibitor I-2 (Calbiochem).
Kinase Assays
For in vitro kinase assays of HeLa G1 extracts or S phase nuclear extracts, samples of 10 μl were preincubated with various kinase or phosphatase inhibitors as indicated for 30 min at 30°C. Samples of 5 μl were used for in vitro degradation assays; samples of 2 μl were used for kinase assays and diluted in kinase buffer (20 mM HEPES-NaOH, pH 7.4, 10 mM MgCl2, 1 mM DTT) plus 0.2 mM ATP, 7.5 μg of histone H1 (Sigma), 10 μM PKC inhibitor, and 0.5 μl of [γ-32P]ATP (6000 Ci/mmol) to a final volume of 20 μl. The reaction was incubated for 30 min at room temperature and stopped by addition of 25 μl of SDS sample buffer. Fifteen-microliter samples were analyzed by SDS-PAGE followed by autoradiography.
For immunoprecipitation of CDK activity, 10 μl of S phase nuclear extracts preincubated with DMSO or 20 nM staurosporine were diluted with NP40 buffer (20 mM HEPES-NaOH, pH 7.4, 10 mM EDTA, 175 mM NaCl, 0.7% NP40, plus complete protease inhibitors) to a final volume of 50 μl. After centrifugation for 20 min at 14,000 × g, the supernatant was precleared using 10 μl of protein A-Sepharose for 45 min at 4°C. The precleared supernatant was then incubated with 3 μl of anti-cyclin A antibodies (100 μg/ml) for 90 min at 4°C followed by the addition of 10 μl of protein A-Sepharose for an additional 90 min Immunocomplexes were harvested by centrifugation, washed three times in NP40 buffer and once in kinase buffer, and resuspended in 20 μl of kinase buffer plus 0.2 mM ATP, 7.5 μg of histone H1, 10 μM PKC inhibitor, and 0.5 μl of [γ-32P]ATP (6000 Ci/mmol). The reaction was incubated for 30 min at room temperature and stopped by addition of 25 μl of SDS sample buffer. Ten-microliter samples were analyzed by SDS-PAGE followed by autoradiography.
Microinjection
PtK1 cells were cultured on inscribed glass coverslips, and microinjections were carried out as previously described (Gorbsky et al., 1998). Briefly, 18-mm holes were cut into 60-mm plastic tissue culture dishes. A 22-mm coverslip was sealed to the inside of the dish with silicone grease. Chambers were sterilized by inversion on a UV transilluminator for 15 min, and cells were grown on the coverslips for 2–3 d. Medium in the chamber was overlaid with light mineral oil before microinjection to prevent evaporation and change in pH. The chamber was placed on a prewarmed stage of a Nikon (Tokyo, Japan) Diaphot inverted microscope. Stage temperature was maintained at 36–37°C with a warm air curtain incubator (Sage, Boston, MA). Microinjections were performed with a micromanipulator (Narishige, Sea Cliff, NY) using freshly pulled glass microneedles. Cells were injected with wild-type UbcH10 (25 mg/ml; 1.27 mM), mutant UbcH10 (20 mg/ml; 1.02 mM), mutant Ubc3 (14 mg/ml; 0.524 mM), mutant UbcH5b (14 mg/ml; 0.802 mM) or buffer (50 mM HEPES-NaOH, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT). Cell were injected to no more than 5% of their total volume. Cells were injected using phase optics with a 40 × 0.55 numerical aperture long working distance objective and 0.3 numerical aperture extra long working distance condenser. Cells were monitored through mitosis by phase-contrast microscopy. Images were collected with a digital cooled charge-coupled device camera (Photometrics, Tucson, AZ) controlled by a computer equipped with Metamorph software (Universal Imaging, Media, PA). Mitotic stages were defined as previously described (Kallio et al., 1998) Most cells were injected in prophase and prometaphase. To avoid DNA damage, prophase cells were injected in the cytoplasm, and prometaphase cells were injected away from the chromosomes. Metaphase was defined as the time at which all kinetochores had assembled to within 1.5 μm of the spindle midplane. Statistical analysis was performed with SigmaStat (Jandel Scientific, Corte Madera, CA).
RESULTS
Extracts of G0, G1, S, G2, and Nocodazole-arrested M Phase Cells Retain the Cell Cycle Stage–specific Differences in Ubiquitin-dependent Cyclin B Proteolysis
In mammalian somatic cells, ubiquitin-dependent destruction of mitotic cyclin B begins near the end of mitosis, continues into G1, and ceases around the time of the G1–S transition (Brandeis and Hunt, 1996) Concentrated extracts of G1 and S phase cells reproduce these stage-specific differences in cyclin B destruction (Brandeis and Hunt, 1996; Arvand et al., 1998; Bastians et al., 1998; Nguyen et al., 1999). We sought to determine whether the stage-specific patterns of destruction activity toward cyclin B and other mitotic targets could also be reproduced in extracts prepared from cells taken at these and other phases of the cell cycle: G0 and G1 (during which cyclin destruction continues), S and G2 (during which cyclins are stable and accumulate), and nocodazole-arrested M phase (during which checkpoint pathways inhibit the destruction of cyclin B) (Murray, 1995; King et al., 1996; Townsley and Ruderman, 1998). Unfortunately, the small number of naturally synchronous M phase cells that can be obtained by mitotic shake off are insufficient for making workable amounts of unperturbed M phase cells.
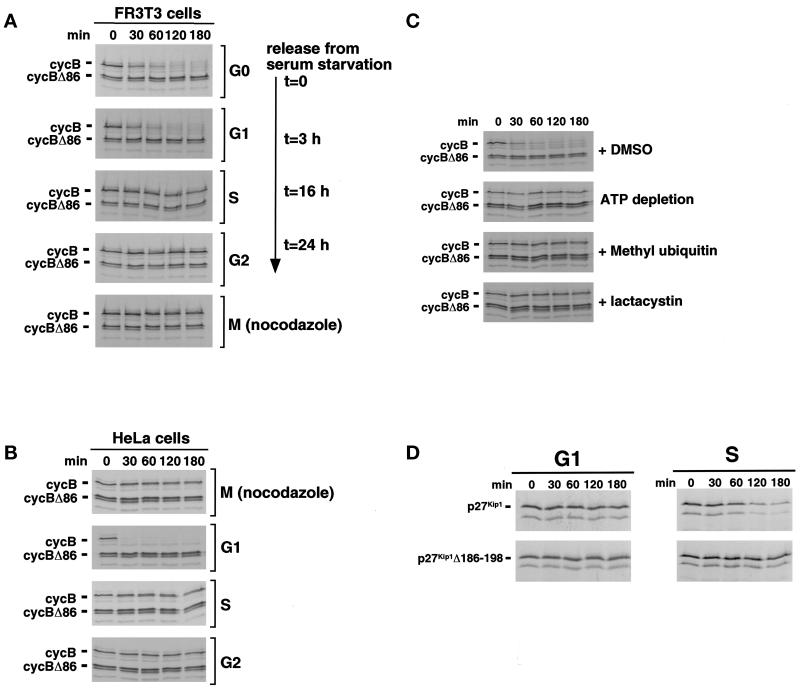
Rat fibroblasts were synchronized in G0 by serum withdrawal and stimulated to reenter the cell cycle synchronously by the addition of serum. Cells were collected in G0, G1, early S, G2, or after nocodazole-arrest in M phase. Positions within the cell cycle were confirmed by FACS analysis (our unpublished results). Extracts were prepared and assayed for the ability to degrade proteins that were provided as [35S]methionine-labeled in vitro translation products. As seen previously (Brandeis and Hunt, 1996; Bastians et al., 1998; Nguyen et al., 1999), cyclin B was destroyed in G1 extracts and stable in S phase extracts, and destruction was dependent on the presence of an N-terminal D box (Figure 1A). When the activities of the other extracts were tested, we found that cyclin B was also rapidly degraded in G0 extracts and stable in G2 extracts; extracts of nocodazole-arrested M phase cells did not degrade cyclin B (Figure 1A). Similar results were obtained with extracts of human (HeLa) cells (Figure 1B). Degradation of cyclin B in somatic cell G1 extracts was completely blocked by inhibitors of the ubiquitin–proteasome pathway, including methyl ubiquitin, lactacystin, and ATP depletion (Figure 1C). All subsequent in vitro data were obtained using HeLa cell extracts.
