Abstract
Poly(C)-binding proteins (PCBPs) are important regulatory proteins that contain three KH (hnRNP K homology) domains. Binding poly(C) D/RNA sequences via KH domains is essential for multiple PCBP functions. To reveal the basis for PCBP-D/RNA interactions and function, we determined the structure of a construct containing the first two domains (KH1-KH2) of human PCBP2 by NMR. KH1 and KH2 form an intramolecular pseudodimer. The large hydrophobic dimerization surface of each KH domain is on the side opposite the D/RNA binding interface. Chemical shift mapping indicates both domains bind poly(C) DNA motifs without disrupting the KH1-KH2 interaction. Spectral comparison of KH1-KH2, KH3, and full-length PCBP2 constructs suggests that the KH1-KH2 pseudodimer forms, but KH3 does not interact with other parts of the protein. From NMR studies and modeling, we propose possible modes of cooperative binding tandem poly(C) motifs by the KH domains. D/RNA binding may induce pseudodimer dissociation or stabilize dissociated KH1 and KH2, making protein interaction surfaces available to PCBP-binding partners. This conformational change may represent a regulatory mechanism linking D/RNA binding to PCBP functions.
Poly(C)-binding proteins (PCBPs)2 are KH (hnRNP-K-homology) domain-containing proteins that specifically recognize poly(C) D/RNA sequences (1, 2). There are five PCBPs in mammalian cells: PCBP1-4 and hnRNP K. Each PCBP contains three KH domains: two consecutive domains at the N terminus and a third domain at the C terminus; an intervening sequence of variable length is present between the second and third domains (Fig. 1A).
FIGURE 1.
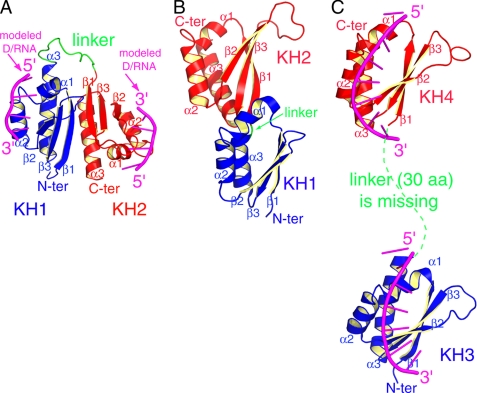
Sequence and structure of PCBP2 KH1-KH2. A, domain structure of PCBP2. Similar domain structures are observed in other PCBPs. B, sequence alignment of the KH1-KH2 domains from human PCBPs. Secondary structures for PCBP2 KH1-KH2 were based on the NMR structure. Residues that define the hydrophobic interfaces for KH1-KH2 interaction are marked with * signs above the one-letter codes. The conserved arginine residues involved in specific recognition of two consecutive cytosine bases are marked with # signs. C, left: backbone traces of the NMR ensemble of the 12 lowest-energy conformers of PCBP2 KH1-KH2. The orange ovals indicate the three less well-defined loop regions: the variable loops within the KH1 and KH2 domains, and the linker between the two domains. Middle and right, two different views of a ribbon presentation of the lowest energy structure of PCBP2 KH1-KH2. The KH1 and KH2 domains are colored blue and red, respectively, the linker is colored green. The secondary structural elements are labeled.
PCBPs regulate gene expression at various levels, including transcription, mRNA processing, mRNA stabilization, and translation, among others. For example, specific binding of hnRNP K and PCBP1 to the single-stranded pyrimidine-rich promoter sequence of the human c-myc gene and mu-opioid receptor (MOR) gene, respectively, activates transcription (3, 4).
Binding of PCBP1 or PCBP2 to cellular mRNAs harboring tandem poly(C) motifs within the 3′-UTRs stabilize these mRNAs, including α-globin, β-globin, collagen-α1, tyrosine hydroxylase, erythropoietin, rennin, and hTERT mRNAs (5-13). In the case of α-globin mRNA, it was established that the stoichiometry of the RNA-protein complex (the α-complex) is 1:1, and a minimum RNA sequence of 20-nt (5′-CCCAACGGGCCCUCCUCCCC-3′) is necessary and sufficient for forming the complex (14).
Interaction of two PCBPs, hnRNP K, and PCBP1/2, with a multiply tandem C-rich sequence (differentiation control element, DICE.) within the 3′-UTR of 15-lipoxygenase (LOX) mRNA leads to translational silencing of the mRNA in erythroid precursor cells (15-17). DICE contains 10 gapless C-rich repeats. The sequence for one repeat is 5′-CCCCACCCUCUUCCCCAAG-3′. A minimum of two repeats is required for efficient translational suppression.
PCBPs can also activate translation of cellular mRNAs. For example, binding of PCBP1 to an 18-nt C-rich sequence (5′-CUCCAUUCCCACUCCCU-3′) within the 5′-UTR of folate receptor mRNA up-regulates its translation (18). Binding of PCBP1/2 to the acute box cis-element in human heavy ferritin mRNA 5′-UTR also enhances translation (19).
Besides cellular mRNAs, PCBPs also participate in regulating critical viral RNA functions. Binding of PCBP1/2 to two cisacting C-rich sequence-containing RNA elements within the 5′-UTR of poliovirus mRNA (also the genomic RNA) is critical for regulation of cap-independent translation and replication of the viral RNA (20-24).
The mechanistic details are not well understood for any of the PCBP functions. What emerges as a common feature is the binding of PCBPs to C-rich sequence motifs (often present in tandem) of the target D/RNAs. The molecular basis of PCBP KH domains-D/RNA interactions has been revealed by a number of crystal structures of individual KH1 or KH3 domain from PCBPs in complex with C-rich D/RNA sequences (25-28). However, there are no structures with KH2.
Little is known about the events subsequent to any KH domain-D/RNA interaction. Pertinent to this point, there are no structures of PCBP constructs containing more than one KH domain. While studies suggest that protein-protein interactions occur and are vital for function, e.g. see Ref. 28), knowledge about how PCBPs engage in protein-protein interaction is also very limited. While a protein-D/RNA interaction seems essential for initiating the sequence of events, protein-protein interactions most likely are responsible for connectivity to various functions in most cases. It is therefore important to explore the missing link between protein-D/RNA and protein-protein interactions.
To address these critical issues regarding the mechanisms of PCBP functions, we have determined the solution structure of the N terminus-half of human PCBP2 containing the KH1 and KH2 domains (corresponding to residues 11-169 of the full-length protein, referred to as PCBP2 KH1-KH2 herein). The sequence of the construct, as well as homologous sequences from other PCBPs, is shown in Fig. 1B. We have also investigated D/RNA binding properties of the KH1-KH2 construct by chemical shift mapping. Spectral comparison of the KH1-KH2, KH3, and full-length protein constructs provides insights into how the three KH domains might be arranged in the full-length protein. Based on results of our studies and available biochemical data, we propose possible regulatory mechanisms for PCBP functions that involve the interplay of D/RNA binding and conformational rearrangement of the KH domains.
EXPERIMENTAL PROCEDURES
Sample Preparation—Three protein constructs of human PCBP2 were prepared containing: KH1-KH2 (residues 11-169 of the full-length sequence), KH3 (residues 285-359), and the “full-length” (residues 11-359). The proteins were expressed with N-terminal His tag (MKH6K; all but the last K could be removed by TAGzyme from Qiagen). The proteins were overexpressed in the BL21(DE3) strain of Escherichia coli (Stratagene). The preparation of properly folded PCBP2 KH1-KH2 proteins involved refolding from denatured proteins obtained from inclusion bodies. Methods for refolding and isotope labeling of the KH1-KH2 construct have been described previously (28). For most samples, the final NMR buffer contains 50 mm deuterated sodium acetate (pH 5.4), 2 mm dithiothreitol, 0.1 m EDTA, in either 90% H2O/10% D2O or 100% D2O. Preparation of the KH3 construct has also been described previously (27). For the full-length construct, protein expression was induced with 0.1 mm isopropyl-β-d-thiogalactopyranoside at A600 = 0.6-0.8 at 10 °C for 48 h before harvest. The His-tagged proteins present in the supernatant were purified using Ni-nitrilotriacetic acid resin. The purified proteins were concentrated and buffer-exchanged by centrifugation. DNA oligonucleotides were purchased from Integrated DNA Technology. Samples of the KH1-KH2-DNA complexes were prepared by titrating a solution of DNA into a solution of the KH1-KH2 protein (molar ratio 2:1) at low concentration (∼10 μm), and subsequently concentrating to ∼0.5 mm. It was found that mixing the DNA with protein and subsequent concentration of the complex led to some precipitation, presumably due to nonspecific aggregation. For this reason, the accurate DNA/protein molar ration for the NMR sample could not be determined.
NMR Spectroscopy and Structure Refinement—Data for structure determination of KH1-KH2 were collected on three isotopically labeled samples: uniformly 13C/15N, 13C/15N with 60% random fractional deuteration, and 13C/15N/2H/(Ile/Leu/Val)-methyl-protonated KH1-KH2. For most of the NMR experiments, the MKH6K-tag was not removed. All NMR experiments were performed on a Varian Inova spectrometer, operating at 600 MHz for protons, equipped with a cryoprobe. Spectra were processed with NMRPipe/NMRDraw (29) and analyzed with SPARKY (30). All NMR experiments were carried out at 25 °C. 1H, 15N, and 13C resonance assignments were achieved using standard double and triple resonance experiments. Interproton distance restraints were derived from a number of 15N or 13C-separated NOESY three-dimensional experiments. NOE-derived inter-proton distance restraints were classified into four categories with a lower bound of 1.8 Å and an upper bound of 2.7, 3.5, 5.0, and 6.0 Å corresponding to strong, medium, weak, and very weak NOEs, respectively. Torsion angle restraints were derived from TALOS (31). Generic hydrogen bond distance restraints were utilized for regions of regular secondary structure that were based on characteristic NOE patterns and chemical shifts characteristic of the secondary structure. Structure refinement was carried out by simulated annealing in torsion angle space using established procedures implemented in the program XPLOR-NIH (32). NMR restraint violations and structure quality were analyzed via the programs AQUA and PROCHECK_NMR (33). The final ensemble of structures contains the twelve lowest energy conformers, from 50 randomized initial structures; all calculated structures converge to the same fold.
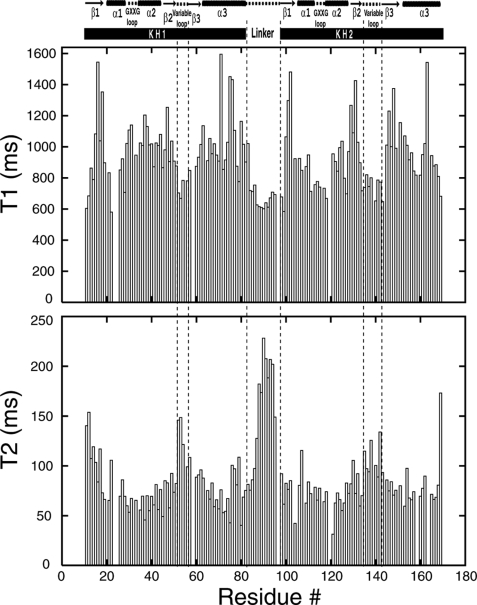
For the measurements of the longitudinal relaxation time (T1) and transverse relaxation time (T2) of the backbone amide 15N nuclei, a series of 15N-HSQC spectra with varied relaxation delays were used (10, 60, 110, 280, 440, 610, 880, 1320 ms for T1; 10, 30, 50, 70, 90, 110, 130, 150 ms for T2). The relaxation experiments were performed on a 15N-labeled sample at 298 K. Relaxation parameters (reported in Fig. 2) were calculated by fitting peak heights measured with the program SPARKY (30). Structure figures were generated using PyMol (DeLano, W. L. The PyMOL Molecular Graphics System (2002).
FIGURE 2.