Figure 1.
Extracts of G0, G1, S, G2, and nocodazole-arrested M phase cells retain their stage-specific differences in D box-dependent cyclin B proteolysis. (A) FR3T3 cells were arrested in G0 by serum starvation and released into the cell cycle by addition of 10% serum. Cells were harvested after 0 (G0), 3 (G1), 16 (S), and 24 (G2) h after release or after growth in the presence of nocodazole [M (nocodazole)]. Cell extracts were prepared, supplemented with an ATP-regenerating system, and incubated for the indicated times with radiolabeled cyclin B or D box-deficient cyclin B Δ86 proteins translated in vitro. Products were analyzed by SDS-PAGE followed by autoradiography. (B) HeLa cells were synchronized in M, G1, S, and G2 as described in MATERIALS AND METHODS. Degradation activity toward radiolabeled cyclin B or cyclin B Δ86 proteins was assayed as described in A. (C) HeLa cell G1 extracts were incubated with DMSO as a control, with 10 mM glucose and 1 μg/μl hexokinase to deplete ATP, with 1 μg/μl methyl ubiquitin to inhibit polyubiquitination, or with 100 μM proteasome inhibitor lactacystin before monitoring the degradation of radiolabeled cyclin proteins as described above. (D) Extracts of HeLa cells synchronized in G1 or S phase were incubated with radiolabeled full-length or C-terminally truncated p27Kip1 translation products and assayed as above.
An important control establishes that the lack of cyclin destruction in S phase extracts was not due to the simple lack of active components of the ubiquitin-dependent proteolysis system in those extracts. The CDK inhibitor p27Kip1 (p27) is stable in G1 cells and becomes unstable in S phase cells, where it is degraded through the ubiquitin–proteasome pathway (Pagano et al., 1995; Montagnoli et al., 1999; Nguyen et al., 1999; Shirane et al., 1999). This stage-specific difference in stability can be reproduced in extracts of G1 and S phase cells (Brandeis and Hunt, 1996; Vlach et al., 1997; Bastians et al., 1998; Montagnoli et al., 1999; Nguyen et al., 1999; Shirane et al., 1999). As shown in Figure 1D, the G1 and S phase extracts used here also behaved in this way. In particular, p27 was actively degraded in S phase extracts, indicating that the general components of the ubiquitin-dependent proteolysis are present and active. Furthermore, removal of C-terminal residues 186–198, which are required for its phosphorylation-dependent destruction during S phase (Sheaff et al., 1997; Montagnoli et al., 1999; Nguyen et al., 1999), blocked p27 destruction in S phase extracts, indicating that both the regulatory and recognition systems responsible for the S phase ubiquitin-dependent proteolysis of p27 are active in S phase extracts (Figure 1D). These results establish that the extracts described here retain the cell cycle stage-specific differences in the proteolysis of cyclin B, including the M phase checkpoint inhibition of cyclin B destruction that operates in vivo.
Maintenance of Cyclin B Destruction in G1 Requires the Activity of PP2A or a PP2A-like Protein Phosphatase
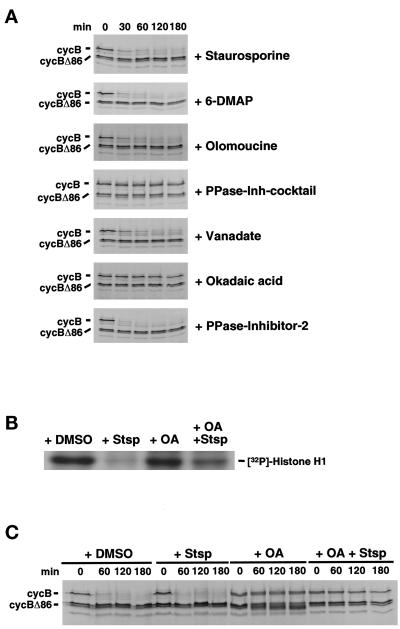
APC/C activation requires both phosphorylation and interaction of the APC/C with the noncatalytic regulators cdc20 and cdh1, which appear to regulate substrate specificity by the APC/C (for review, see King et al., 1996; Peters, 1998; Townsley and Ruderman, 1998). To ask whether the maintenance of cyclin destruction activity in G1 requires ongoing phosphorylation, the effects of various kinase and phosphatase inhibitors on cyclin B destruction were tested in G1 extracts. As shown in Figure 2A, destruction activity was not blocked by the addition of staurosporine, a general kinase inhibitor, or by 6-DMAP or olomoucine, inhibitors of cyclin-dependent kinases. By contrast, addition of a mix of phosphatase inhibitors that inhibit both tyrosine and serine/threonine protein phosphatases blocked cyclin destruction completely. Vanadate, an inhibitor that blocks tyrosine phosphatases, did not inhibit destruction, but okadaic acid (OA, an inhibitor of serine/threonine phosphatases of type 1 and 2A) did. Heat-stable inhibitor I-2, which specifically inhibits type 1 phosphatases, did not block destruction of cyclin B in G1 extracts. These results suggest that a type 2A phosphatase is involved in the maintenance of destruction activity in G1.
Figure 2.
A protein phosphatase of type 2A is required to maintain cyclin degradation activity in G1. (A) HeLa cell G1 extracts were preincubated with various kinase inhibitors (20 nM staurosporine, 5 mM 6-DMAP, 50 μM olomoucine) or phosphatase inhibitors (inhibitor mix, see MATERIALS AND METHODS; 1 mM sodium orthovanadate, 1 μM OA, 200 nM heatstable inhibitor I-2) as indicated before the addition of radiolabeled cyclin B protein and assayed as in Figure 1. (B) Histone H1 kinase activity in Hela cell G1 extracts treated with kinase or phosphatase inhibitors or both. Hela cell G1 extracts preincubated with DMSO, 20 nM staurosporine (Stsp), 1 μM OA, or 1 μM OA plus 20 nM stauroporine were subjected to histone H1 kinase assays. Phosphorylated histone H1 was resolved on 20% SDS-PAGE followed by autoradiography. (C) OA does not activate a kinase activity that is responsible for inactivating the cyclin B degradation activity in G1 extracts. HeLa cell G1 extracts used in B were preincubated with DMSO (as a control), 20 nM staurosporine (Stsp), 1 μM OA, or 1 μM OA acid plus 20 nM staurosporine before assaying the cyclin B degradation activity as decribed previously.