Plots of backbone amide 15N T1 (top) and T2 (bottom) relaxation times as a function of residue number in PCBP2 KH1-KH2. The NMR-derived secondary structures of the protein construct are shown above the relaxation parameter plots. Dashed vertical lines highlight the three flexible loop regions (the variable loops of KH1 and KH2, as well as the linker between KH1 and KH2).
RESULTS
PCBP2 KH1-KH2 Exists as a Monomer in Solution—The PCBP2 KH1-KH2 protein (with a removable N-terminal His tag) was expressed in inclusion bodies. Properly folded proteins suitable for structural study were obtained by refolding. To investigate whether refolded KH1-KH2 is in monomeric or multimeric states, we employed the following method. Two KH1-KH2 samples, one unlabeled and the other 15N-labeled, were prepared by refolding. For the 15N-labeled sample, the His tag was removed. Equal amounts of unlabeled (with His tag) and 15N-labeled (without His tag) samples were mixed and denatured in 8 m urea. The denatured protein mixtures were then refolded. The refolded proteins were recovered from the refolding solution by Ni-NTA resin. If the refolded proteins were in multimeric states, the sample recovered by the resin would contain both the His-tagged unlabeled and tag-removed 15N-labeled proteins. SDS-PAGE electrophoresis and NMR experiments showed that only the His-tagged unlabeled protein was present in the recovered sample, indicating that no intermolecular protein-protein interaction took place during refolding: the refolded KH1-KH2 is in a monomeric state. Results from gel filtration chromatography also suggested that KH1-KH2 exists as a monomer (data not shown).
Structure and Dynamics of PCBP2 KH1-KH2—Nearly complete 1H/13C/15N chemical shifts assignments for PCBP2 KH1-KH2 were obtained and deposited in the BioMagResBank (BMRB accession number 15049). The structure was determined using distance restraints derived from NOE measurements, torsion angle restraints derived from TALOS, as well as generic hydrogen bond restraints for regular secondary structure elements. Structure calculation statistics are summarized in Table 1. The structure ensemble and two views of the lowest energy structure as a representative are shown in Fig. 1C.
TABLE 1.
NMR and structure determination statistics for PCBP2 KH1-KH2
| NOE-derived distance restraints | 1319 |
| Intra-residue | 297 |
| Sequential | 495 |
| Medium range (i+2 to i+5) | 317 |
| Long range (>i+5) | 210 |
| Total hydrogen bond restraintsa | 170 |
| Total torsion angle restraints | 256 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.0019 ± 0.0008 |
| Angles (°) | 0.3376 ± 0.0081 |
| Impropers (°) | 0.2228 ± 0.0115 |
| Backbone R.M.S.D. from mean structure (Å)b | |
| Secondary structure region | 0.85 |
| All but the linker | 1.18 |
| Ramachandran plot analysisc | |
| Residues in most favored regions (%) | 83.2 |
| Residues in additional allowed regions (%) | 13.1 |
| Residues in generously allowed regions (%) | 2.4 |
| Residues in disallowed regions (%) | 1.3 |
Includes two restraints per hydrogen bond.
Averaged over the 12 accepted structures.
Checked by PROCHECK_NMR (33).
In PCBP2 KH1-KH2, both the KH1 and KH2 domains adopt the classical type-I KH fold, which consists of three α-helices and three β-strands arranged in the order β1-α1-α2-β2-β3-α3. The conserved GXXG invariable loop (30GKKG and 114GKGG for KH1 and KH2, respectively) is located between α1 and α2; the variable loop (Ser50-Pro55 and Ala134-Thr142 for KH1 and KH2, respectively) is between β2 and β3 (Fig. 1, B and C). The three β-strands form an antiparallel β-sheet, with a spatial order of β1-β3-β2; the three α-helices are packed against one side of the β-sheet. Regions of the secondary structures and GXXG loops are well defined. But the variable loops within the two KH domains and the linker between the two KH domains are not. These loop regions of the molecule most likely are relatively flexible (Fig. 1C).
Our previous work on crystal structures of the PCBP2 KH1 domain in complex with D/RNAs (26, 28) revealed that the KH1 domain could engage in KH1-KH1 homodimerization. The protein interaction interface is a large and continuous hydrophobic surface comprised of residues from the β1-strand and α3-helix. It is clear from the present structure that the KH2 domain also has a hydrophobic surface comparable to that of the KH1 domain (Fig. 3A, left and middle). The two hydrophobic surfaces of the KH1 and KH2 domains interact with each other, resulting in an intramolecular KH1-KH2 association that buries 1528 Å2 of molecular surface from the two KH domains. The buried surface area is significantly larger than the estimated minimal area of 1200 Å2 required for a stable protein-protein complex (34, 35).
FIGURE 3.
The dimerization interfaces of the KH1 and KH2 domains. A, surface representations for the protein-interacting interfaces of the KH1 (left) and KH2 (middle) domains. For comparison, the corresponding surface of the KH3 domain (PDB ID: 2P2R) is also shown (right). The surfaces are colored as followed: C/S and C/S-bound hydrogen atoms are green, N and N-bound hydrogen atoms are blue, O and O-bound hydrogen atoms are red. Note the large, contiguous hydrophobic surface area (green) of the KH1 and KH2 domains. On the surfaces of the KH1 and KH2 domains, the β1-strand and α3-helix from the interacting KH2 and KH1 domains are also shown as ribbon and sticks (in orange; only side chains involved in direct KH1-KH2 contacts are shown. The positively charged functional groups of the lysine and arginine residues are colored deep blue). B, stereo-view of the dimerization interfaces defined by the anti-parallel positioning of strand β1 and helix α3 of the two domains. The KH1 and KH2 domains are colored blue and red, respectively. The side chains of the residues located at the dimerization interface are shown as sticks; protons are not shown for clarity. The view is similar to that in the middle panel of Fig. 2A.
The KH1 and KH2 domains are arranged in a head-to-toe manner. The protein-protein interaction is defined by the anti-parallel positioning of the longest α-helix (α3) and β-strand (β1) in the protein domains (Fig. 3B). A number of hydrophobic residues from these structural elements are involved in hydrophobic protein-protein interaction (Fig. 3, A and B). KH1-KH2 association also brings the two three-stranded antiparallel β-sheets of the KH domains together in such a way that a six-stranded antiparallel β-sheet is formed (Fig. 1C, right). Amideamide NOEs between Arg17 and Val103, Leu19, and Arg101 were observed (28), characteristic of antiparallel β-strands.
While hydrophobic forces are clearly the driving impetus for KH1 interaction with KH2, other factors, such as shape complementarities, hydrogen bonds, and electrostatic interactions may also play a role. There are two generic hydrogen bonds between backbone carbonyl and amide groups of the Arg17/Val103 and Arg101/Leu19 pairs of the interdomain antiparallel β-sheet. The guanidino groups of Arg17/Arg101, and the ε-amino group of Lys160 may mediate electrostatic interactions between the KH1 and KH2 domains (Fig. 3, A and B). Substantial interactions between the KH1 and KH2 domains show that both domains harbor a genuine protein interaction interface.
Structures of type-I KH domain-D/RNA complexes (25, 27, 28, 36-40) show that type-I KH domains recognize their D/RNA targets using a common binding interface, which is mainly defined by helices α1, α2, and strand β2. This D/RNA binding interface is located on the molecular surface opposite the protein interaction interface. Interaction of the KH1 and KH2 domains results in placement of the two D/RNA binding interfaces of the KH1 and KH2 domains on opposite molecular surfaces in the structure of PCBP2 KH1-KH2, far from each other (Fig. 4A). Modeling of the bound D/RNA can readily be achieved by comparison with the complexes entailing a single KH domain.
FIGURE 4.
Comparison of three structures of tandem type-I KH domains. A, NMR structure of PCBP2 KH1-KH2 from the present study, with modeled poly(C) DNAs bound in the nucleic acid binding grooves. B, crystal structure of FMRP KH1-KH2 (PDB ID: 2QND). C, NMR structure of FUB KH3-KH4 in complex with a single-stranded DNA (PDB ID: 1J4W). The N-terminal and C-terminal KH domains are colored blue and red, respectively. The linker region between the two KH domains is colored green. For FUB KH3-KH4 in complex with DNA and PCBP2 KH1-KH2 with modeled DNAs, the DNAs are colored magenta.
The 14-residue long linker (Ser84 to Pro97) between the KH1 and KH2 domains does not assume a regular secondary structure (Fig. 1C). This region of the molecule is the most ill-defined due to lack of constraints. However, chemical shifts for all of the linker residues were assigned. Indeed, residues from the linker region in general yielded stronger resonance signals than residues from the KH domains, indicating that the linker region probably experiences a higher degree of internal mobility.
To characterize the backbone dynamics of the PCBP2 KH1-KH2 construct, we measured the 15N longitudinal and transverse relaxation times (T1 and T2) of the protein construct. The experimental T1 and T2 values for 131 residues are shown in Fig. 2. The construct contains 158 native residues including 6 prolines. T1 and T2 values for 21 other residues could not be determined due to either absence of cross-peak or spectral overlap.
It is apparent that the measured T1 and T2 values indicate a rather non-uniform dynamic behavior of the protein construct. Excluding the terminal residues, there are four stretches of amino acids that show lower-than-average T1 values, corresponding to the variable loop of KH1, the linker between KH1 and KH2, the GXXG and variable loops of KH2, respectively (Fig. 2, top). Most residues from the two variable loops and the linker also show higher-than-average T2 values (Fig. 2, bottom). These data clearly indicate that the two variable loops within the KH domains and the linker region between the KH domains experience higher backbone mobility than other parts of the molecule. Residues in the linker region exhibit some of the lowest values of T1 and highest values of T2. A stretch of 10 amino acids (Ser86 to Arg95) within the linker has an average T2 value of 176 ms, doubling the average T2 value for the entire protein construct (88 ms). Overall, the relaxation data show that the linker region between the two KH domains is the most flexible part of the molecule. Within the two KH domains, the variable loop in each of the domains experiences increased internal molecular motion. Residues from the other secondary structural elements of the two KH domains in general show higher T1 and lower T2 values than the averages, indicating that these secondary structural elements are tightly packed in the individual KH domains, with reduced molecular mobility.
Comparison with the Two Previous Structures of Tandem KH Domains—There are two published structures of tandem type-I KH domains: the KH3-KH4 domains of the FUSE-binding protein (FBP) in complex with single-stranded DNA (38) and the KH1-KH2 domains of fragile X mental retardation protein (FMRP) (42). In the crystal structure of FMRP KH1-KH2, the linker between the two domains has only one residue (Glu) separating the last α-helix of KH1 and the first β-strand of KH2 (Fig. 4B). Compared with most other published structures of type-I KH domains (25-28, 36-40, 43-48), the KH1 domain in FMRP has a relatively short α3-helix (∼3 helical turns; the typical length is ∼5 helical turns). These structural features may impose some degree of rigidity on the relative orientation between the two KH domains. In solution, FMRP KH1-KH2 exists as a monomer.
In the NMR structure of FBP KH3-KH4 in complex with DNA (Fig. 4C), the relative orientation between the two KH domains should be well defined given the large number of RDC constraints (38). However, 30 amino acids in the linker region between KH3 and KH4 domains, as well as four nucleotides between the two bound DNA motifs were not included in the deposited coordinates, presumably due to lack of assignments or structural restraints. Missing linker regions in both protein and DNA, the actual distance between the two KH domain-DNA complexes could not be determined. Obviously, there is no protein-protein contact between the two KH domains (Fig. 4C).
The relative orientation between the KH1 and KH2 domains in PCBP2 KH1-KH2 is dramatically different from the orientations in the two previously published structures (Fig. 4, A-C). Moreover, stable protein-protein interaction between the two KH domains is observed only in the structure of PCBP2 KH1-KH2. In FMRP KH1-KH2, protein-protein contacts between the two domains are negligible. The difference in the orientation of the two KH domains in the structures also leads to different presentations of the D/RNA binding interfaces, which may correspond to different modes of cooperative target recognition by the KH domains. The two D/RNA binding interfaces in FBP KH3-KH4 have similar orientations, as do the two bound DNA motifs. The two D/RNA binding interfaces in FMRP KH1-KH2 are in close proximity to each other on the same side of the molecular surface (Fig. 4B, the surfaces facing outward). D/RNA motifs bound to the two KH domains should have somehow similar overall orientations.