Inhibition of type 2A phosphatases can lead to the activation of cdc2 and other CDKs, both in vivo and in crude extracts (Picard et al., 1989; Félix et al., 1990; Picard et al., 1991). To test the possibility that OA blocked destruction through activation of a CDK or other kinase that turns off destruction activity, we added OA, either alone or in combination with staurosporine, a kinase inhibitor that blocks CDK activity (Gadbois et al., 1992; Lawrie et al., 1997; Alessi et al., 1998) and is an effective inhibitor of total histone H1 kinase activity in our extracts (Figure 2B). The ability of OA to block destruction activity was not affected by the addition of staurosporine (Figure 2C). Taken together, these results indicate that OA inhibits destruction through a pathway that does not depend on a CDK or other staurosporine-inhibitable kinase.
S and G2 Nuclei Contain a Dominant Activity That Turns Off the G1 Cyclin Destruction Machinery
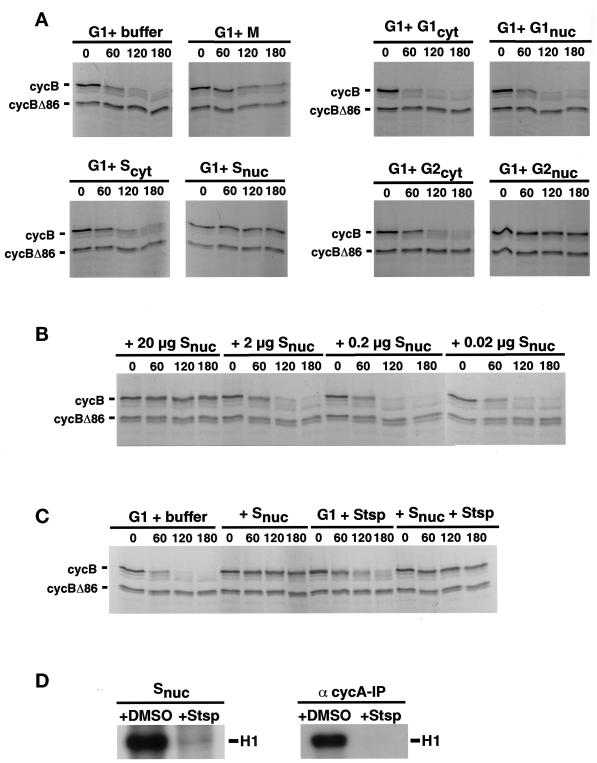
In somatic cells of both mammals and yeast, APC/C activity toward cyclin B is turned on during exit from mitosis, persists during G1 phase, and is turned off near the beginning of S phase. Whole-cell extracts reproduce these stage-specific differences in destruction activity. To ask whether S phase cells contain a dominant activity that can turn off cyclin destruction in G1 extracts, we performed a series of mixing experiments. Cells were synchronized at various cell cycle stages, and extracts of cytoplasmic and nuclear fractions were prepared. These were normalized for protein concentration and assayed for their effect on cyclin B destruction activity in G1 whole-cell extracts. The addition of G1 cytoplasm to G1 whole-cell extracts increased the rate cyclin B destruction (Figure 3A), consistent with our observation that the majority of cyclin B destruction activity was found in the cytoplasmic fraction (our unpublished results). Addition of cytoplasmic extracts from S or G2 phase cells did not affect destruction (Figure 3A). By contrast, nuclear extracts derived from S or G2 phase cells turned off cyclin degradation in G1 extracts (Figure 3A), and this inhibitory activity was dose dependent (Figure 3B). Most importantly, G1 nuclear extracts did not inhibit cyclin destruction activity, demonstrating that the destruction-terminating activity present in S and G2 nuclei is not due to a nonspecific inhibitory component in nuclear preparations. These results indicate that after the completion of G1, cells produce a nuclear, trans-acting activity that can turn off mitotic destruction, and this activity is retained as cells progress through G2.
Figure 3.
Nuclei from HeLa cells synchronized in S and G2 phase contain an inhibitor of cyclin B degradation. (A) An inhibitory activity for cyclin B degradation resides in S and G2 phase nuclei. HeLa cells were synchronized in early mitosis (nocodazole), G1, S, or G2, and nuclear and cytoplasmic extracts were prepared. These extracts (20 μg) or extraction buffer as control were mixed with total cell extracts derived from G1 cells (200 μg). Degradation activity toward cyclin B and cyclin B Δ86 was assayed as in Figure 1. (B) Dose dependency of nuclear inhibitory activity. Different amounts of protein of S phase nuclear extracts were added to HeLa cell G1 extracts, and destruction activity of radiolabeled cyclin B was assayed. (C) The S phase nuclear component responsible for inactivating cyclin B degradation in G1 extracts is not a kinase. HeLa cell G1 extracts were coincubated with buffer (as control), S phase nuclear extract (20 μg), 20 nM staurosporine (Stsp), or S phase nuclear extract pretreated with 20 nM staurosporine. Degradation activity toward cyclin B and cyclin B Δ86 was assayed. (D) S phase nuclear extracts treated with kinase inhibitor exhibit no kinase activity. Nuclear extracts derived from HeLa cells synchronized in S phase used in C were preincubated with DMSO (as control) or 20 nM staurosporine (Stsp), and histone H1 kinase activity was assayed (left panel). The same assay was performed using anti-cyclin A immunoprecipitates from S phase nuclear extracts (right panel).
In Drosophila embryonic cells, cyclin E-associated CDK activity appears to be required for inactivation of cyclin destruction at the G1–S transition (Knoblich et al., 1994). To investigate whether the inhibitory component present in S and G2 nuclei is a CDK or CDK activator, we asked whether the destruction-terminating activity present in S phase nuclei could be inactivated by the addition of kinase inhibitors. The ability of S phase nuclear extract to turn off destruction activity was not affected by the addition of staurosporine (Figure 3C), under conditions in which staurosporine was found to inhibit histone H1 kinase activity, both in extracts and in cyclin A-CDK2 immunoprecipitates (Figure 3D). Similar results were obtained with 6-DMAP (our unpublished results), a more specific CDK inhibitor (Alessi et al., 1998). These results indicate that the nuclear activity responsible for terminating cyclin B destruction activity is unlikely to be a CDK. Indeed, one possibility is that the nuclear inhibitor is the same component that is activated by inhibition of the OA-sensitive phosphatase required to maintain destruction activity in G1 extracts (Figure 2). We cannot, however, exclude the possibility that CDK activity is required transiently at or near the time of the G1–S transition to activate the inhibitor of destruction.