In contrast to the two previous structures, the two D/RNA binding interfaces in PCBP2 KH1-KH2 are far from each other, on opposite sides of the molecular surface; the two D/RNA binding interfaces are also oriented opposite to one another. D/RNA motifs bound to the KH1 and KH2 domains in the structure of PCBP2 KH1-KH2 would therefore have a ∼180° difference in their backbone orientations (Fig. 4A). It should be noted that the structures of PCBP2 KH1-KH2 and FMRP KH1-KH2 are both for the proteins without ligands. Binding D/RNA targets may lead to conformational changes on the relative spatial relationship of the KH domains. While the very short linker in FMRP KH1-KH2 would limit the achievable conformational space, the 14 amino acid linker in PCBP2 KH1-KH2 would allow a relatively large expanse of conformational space to be explored. Comparison of the available tandem KH domain structures suggests that different properties of the KH domains, linkers, and relative orientations between the tandem KH domains may correspond to different modes of nucleic acid recognition and regulatory mechanisms.
Interaction of PCBP2 KH1-KH2 with Poly(C) Motifs—In the structure of PCBP2 KH1-KH2, both KH domains have their D/RNA binding interfaces fully exposed (Fig. 4A). To test whether the KH domains in PCBP2 KH1-KH2 can bind poly(C) sequences, we titrated PCBP2 KH1-KH2 with 7-nt and 12-nt DNA oligomers containing one or two C-rich motifs (5′-AACCCTA-3′ and 5′-AACCCTAACCCT-3′). The sequences correspond to 1 and 2 repeats of the human C-rich strand telomeric DNA. Our previous studies showed that both of these DNA sequences were able to bind to the KH1 and KH3 domains of PCBP2 (26-28, 40). Both DNA oligomers caused resonance broadening and chemical shift changes of PCBP2 KH1-KH2. We were able to assign most of the backbone 1H/15N/13C resonances of PCBP2 KH1-KH2 in the presence of the 7-nt DNA. Severe resonance broadening caused by titration with the 12-nt DNA. It is possible that the 12-nt DNA binds to two PCBP2 KH1-KH2 molecules simultaneously, leading to aggregation.
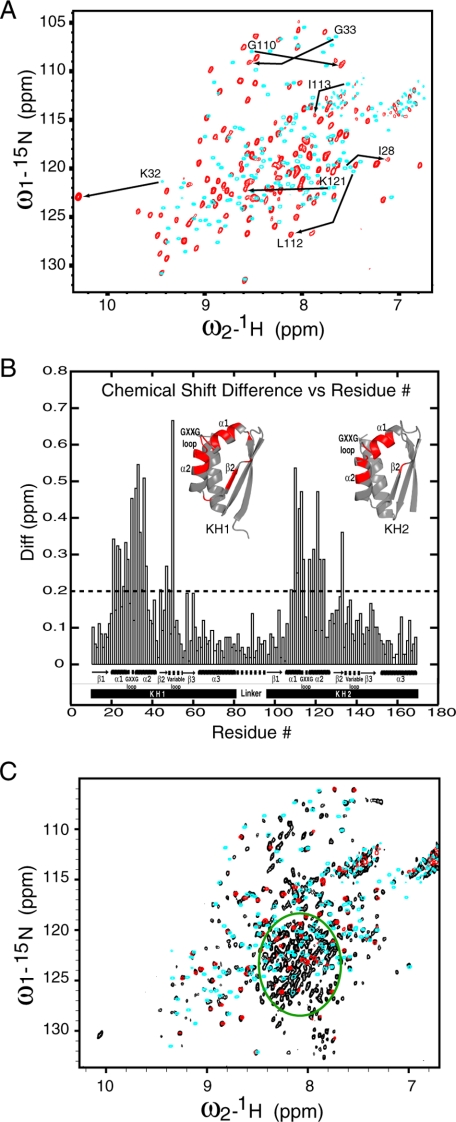
As shown in Fig. 5, A and B, the 7-nt poly(C) DNA caused some significant chemical shift changes for both the KH1 and KH2 domains. These changes are mapped to residues that define the D/RNA binding groove; residues located on the protein interaction interfaces experienced little change in chemical shifts. These data indicate that both the KH1 and KH2 domains can interact with the 7-nt poly(C) DNA using the conserved D/RNA binding interface; binding of the DNA does not interfere with protein-protein interaction between the KH1 and KH2 domains.
FIGURE 5.
A and B, chemical shift changes of PCBP2 KH1-KH2 caused by DNA binding. A, overlay of two 15N-TROSY-HSQC spectra of PCBP2 KH1-KH2 in its free form (cyan) and DNA-bound form (red). The spectra were recorded at 25 °C in a 90% H2O/10% D2O buffer containing 50 mm sodium acetate (pH 5.4), 1 mm dithiothreitol. Examples of significant chemical shift changes for residues of the KH1 and KH2 domains are indicated by arrows. B, composite chemical shift differences (in ppm) between DNA-bound and free KH1-KH2 plotted against residue numbers. The composite difference is calculated as: ((ΔH)2 + (0.154 ×ΔN)2 + (0.256 ×ΔCA)2)1/2, where Δ is the chemical shift difference value in ppm (41). The amide proton, amide nitrogen, and Cα carbon chemical shift differences are calculated as: Δ= value in DNA bound form - value in free form. Significant chemical shift changes (bigger than the dashed cut-off line) are mapped to the structures of the KH1 and KH2 domains in red. C, spectral comparison of the full-length PCBP2 with the KH1-KH2 and KH3 domains. Shown is an overlay of three 15N-TROSY-HSQC spectra for the full-length PCBP2 construct (black), the KH1-KH2 construct (cyan), and the KH3 construct (red). The green circle denoting a crowded region in the spectrum of the full-length PCBP2 construct shows little chemical shift dispersion relative to unfolded protein.