Intriguingly, despite the stability of cyclin B in nocodazole-arrested cells (Hunt et al., 1992) and in extracts of such cells (Figure 1), extracts of nocodazole-arrested cells did not turn off destruction in G1 extracts (Figure 3A). It is possible that such extracts do contain an inhibitor of destruction but that it is sufficiently diluted when the nuclear contents are released upon nuclear envelope breakdown that it can no longer inhibit after the further 1:10 dilution that occurs when it is mixed with G1 extract. Another possibility is that soluble inhibitors present in G2 nuclei are replaced by inhibitors that are resistant to extraction during checkpoint arrest. Several of the MAD and BUB gene products, components of the checkpoint pathway that blocks APC/C activation before chromosome congression or during spindle damage, become associated with chromosomal or spindle structures during checkpoint arrest (reviewed by Rudner and Murray, 1996; Rieder and Salmon, 1998). In cells containing two spindles, anaphase onset in one spindle is not inhibited by a second, checkpoint-arrested spindle unless the two spindles physically interact (Rieder et al., 1997). Thus, although we do not know the fate of the G2 nuclear inhibitors once cells have entered M phase, the inability of nocodazole-arrested M phase extracts to turn off destruction in G1 extracts is consistent with observations made in vivo.
Coordinate Degradation of Mitotic Regulatory Proteins
A small set of proteins identified in various systems are now known to be substrates of D box-dependent, APC/C-mediated degradation during exit from mitosis. These include cyclins A and B (Glotzer et al., 1991; Luca et al., 1991), geminin H, a Xenopus protein that can block activation of DNA synthesis in cell-free systems (McGarry and Kirschner, 1998), the anaphase inhibitors Cut2p in fission yeast (Funabiki et al., 1997) and Pds1p in budding yeast (Cohen-Fix et al., 1996), and Ase1p, a protein required for spindle elongation during telophase in budding yeast (Juang et al., 1997). Although it is now well established that components of the destruction machinery are highly conserved among eukaryotes, less is known about conservation of target recognition or conservation of the pathways that determine the timing of destruction. It was thus of interest to determine whether these proteins could be recognized by the HeLa cell G1 destruction machinery and, if so, whether their destruction would be turned off during S phase.
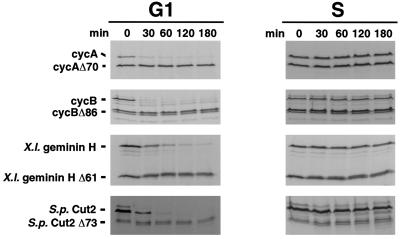
To test this, the stabilities of radiolabeled in vitro translation products were assayed in extracts of G1 or S phase cells (Figure 4). Like cyclin A and B, geminin and Cut2p were rapidly degraded in G1 extracts; their destruction depended on the presence of an N-terminal D box; and these proteins were stable in extracts of S phase cells. These results indicate that, in at least some cases, destruction recognition elements are sufficiently conserved to allow efficient, stage-specific destruction.
Figure 4.
D box-dependent in vitro degradation of cyclin A, cyclin B, Xenopus geminin H, and S. pombe Cut2p is active in G1 and inactive in S phase. HeLa cells were synchronized in G1 or S phase. Cell extracts were prepared, supplemented with an ATP-regenerating system and 2.5 μM purified recombinant wild-type UbcH10, and then assayed for their destruction activity toward various in vitro translated proteins as indicated.
By contrast, neither of the budding yeast proteins Pds1p nor Ase1p was degraded in G1 extracts (our unpublished results), despite the fact that both contain D boxes and are APC/C targets in budding yeast. At present, it is not known whether their failure to be degraded reflects an incompatibility between heterologous systems or the lack of substrate-specific cofactors necessary for their destruction.
In Vitro, Dominant Negative UbcH10 Blocks Destruction of Cyclin A, Cyclin B, Geminin, and Cut2p
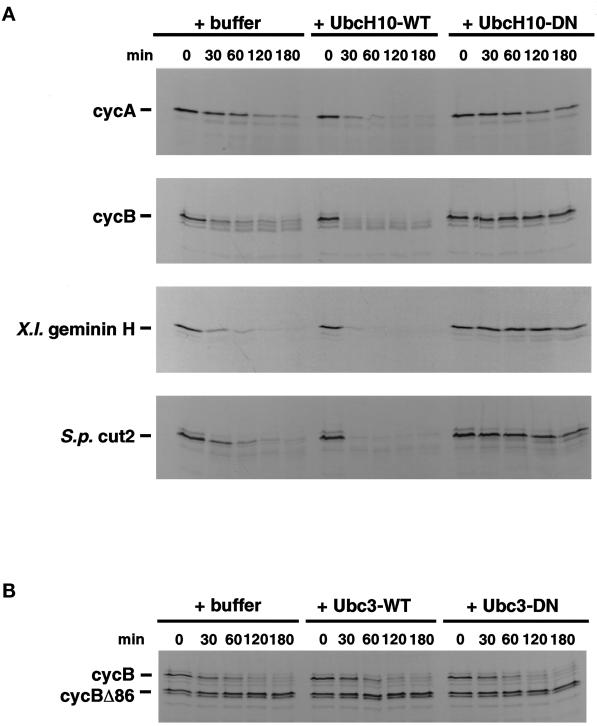
UbcH10, the human homologue of clam E2-C, Xenopus Ubc-x, S. pombe ubcP4, is required for APC/C-dependent ubiquitination and degradation of mitotic cyclins A and B in animal cells and for anaphase onset in fission yeast (for review, see Townsley and Ruderman, 1998). To ask whether UbcH10 is also involved in the destruction of Xenopus geminin and S. pombe Cut2p, the effects of adding either additional wild-type UbcH10 protein or dominant negative UbcH10 protein were examined. As shown in Figure 5A, addition of wild-type UbcH10 accelerated destruction of cyclin A, cyclin B, geminin, and Cut2p, suggesting that UbcH10 is rate limiting in G1 extracts. By contrast, addition of dominant negative UbcH10 blocked destruction of cyclin B. Nonspecific effects were ruled out by observations using a different Ubc (human Ubc3, the homologue of budding yeast Ubc3/Cdc34). Addition of wild-type Ubc3/Cdc34 failed to accelerate destruction of cyclin B, and addition of dominant negative Ubc3/Cdc34 failed to block its destruction (Figure 5B). This result further supports the idea that the APC/C-UbcH10 pathway is specialized for the coordinate degradation of a set of mitotic targets, and that many of the recognition, catalytic, and regulatory components of this pathway have remained highly conserved during evolution. It also shows that UbcH10, originally identified as the Ubc required for mitotic cyclin destruction, is involved in the proteolysis of other mitotic targets.
Figure 5.