There was no experimentally determined structure of a PCBP KH2 domain prior to this study. It has been reported that KH2 domains from PCBP1, PCBP2, and hnRNP-K, when expressed as an individual domain from E. coli, could not bind poly(C) homo-oligomers as judged by gel shift assay (49). The present study not only provides the first structure of a PCBP KH2 domain, but also shows that it can interact with poly(C) motifs, most likely in a mode very similar to KH1-D/RNA interaction. The ability of the KH2 domain to recognize the poly(C) motif is fully expected. The two arginine residues in KH1 and KH3 domains responsible for specific recognition of the two central cytosine bases within the poly(C) motifs (25-28) are conserved in the KH2 domains (Fig. 1B, labeled with # signs). Because D/RNA targets of PCBPs often contain multiple poly(C) motifs, clarifying the D/RNA binding specificity of the KH2 domain is critical in understanding how the full-length PCBPs may interact with the D/RNA targets, and how D/RNA-protein interaction may regulate the functions of PCBPs.
Spatial Arrangement of the Three KH Domains in Full-length PCBP2—We have previously determined the solution structure of PCBP2 KH1 domain (40) and crystal structures of PCBP2 KH1 and KH3 domains in complex with poly(C) D/RNA oligomers (26-28). With results from the current study, it is clear that all three KH domains in PCBP2 adopt the same characteristic type-I KH domain fold and have the ability to bind poly(C) sequences using similar D/RNA interacting interfaces. Interestingly, while KH1 and KH2 domains both have a hydrophobic molecular surface involved in protein-protein interaction, the corresponding molecular surface in the KH3 domain is much more hydrophilic (Fig. 3A, right). In all three previous crystal structures of PCBP2 KH1-D/RNA complexes we determined (26, 28), the KH1 domain self-associated via the hydrophobic molecular surface to form a homodimer. In contrast, the KH3 domain existed as a monomer in the crystal structure of KH3-DNA complex (27). In solution, the KH1 domain and KH3 domain should also exist as dimer and monomer, respectively, as suggested by results from gel filtration chromatography (data not shown). Different characteristics of the KH domains, in conjunction with their relative locations in the primary sequence, may decide how the three KH domains assemble in the full-length PCBP2 protein.
To gain insight into the spatial arrangement of the three KH domains in full-length PCBP2, we compare the 15N-HSQC spectra of full-length PCBP2, KH1-KH2, and KH3 constructs. As shown in Fig. 5C, nearly all cross-peaks for back-bone amide N-H correlations from the KH3 construct overlay precisely with cross-peaks from the full-length PCBP2, indicating that the chemical environment experienced by KH3 in the context of the full-length protein is very similar to that experienced by the isolated KH3 domain. The KH3 domain in the full-length protein most likely does not interact with other parts of the protein in a significant way. The fact that all KH3 cross-peaks could be found in the spectrum of the full-length PCBP2 protein also indicates that the KH3 domain as a whole experiences a higher degree of molecular mobility than the KH1-KH2 domains (see below for more details about the KH1-KH2 domains).
For the KH1-KH2 construct, more than 86% of the cross-peaks for backbone amide N-H correlations (130 of the 151 observable cross-peaks) either overlay precisely or are very close to (chemical shift differences <0.2 ppm for 15N and <0.02 ppm for 1H) cross-peaks from the full-length PCBP2 construct. There are two possible reasons why ∼14% of the cross-peaks from the KH1-KH2 construct do not overlap corresponding cross-peaks in the spectrum of the full-length PCBP2 construct. First, the corresponding cross-peaks may not appear in the spectrum of the full-length construct. The spectrum of the PCBP2 construct was acquired on a 15N-labeled/non-deuterated sample (we were not able to obtain a deuterated sample, probably due to the need to induce protein expression at a very low temperature, and the very low expression level of soluble proteins). The PCBP2 construct is a large molecule (>37 kDa) by NMR standards. Without deuteration, we might expect that some of the cross-peaks could not be observed in the 15N-HSQC spectrum of this construct. As shown in Fig. 2, even in the isolated KH1-KH2 construct, some of the residues from the regular secondary structural elements of the two KH domains already show relatively short T2 values (∼50 ms). The T2 values of these residues should decrease even further in the context of the full-length construct, since the molecular mass of the full-length PCBP2 construct is more than double that of the KH1-KH2 construct. Second, the corresponding cross-peaks may indeed experience chemical shift changes with concomitant line broadening in the 15N-HSQC spectrum of the full-length PCBP2 construct due to transient interactions of those residues.
No matter what the scenario is, substantial spectral similarities between the PCBP2 KH1-KH2 construct and the full-length PCBP2 construct strongly suggest that the KH1-KH2 interaction evident in the solution structure of PCBP2 KH1-KH2 almost certainly is present in the full-length PCBP2. It is important to note that this kind of KH1-KH2 interaction need not be intra-molecular. It is not difficult to envisage a scenario in which the KH1 and KH2 domains of one PCBP2 molecule interact with the respective KH2 and KH1 domains of another PCBP2 molecule, resulting in a PCBP2 homodimer containing two sets of KH1-KH2 interactions. In our hand, the full-length PCBP2 construct eluted from a gel-filtration column with molecular mass corresponding to a monomer (data not shown). Presence of an intramolecular KH1-KH2 interaction in the full-length PCBP2 construct is most consistent with our NMR and chromatographic data.
As shown in Fig. 5C, many HSQC cross-peaks from full-length PCBP2 have no matching cross-peaks in spectra from KH1-KH2 and KH3. Most of these cross-peaks are found in a crowded region (green circle) with little chemical shift dispersion. This implies that the residues are in an unstructured region, undoubtedly corresponding to the ∼120-amino acid intervening sequence between the KH2 and KH3 domains. In the absence of a structure for a full-length PCBP protein, our NMR data provide critical insights into the spatial arrangement of the three KH domains in the full-length PCBP2 protein.
DISCUSSION
Prior to the present study, no structure had been reported for protein-protein interactions between two different type-I KH domains or between a type-I KH domain and another protein. The present structure confirms our hypothesis (28) that some type-I KH domains can harbor a genuine protein-protein interaction interface in addition to the established D/RNA binding interface; these KH domains can assume dual functional roles in protein-protein and protein-D/RNA interactions. Sequence conservation among the PCBP proteins (Fig. 1B) indicates that the KH1 and KH2 domains in other PCBPs should possess properties similar to the PCBP2 KH1 and KH2 domains.
Most of the identified D/RNA targets for PCBPs harbor repetitive C-rich motifs. In several cases where the minimum binding sequence has been determined, three C-rich motifs are present, with a few intervening residues between the motifs. Examples include: α-globin mRNA, 5′-CCCAACGGGCCCUCCUCCCC-3′ (14) (potential C-rich motifs with the four core recognition residues for KH domain binding are underlined); folate receptor mRNA, 5′-CUCCAUUCCCACUCCCU-3′ (18); 15-LOX mRNA, 5′-CCCCACCCUCUUCCCCAAG-3′ (15-17). We have established that all three KH domains of PCBP2 have the ability to bind poly(C) motifs, so it is very likely that all three KH domains contribute to D/RNA binding when the target contains tandem poly(C) motifs. A previous study on Drosophila P-element somatic inhibitor protein (PSI) showed that all of its four tandem KH domains were necessary for maximal binding affinity, and the four KH domains bound the RNA substrate cooperatively (50). Such cooperative binding would increase both specificity and affinity of the protein-RNA interaction.
We modeled how the PCBP2 KH domains might cooperatively interact with D/RNA targets containing multiple triple poly(C) motifs. It became readily apparent that the presence or absence of the KH1-KH2 intramolecular pseudodimer has a profound impact on possible modes of cooperative binding. Formation of the KH1-KH2 pseudodimer places the D/RNA interacting interfaces of the two domains on opposite molecular surfaces (Fig. 4A). Given the short intervening sequence (typically 2-3 nucleotides) between two tandem poly(C) motifs, it is impossible for the KH1 and KH2 domains to bind two tandem poly(C) motifs simultaneously without dissociation of the KH1-KH2 pseudodimer. It is stereochemically possible for the KH1-KH2 pseudodimer to bind the two terminal poly(C) motifs and the KH3 domain to bind the middle motif (Fig. 6A). However, this may not be energetically favorable. It is known from previous structures of KH domain-D/RNA complexes that the core motifs recognized by the KH domains consist of four residues with a characteristic conformation; base-stacking is also frequently observed for flanking residues (25, 27, 28, 36-40). Keeping the tetranucleotide core motifs in their characteristic bound conformations, not to mention the typically stacked flanking residues, would stress the intervening nucleotides to assume unusual backbone orientations. This binding mode, if it really occurred, would also mask the protein-protein interaction interface of the KH1 and KH2 domains from interaction with other protein partners. This model thus cannot explain the dependence of PCBP functions on D/RNA binding.
FIGURE 6.
Schematic diagrams illustrating possible modes of cooperative D/RNA binding by the PCBP2 KH domains. The KH1, KH2, and KH3 domains in PCBP2 are all capable of binding poly(C) D/RNA motifs. The KH1 and KH2 domains also harbor a protein-protein interacting interface. The D/RNA target contains three tetranucleotide C-rich motifs (represented as black triangles); the intervening sequences between the C-rich motifs typically have 2-3 nucleotides. The presence or absence of the KH1-KH2 intra-molecular pseudodimer has a profound impact on possible modes of cooperative binding. A, in the presence of a KH1-KH2 pseudodimer, cooperative binding requires the KH1 and KH2 domains to bind the two terminal C-rich motifs, and the KH3 domain to bind the middle C-rich motif. The question mark indicates that structural features (if any) of the intervening sequence between the KH2 and KH3 domains are not known. B, KH1, KH2, and KH3 domains bind to the 3′-end, middle, and 5′-end C-rich motifs, respectively. Dissociation of the KH1-KH2 pseudodimer exposes the protein-interacting interfaces of the KH1 and KH2 domains for interaction with other proteins that are functional partners of PCBP2. C, dissociated KH1 and KH2 domains could also interact with the KH2 and KH1 domains of another PCBP protein, leading to homo- or heterodimerization of PCBPs.
An alternative mode of cooperative binding is based on disruption of the KH1-KH2 pseudodimer. Without the constraints imposed by KH1-KH2 pseudodimer formation, there would be different ways to match the three KH domains with the three poly(C) motifs. The most straightforward would be for the KH domains and poly(C) motifs to arrange in a sequential manner with the KH1, KH2, and KH3 domains recognizing the 3′-end, middle, and 5′-end poly(C) motifs, respectively (Fig. 6, B and C). (It is not necessary for the three KH domains to assume the simple linear orientation as the schematic drawing might suggest.) This binding mode imposes minimal stress on the linker regions of both the protein and bound D/RNA. The protein-interacting interfaces of both the KH1 and KH2 domains are fully exposed, enabling them to engage in protein-protein interactions with other protein partners of PCBP2 (Fig. 6B) or dimerization with another PCBP protein (Fig. 6C). This mode of cooperative binding suggests possible regulatory mechanisms for PCBP functions. Binding tandem poly(C) motifs may induce dissociation of the KH1-KH2 intramolecular pseudodimer, but they certainly should stabilize the KH1 and KH2 domains once the pseudodimer is dissociated. As a result, the protein-interacting surfaces of the KH1 and KH2 domains become available for interaction with other proteins. Of course, this could provide the connection to regulating various cellular functions.
Binding D/RNA targets by the KH domains of PCBPs is an essential event in the mechanisms of most (if not all) PCBP functions. However, the subsequent events that link D/RNA binding to various functional consequences are not known. Because no other functional protein motifs are present in PCBPs except the three KH domains, PCBPs most likely act as adaptor or recruiter proteins that require other proteins for function, at least in most cases. For example, it was shown that poly(A)-binding protein (PABP) was critical for the mRNA-stabilizing function of PCBP1/2 in the α-complex. PCBP1/2 interacts with PABP directly, but the interaction is dependent on RNA binding to PCBP1/2. It was suggested that RNA binding might expose a protein interaction domain (51). Results from our present study reveal attractive possible mechanisms to link cooperative D/RNA binding to protein-protein interaction. Dual D/RNA and protein binding functionalities of two tandem KH domains and their relative spatial relationship are key to these mechanisms.