The in vitro degradation of cyclin A, cyclin B, Xenopus geminin H, and S. pombe Cut2p involves the function of the “cyclin-selective Ubc” UbcH10. (A) HeLa G1 cell extracts were supplemented with buffer, 2.5 μM purified recombinant wild-type UbcH10 (-WT), or 2.5 μM purified recombinant dominant negative UbcH10 (-DN) and assayed for destruction activity toward radiolabeled cyclin A, cyclin B, Xenopus geminin H, and S. pombe Cut2p as described in Figure 1. (B) As a control, G1 extracts were supplemented with either 2.5 μM wild-type human Ubc3/CDC34 or 2.5 μM dominant negative human Ubc3/CDC34, and degradation of cyclin B was monitored.
In Vivo, Microinjection of Dominant Negative UbcH10 Inhibits Mitotic Progression by Delaying Anaphase Onset
Previous work demonstrated that transient transfection of dominant negative UbcH10 into mammalian cells interfered with destruction of mitotic cyclins and exit from mitosis (Townsley et al., 1997). When dominant negative UbcH10 mRNA was introduced into fertilized frog eggs, numerous cell divisions occurred; by the time of late blastula, it became evident that injected cells had a retarded rate of cell division and a pronounced accumulation of metaphase figures. In both cases, however, limitations of the experimental design made it impossible to determine whether dominant negative UbcH10 interfered with a single stage of mitotic progression or multiple stages. To investigate this point in more detail, UbcH10 protein (wild-type or dominant negative) was injected into PtK1 cells during prophase or prometaphase or after anaphase onset. PtK1 cells retain their flattened morphology during mitosis, allowing progression through the different mitotic stages to be accurately recorded in individual cells in real time using phase-contrast microscopy.
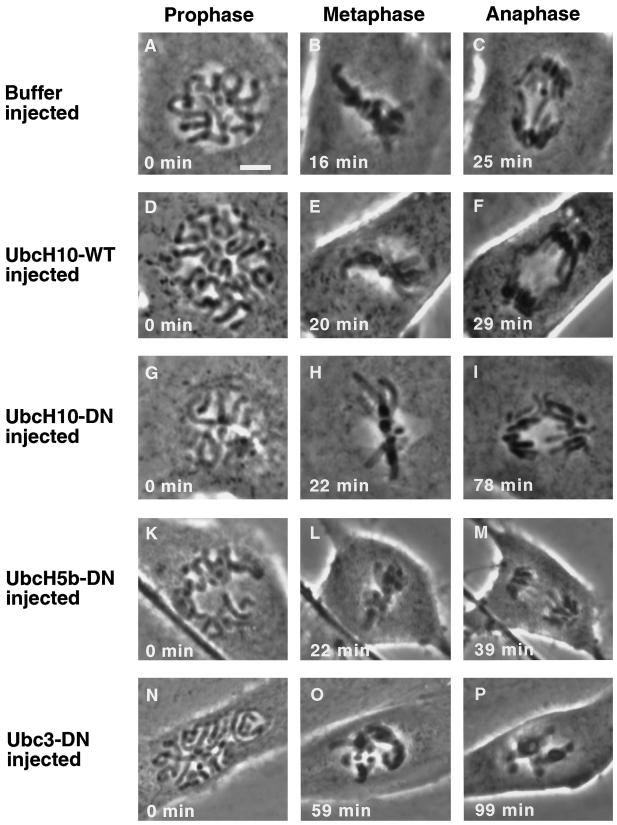
Cells injected with wild-type UbcH10 (Figure 6, D–F, UbcH10-WT) early in mitosis showed no apparent effects and exhibited no differences in either the morphology or timing of mitotic progression when compared with cells injected with buffer (Figure 6, A–C). Both sets of cells passed through mitosis at rates similar to uninjected cells. On average, uninjected cells or those injected with either buffer or UbcH10-WT required ∼20 min to reach metaphase after nuclear envelope breakdown and initial bipolar attachment of all the chromosomes. In all three cases, cells initiated anaphase after ∼9 min in metaphase (Figure 6 and Table 1) and completed cytokinesis ∼20 min later (our unpublished results).
Figure 6.
Microinjection of purified dominant negative UbcH10 protein delays anaphase onset in PtK1 cells in vivo. Phase-contrast images are shown of PtK1 cells injected either buffer (A–C), wild-type UbcH10 protein (D–F), dominant negative UbcH10 (G–I), dominant negative UbcH5b (K–M), or dominant negative human Ubc3/CDC34 (N–P). All cells were injected in prophase (2–6 min before capture of the image), and time was determined when chromosomes aligned at the metaphase plate (metaphase) and separated subsequently (anaphase). Bar, 5 μm.
Table 1.
Effects of different Ubcs on progression through mitosis
| Injection | Average (SD) | Range | Number of cells |
|---|---|---|---|
| Buffer | 8.7 (4.2) | 6–11.5 | 8 |
| Wild-type UbcH10 | 9.9 (4.1)a | 5–18.5 | 19 |
| Mutant UbcH10 | 53.8 (17.2)b | 25–104 | 37 |
| Mutant UbcH5b | 17.7 (5.13)a | 11–28 | 14 |
| Mutant Ubc3/CDC34 | >100c | >100c | 17 |
a Not significantly different from cells injected with buffer.
b Significantly different from other values; P < 0.001.
Eleven of 17 cells did not established a metaphase plate during observation period (120 min); six cells arrested at metaphase.
Injection of dominant negative UbcH10 (UbcH10-DN) did not noticeably affect the timing of chromosome congression to the metaphase plate but did cause a dramatic delay in anaphase onset. Cells injected with UbcH10-DN remained with chromosomes aligned at the metaphase plate for an average of 54 min before anaphase onset occurred (Figure 6, G–I), compared with 9 min for controls (Figure 6, A–F, and Table 1). Injection of UbcH10-DN resulted in metaphase arrest, even when cells were injected in late prometaphase just before all chromosomes were aligned; cells injected with UbcH10-DN after anaphase onset completed mitosis and cytokinesis normally (our unpublished results). By contrast, cells injected with a control protein, a dominant negative version of UbcH5b, which has no known role in mitosis (Scheffner et al., 1994; Coux and Goldberg, 1998; Gonen et al., 1999), proceeded through mitosis without any obvious delays or defects (Figure 6, K–M). The results, which are summarized in Table 1, indicate that 1) UbcH10 activity is not required for chromosome congression; 2) its first major detectable execution point during mitosis lies between the completion of chromosome alignment and initiation of anaphase; and 3) subsequent events can occur in the absence or reduced amounts of UbcH10-mediated ubiquitination. Because cyclin B destruction is required for completion of mitosis (Glotzer et al., 1991; Luca et al., 1991; Holloway et al., 1993), these results are also consistent with previous observations that cyclin B destruction is completed by the time of anaphase onset or very shortly thereafter. Finally, taken together with the demonstration that dominant negative UbcH10 blocks destruction of Cut2p in vitro (Figure 5), these results also argue for the existence of a functionally equivalent anaphase inhibitor whose APC/C- and UbcH10-dependent destruction is required for anaphase onset in mammalian cells.
Dominant Negative Ubc3/Cdc34 Interferes with Chromosome Congression
Injection of another presumptive control protein, dominant negative Ubc3, also had a striking effect on mitotic progression. Ubc3 is the human homologue of the budding yeast protein Cdc34. The best understood role of Ubc3/Cdc34 is in regulation of the G1–S transition. In yeast, it is required for the ubiquitin-dependent proteolysis of the S phase CDK inhibitor Sic1p and the resulting activation of cyclin/CDK complexes that catalyze entry into S phase (reviewed by Patton et al., 1998). In Xenopus embryos it is required for activation of cyclin E/CDKs and entry into S phase (Yew and Kirschner, 1997). In cells injected with dominant negative Ubc3 (Ubc3-DN) at the beginning of prophase, chromosomes failed to associate properly with the mitotic spindle or congress to the metaphase plate (Figure 6, N–P, and Table 1). Instead, they continued to condense, and cells arrested in prometaphase (11 cells) or reached a metaphase-like configuration only after considerable delay (six cells). None of the cells was observed to begin anaphase and complete mitosis, despite the presence of a mitotic spindle (our unpublished results). These observations suggest that, in addition to its role in G1–S, Ubc3/Cdc34 also functions during mitosis in mammalian cells.
It should be emphasized that the mitotic response of cells to injection of dominant negative Ubc3/Cdc34 was quite different from that seen with dominant negative UbcH10; Ubc3-DN blocked congression of chromosomes to the metaphase plate, whereas UbcH10-DN blocked separation of sister chromatids at anaphase onset. These results further support the conclusion that UbcH10 is required for the very selective ubiquitin-dependent degradation of an inhibitor of anaphase onset.
DISCUSSION
The results presented here establish the following main points. 1) Extracts of G0, G1, S, G2, and checkpoint-arrested M phase mammalian cells retain their stage-specific differences in mitotic cyclin destruction. 2) Like cyclin A and B, destruction activity toward two other mitotic targets of the APC/C (Xenopus geminin and S. pombe Cut2p) remains active during G1, continues to depend on the presence of a D box in the target protein during G1, requires the specialized ubiquitin carrier protein UbcH10, and is terminated by G1/S. 3) Destruction activity in G1 depends on the continuing activity of a PP2A-like phosphatase. and 4) The cessation of destruction activity at the end of G1 is due to the appearance of a dominant inhibitory activity that is localized in nuclei of S and G2 phase cells. This destruction-terminating activity appears not to be a CDK or other kinase. 5) Functional roles for a presumed mammalian anaphase inhibitor are supported by experiments showing that dominant negative UbcH10 blocks destruction of Cut2p in vitro and blocks anaphase onset in vivo. 6) Results obtained with dominant negative Ubc3/Cdc34, whose role in G1/S control is well established and has been implicated in kinetochore function in yeast, argue that Ubc3/Cdc34 is required also for congression of chromosomes to the metaphase plate in mammalian cells.
Cell-free systems from embryonic cells and genetic studies with fungi have identified many of the proteins responsible for driving progression through the cell cycle and components of checkpoint pathways that regulate major cell cycle transitions catalyzed by those proteins. Extracts of mammalian somatic cells that reproduce the cell cycle stage–specific differences in mitotic cyclin destruction occurring during the cell cycle provide important opportunities to investigate aspects of regulated APC/C activation, inactivation, and substrate recognition and to determine which aspects are generally applicable versus those that are specific to mammalian cells, to the somatic cell cycle, or to specialized cell types. Furthermore, the potential of these extracts is not limited to studies of the mitotic destruction machinery; as shown here and elsewhere (Brandeis and Hunt, 1996; Montagnoli et al., 1999; Nguyen et al., 1999; Shirane et al., 1999), G1 and S phase extracts also reproduce the stage-specific ubiquitin-dependent proteolysis of the CKI p27Kip1 seen in vivo.
A growing set of mitotic targets of the APC/C has now been identified, but less is known about their continued susceptibility to destruction during G1 progression, especially in mammalian somatic cells. Furthermore, several of these targets have been identified in only one type of organism, raising questions about the degree of conservation of substrate recognition and regulated destruction. Although the set of targets examined here was small, we can make two conclusions. First, some but not all are recognized by the destruction activity that remains active in G1 extracts. The two budding yeast proteins, Pds1p and Ase1p, were not destroyed despite the presence of obvious D box motifs that are known to be recognized by endogenous APC/C activity in yeast and, in the case of Pds1p, by frog egg APC/C (Cohen-Fix et al., 1996; Funabiki et al., 1997). Lack of destruction could be explained if recognition elements are not sufficiently conserved to allow destruction by extracts of human cells or if additional essential components are absent from human cells or extracts derived from them. The degradation of Prc1, a presumptive human homologue of Ase1p (Jiang et al., 1998), has not yet been tested. Second, destruction of human cyclin A and B, Xenopus geminin, and fission yeast Cut2p appears to coordinately regulated. For each, destruction continues through G1, G1 destruction remains dependent on both an intact D box and UbcH10 activity, and destruction is terminated by the end of G1.
As a set, the continuing destruction of these four proteins during G1 is probably important. Previous work has shown that, in HeLa cells, geminin accumulates during S, G2, and M, is degraded during mitosis, and is absent during G1 (McGarry and Kirschner, 1998). In vitro, geminin blocks formation of prereplication complexes but does not inhibit elongation. It has been proposed that at the G1–S transition, the levels of geminin accumulation are initially too low initially to block initiation; as S phase progresses, geminin accumulates to levels sufficient to block further assembly of prereplication complexes, but because it does not block elongation, replication continues to completion. Thus, switching off geminin destruction at G1–S may be part of the mechanism that limits DNA synthesis to a single round per cell cycle.
In budding yeast, continuing activity of the APC/C during G1 is essential to prevent premature initiation of DNA synthesis (Irniger and Nasmyth, 1997). In animal cells, cyclin A is a positive, essential, and rate-limiting regulator of S phase that begins to accumulate around the time of the G1–S transition (Fang and Newport, 1991; Pagano et al., 1992; Resnitzky et al., 1995). Termination of cyclin A destruction is thus required by this time to allow S phase progression; termination earlier in G1 could result in inappropriately premature activation of DNA synthesis. Cyclin B accumulation begins later, during late S–G2, and cyclin B/cdc2 complexes are kept inactive during G2 by inhibitory phosphorylation of cdc2 until the completion of G2. Although the consequences of premature cyclin B accumulation in mammalian cells are not known, injection of cyclin B into G2-arrested oocytes triggers entry into M phase (Westendorf et al., 1989), suggesting that both the strong transcriptional control of cyclin B mRNA levels (Brandeis and Hunt, 1996) and proteolytic control of cyclin B protein levels are used to avoid premature entry into M phase.
Fission yeast Cut2p is an inhibitor of anaphase onset whose D box- and APC/C-dependent destruction is required for sister chromosome separation (Funabiki et al., 1996a,b, 1997). In S. pombe cells in vivo, Cut2p destruction remains active during G1 arrest (Funabiki et al., 1996b). As shown here, destruction toward Cut2p remains active in HeLa G1 extracts and is turned off in S phase extracts (Figure 4). In view of emerging information about the regulation of sister chromatid cohesion in yeast, termination of Cut2p destruction at the end of G1 is likely to be important for the establishment of sister chromatid cohesion during S phase. In fission yeast, Cut2p binds and inhibits Cut1p; the APC/C-dependent destruction of Cut2p releases Cut1p, which then initiates loss of sister chromatid cohesion (Kumada et al., 1998). In budding yeast, Pds1p (the presumptive functional homologue of Cut2p) binds and inhibits Esp1p (McGrew et al., 1992); the APC/C-dependent destruction of Pds1p releases Esp1p, which then induces loss of cohesion by inducing dissociation of cohesin proteins from the linked sister chromatid pairs (Ciosk et al., 1998). In budding yeast, the chromosome separation defect of Pds1 mutants occurs around the G1–S boundary, indicating that Pds1p begins to function at the time when cohesion of newly replication sister chromatid segments must begin (Yamamoto et al., 1996a). If Cut2p is indeed the functional homologue of Pds1p, then its destruction should be shut off at G1–S along with other “mitotic” target proteins: termination of Cut2p destruction must occur by the end of G1 so that cohesion of newly replicated sisters can be established.
Our findings that 1) Cut2p is degraded in mitotic and G1 extracts of mammalian cells, 2) dominant negative UbcH10 blocks destruction of Cut2p in human G1 extracts, and 3) injection of dominant negative UbcH10 blocks anaphase onset in PtK1 cells in vivo strongly support the existence of a functional mammalian homologue of Cut2p. Such a protein was reported very recently, after the completion of the experiments described here, confirming and extending our work. Zou et al. (1999) identified a protein called PTTG, also termed securin, that binds to Esp1p and is degraded around the time of anaphase onset. PTTG/securin destruction is D box dependent, and nondegradable versions block sister chromatid segregation in vitro in Xenopus egg extracts. Based on our results, we would predict that PTTG/securin destruction would be blocked by UbcH10-DN, and injection of nondegradable PTTG/securin into mammalian somatic cells would block anaphase onset in vivo.
What terminates destruction activity toward mitotic targets at the end of G1? Our results demonstrate that by the beginning of S phase, cells contain a dominant activity that can shut off destruction activity in G1 extracts and that this activity is localized in nuclei. In budding yeast, the appearance of active G1 cyclin/Cdc28 complexes are required to terminate mitotic proteolysis, but the direct targets of this activity are not known (Amon et al., 1994). Cyclin E/CDK activity has been implicated in Drosophila (Knoblich et al., 1994), but little is known about its requirement in mammalian somatic cells. Our results show that a PP2A-like phosphatase activity is required to maintain destruction during G1. Thus, a component that switches off that phosphatase activity could be responsible. Because the addition of OA, which both inhibits PP2A-like phosphatases and inhibits destruction activity, can stimulate cyclin/CDK activity in crude extracts, it seemed possible that the appearance of a G1–S cyclin/CDK activity could be responsible for terminating destruction in mammalian somatic cells. Two results, however, do not support this idea. First, as shown here, the destruction-terminating activity is not inhibited by staurosporine or 6-DMAP, which are very effective CDK inhibitors. Second, the addition of pure cyclin E/CDK2 to G1 extracts does not inhibit their destruction activity (cited by Brandeis and Hunt, 1996). The identity and target of both the PP2A-like phosphatase, which is required to maintain destruction activity during G1, and the nuclear activity that terminates destruction at the end of G1 are important goals for future work.
A large body of work over the past 10 years resulted in the identification a complex system devoted to the ubiquitin-mediated proteolysis of a small set of key mitotic target proteins during exit from mitosis, identification of the recognition determinants of those proteins, and a growing biochemical understanding of how the core ubiquitinating system (the APC/C and UbcH10) is turned on to initiate anaphase onset. There is, however, growing evidence that another ubiquitination system involving Ubc3/Cdc34, and possibly Ubc3/Cdc34-mediated proteolysis, is important for an earlier mitotic event, namely, chromosome congression. The best understood role of Ubc3/Cdc34 in cell cycle progression is at the G1–S transition, at which, in association with a Skp1, Cdc53, F-box protein (SCF) complex, it catalyzes the ubiquitination (and thus proteolysis) of Sic1p, the inhibitor of S phase cyclin/CDK complexes, resulting in CDK activation and entry into S phase (reviewed by Krek, 1998; Patton et al., 1998; Peters, 1998). More recently, Ubc3/Cdc34 has been shown to be required for the G2–M transition in both yeast and animal cells, where it promotes the ubiquitination of Swe1p and wee1, respectively (Kaiser et al., 1998; Michael and Newport, 1998). These kinases phosphorylate mitotic CDKs at inhibitory positions; their Cdc34-dependent degradation allows activation of the CDKs by the phosphatase CDC25. Studies of kinetochore function in yeast have revealed a requirement for Ubc3/Cdc34 in that process as well. Kinetochores are the sites at which spindle microtubules attach to replicated chromosomes, and these attachments are essential for chromosome congression to the metaphase plate and separation of sister chromatids during anaphase (reviewed by Rieder and Salmon, 1998; Skibbens and Hieter, 1998; Dobie et al., 1999). Best defined in yeast, this region consists of centromeric DNA plus associated protein complexes. One such complex, CBF3, consists of four proteins. One of these, Cbf2p/Ndc10p, appears to be ubiquitinated by Ubc3/Cdc34 both in vivo and in vitro (Yoon and Carbon, 1995). Certain temperature-sensitive alleles of a Cdc34-associated protein, Skp1, arrest in G2–M and are viable but show increased chromosome loss at the semipermissive temperature (Bai et al., 1996; Connelly and Hieter, 1996). A third component, p58/ctf13, is an unstable, F box-containing protein whose degradation requires Ubc3/Cdc34 (Kaplan et al., 1997; Russell et al., 1999). None of these results, however, distinguish whether Cdc34 acts during prometaphase or acts earlier in the cell cycle. Our finding that injection of dominant negative Ubc3/Cdc34 into early prophase mammalian cells dramatically interferes with chromosome congression argues strongly that Ubc3/Cdc34 activity is required during early mitosis for kinetochore-mediated chromosome congression to the metaphase plate. Whether this depends on formation of stable, monoubiquitinated targets or the degradation of multiubiquitinated targets is an important question for future work.
ACKNOWLEDGMENTS
We thank Mark Rolfe, Michele Pagano, David Pellman, Mitsuhiro Yanagida, Tom McGarry, Mark Kirschner, Doug Koshland, and Orna Cohen-Fix for plasmids and Vincent Chau and Seth Sadis for Ubc5b protein. We thank Heike Krebber and members of the Ruderman laboratory for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (H.B.) and National Institutes of Health (J.V.R.).
REFERENCES
- Adams PD, Lopez P, Sellers WR, Kaelin WG. Fluorescence-activated cell sorting of transfected cells. Methods Enzymol. 1997;283:59–72. doi: 10.1016/s0076-6879(97)83007-8. [DOI] [PubMed] [Google Scholar]
- Alessi F, Quarta S, Savio M, Riva F, Rossi L, Stivala LA, Scovassi AI, Meijer L, Prosperi E. The cyclin-dependent kinase inhibitors olomoucine and roscovitine arrest human fibroblasts in G1 phase by specific inhibition of CDK2 kinase activity. Exp Cell Res. 1998;245:8–18. doi: 10.1006/excr.1998.4216. [DOI] [PubMed] [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvand A, Bastians H, Welford S, Ruderman J, Denny C. EWS/FLI1 up regulates E2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene. 1998;17:2039–2045. doi: 10.1038/sj.onc.1202129. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bastians H, Townsley FM, Ruderman JV. The cyclin-dependent kinase inhibitor p27KIP1 induces N-terminal proteolytic cleavage of cyclin A. Proc Natl Acad Sci USA. 1998;95:15374–15381. doi: 10.1073/pnas.95.26.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. Sister chromatid cohesion in mitosis. Curr Opin Cell Biol. 1998;10:769–775. doi: 10.1016/s0955-0674(98)80120-8. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J, Kirschner M, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Goldberg AL. Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor kappaB1. J Biol Chem. 1998;273:8820–8828. doi: 10.1074/jbc.273.15.8820. [DOI] [PubMed] [Google Scholar]
- Dobie KW, Hari KL, Maggert KA, Karpen GH. Centromere proteins and chromosome inheritance: a complex affair. Curr Opin Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- Fang F, Newport JN. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase intitiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Cohen P, Karsenti E. cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: evidence from the effects of okadaic acid. EMBO J. 1990;9:675–683. doi: 10.1002/j.1460-2075.1990.tb08159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle, and form large complexes. EMBO J. 1996a;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996b;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadbois DM, Hamaguchi JR, Swank RA, Bradbury EM. Staurosporine is a potent inhibitor of p34cdc2 and p34cdc2-like kinases. Biochem Biophys Res Commun. 1992;184:80–85. doi: 10.1016/0006-291x(92)91160-r. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Chen RW, Murray AW. Microinjection of antibody to Mad2 protein into mammalian cells induces premature anaphase. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen LH, Luca FC, Ruderman JV, Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hunt T, Luca FC, Ruderman JV. The requirements for protein synthesis, and the control of cyclin destruction in the meiotic and mitotic cell cycles of the clam embryo. J Cell Biol. 1992;116:707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely retated human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–885. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- Juang Y-L, Huang J, Peters J-M, McLaughlin ME, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Sia RAL, Bardes EGS, Lew DJ, Reed SI. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes & Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Durk DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degadation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- King R, Peters J, Tugendreich SM, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters J, Kirschner M. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Knoblich J, Sauer K, Jones L, Richardson H, Saint R, Lehner C. Cyclin E controls S phase progression and its down-regulation during Drosophila embyrogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tugendreich S, Fujii M, Jorgensen P-M, Watanabe N, Hoog C, Hieter P, Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1/S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M. Cut1 is loaded onto the spindle by binding to cut2 and promotes anaphase spindle movement upon cut2 proteolysis. Curr Biol. 1998;8:633–641. doi: 10.1016/s0960-9822(98)70250-7. [DOI] [PubMed] [Google Scholar]
- Lawrie AM, Noble ME, Tunnah P, Brown NR, Johnson LN, Endicott JA. Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol. 1997;4:796–801. doi: 10.1038/nsb1097-796. [DOI] [PubMed] [Google Scholar]
- Luca FC, Shibuya EK, Dohrmann CD, Ruderman JV. Both cyclin AΔ60 and BΔ97 are stable and arrest cells in M-phase, but only cyclin BΔ97 turns on cyclin destruction. EMBO J. 1991;10:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- McGrew JT, Goetsch L, Byers B, Baum P. Requirement for ESP1 in the nuclear division of S. cerevisiae. Mol Biol Cell. 1992;3:1443–1454. doi: 10.1091/mbc.3.12.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Koonce MP. Mitosis. Science. 1989;246:622–628. doi: 10.1126/science.2683078. [DOI] [PubMed] [Google Scholar]
- Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes & Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Murray A. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Gitig DM, Koff A. Cell-free degradation of p27kip1, a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Blangy A, Lane HA. Dynamic changes in nuclear architecture during mitosis: on the role of protein phosphorylation in spindle assembly and chromosome segregation. Exp Cell Res. 1996;229:174–180. doi: 10.1006/excr.1996.0356. [DOI] [PubMed] [Google Scholar]
- Osaka F, Seino H, Seno T, Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol Cell Biol. 1997;17:3388–3397. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, McGuire SL, Osmani SA. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell. 1991;67:283–291. doi: 10.1016/0092-8674(91)90180-7. [DOI] [PubMed] [Google Scholar]
- Osmani S, Ye X. Targets of checkpoints controlling mitosis: lessons from lower eukaryotes. Trends Cell Biol. 1997;7:283–288. doi: 10.1016/S0962-8924(97)01086-6. [DOI] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Peters J-M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Picard A, Capony JP, Brautigan DL, Dorée M. Involvement of protein phosphatases 1 and 2A in the control of M phase-promoting factor activity in starfish. J Cell Biol. 1989;109:3347–3354. doi: 10.1083/jcb.109.6.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard A, Labbe J-C, Barakat H, Cavadore J-C, Doree M. Okadaic acid mimics a nuclear component required for cyclin B-cdc2 kinase microinjection to drive starfish oocytes into M phase. J Cell Biol. 1991;115:337–344. doi: 10.1083/jcb.115.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:706–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- Resnitzky D, Hengst L, Reed SI. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Khodjakov A, Paliulis LV, Fortier TM, Cole RW, Sluder G. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc Natl Acad Sci USA. 1997;94:5107–5112. doi: 10.1073/pnas.94.10.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–317. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Howley PM. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff BJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes & Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- Shirane M, Harumiya Y, Ishida N, Hirai A, Miyamoto C, Hatakeyama S, Nakayama K, Kitagawa M. Down-regulation of p27kip1 by two mechanisms, ubiquitin-mediated degradation and proteolytic processing. J Biol Chem. 1999;274:13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F, Ruderman JV. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cell cycle-dependent kinase inhibitor p27Kip1. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells W. The spindle-assembly checkpoint: aiming for a perfect mitosis, every time. Trends Cell Biol. 1996;6:228–234. doi: 10.1016/0962-8924(96)10018-0. [DOI] [PubMed] [Google Scholar]
- Westendorf JM, Swenson KI, Ruderman JV. The role of cyclin B in meiosis I. J Cell Biol. 1989;108:1431–1444. doi: 10.1083/jcb.108.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast Saccharomyces cerevisiae. J Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 1998;8:144–149. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- Yew PR, Kirschner MW. Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science. 1997;177:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]
- Yoon H-J, Carbon J. Genetic and biochemical interactions between an essential kinetochore protein, cbf2p/ndc10p, and the CDC34 ubiquitin-conjugating enzyme. Mol Cell Biol. 1995;15:4835–4842. doi: 10.1128/mcb.15.9.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, King RW, Peters J-M, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]