Accession Numbers—The structure and parameters deposited at the Protein Data Bank have been assigned PDB ID 2JZX and RCSB ID RCSB100500.
The atomic coordinates and structure factors (code 2JZX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The chemical shift assignments reported in this paper have been deposited in the BioMagResBank under BMRB accession number 15049.
This work was supported, in whole or in part, by National Institutes of Health Grant AI46967 (to T. L. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PCBP, poly(C)-binding proteins; KH, K homology; UTR, untranslated region; PDB, Protein Data Bank; R.M.S.D., root mean square deviation.
References
- 1.Makeyev, A. V., and Liebhaber, S. A. (2002) RNA 8 265-278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamarnik, A. V., and Andino, R. (2000) J. Virol. 74 2219-2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomonaga, T., and Levens, D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5830-5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik, A. K., Flock, K. E., Godavarthi, C. L., Loh, H. H., and Ko, J. L. (2006) Brain Res. 1112 33-45 [DOI] [PubMed] [Google Scholar]
- 5.Weiss, I., and Liebhaber, S. (1995) Mol. Cell. Biol. 15 2457-2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chkheidze, A. N., Lyakhov, D. L., Makeyev, A. V., Morales, J., Kong, J., and Liebhaber, S. A. (1999) Mol. Cell. Biol. 19 4572-4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu, J., and Russell, J. E. (2001) Mol. Cell. Biol. 21 5879-5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefanovic, B., Hellerbrand, C., Holcik, M., Briendl, M., Aliebhaber, S., and Brenner, D. (1997) Mol. Cell. Biol. 17 5201-5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindquist, J. N., Kauschke, S. G., Stefanovic, B., Burchardt, E. R., and Brenner, D. A. (2000) Nucleic Acids Res. 28 4306-4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulding, W. R., and Czyzyk-Krzeska, M. F. (1999) J. Biol. Chem. 274 2532-2538 [DOI] [PubMed] [Google Scholar]
- 11.Czyzyk-Krzeska, M. F., and Bendixen, A. C. (1999) Blood 93 2111-2120 [PubMed] [Google Scholar]
- 12.Persson, P. B., Skalweit, A., Mrowka, R., and Thiele, B. (2003) Am. J. Physiol. Regul Integr Comp Physiol 285 R491-R497 [DOI] [PubMed] [Google Scholar]
- 13.Emerald, B. S., Chen, Y., Zhu, T., Zhu, Z., Lee, K.-O., Gluckman, P. D., and Lobie, P. E. (2007) J. Biol. Chem. 282 680-690 [DOI] [PubMed] [Google Scholar]
- 14.Waggoner, S. A., and Liebhaber, S. A. (2003) Exp. Biol. Med. 228 387-395 [DOI] [PubMed] [Google Scholar]
- 15.Ostareck-Lederer, A., Ostareck, D. H., Standart, N., and Thiele, B. J. (1994) EMBO J. 13 1476-1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostareck, D. H., Ostareck-Lederer, A., Wilm, M., Thiele, B. J., Mann, M., and Hentze, M. W. (1997) Cell 89 597-606 [DOI] [PubMed] [Google Scholar]
- 17.Ostareck, D. H., Ostareck-Lederer, A., Shatsky, I. N., and Hentze, M. W. (2001) Cell 104 281-290 [DOI] [PubMed] [Google Scholar]
- 18.Xiao, X., Tang, Y. S., Mackins, J. Y., Sun, X. L., Jayaram, H. N., Hansen, D. K., and Antony, A. C. (2001) J. Biol. Chem. 276 41510-41517 [DOI] [PubMed] [Google Scholar]
- 19.Thomson, A. M., Cahill, C. M., Cho, H.-H., Kassachau, K. D., Epis, M. R., Bridges, K. R., Leedman, P. J., and Rogers, J. T. (2005) J. Biol. Chem. 280 30032-30045 [DOI] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and Andino, R. (1997) RNA 3 882-892 [PMC free article] [PubMed] [Google Scholar]
- 21.Gamarnik, A. V., and Andino, R. (1998) Gene Dev 12 2293-2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsley, T. B., Towner, J. S., Blyn, L. B., Ehrenfeld, E., and Semler, B. L. (1997) RNA 3 1124-1134 [PMC free article] [PubMed] [Google Scholar]
- 23.Blyn, L. B., Swiderek, K. M., Richards, O., Stahl, D. C., Semler, B. L., and Ehrenfeld, E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 11115-11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blyn, L. B., Towner, J. S., Semler, B. L., and Ehrenfeld, E. (1997) J. Virol. 71 6243-6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backe, P. H., Messias, A. C., Ravelli, R. B., Sattler, M., and Cusack, S. (2005) Structure (Camb) 13 1055-1067 [DOI] [PubMed] [Google Scholar]
- 26.Du, Z., Lee, J. K., Tjhen, R., Li, S., Pan, H., Stroud, R. M., and James, T. L. (2005) J. Biol. Chem. 280 38823-38830 [DOI] [PubMed] [Google Scholar]
- 27.Fenn, S., Du, Z., Lee, J. K., Tjhen, R., Stroud, R. M., and James, T. L. (2007) Nucleic Acids Res. 35 2651-2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du, Z., Lee, J. K., Fenn, S., Tjhen, R., Stroud, R. M., and James, T. L. (2007) RNA 13 1043-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomolec. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 30.Goddard, T. D., and Kneller, D. G. (1998) SPARKY, 3.0 Ed., University of California, San Francisco
- 31.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 32.Schwieters, C. D., Kuszewski, J. J., Tjandra, N., and Clore, G. M. (2003) J. Magn. Res. 160 66-74 [DOI] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomolec. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 34.Chothia, C., and Janin, J. (1975) Nature 256 705-708 [DOI] [PubMed] [Google Scholar]
- 35.Jones, S., and Thornton, J. M. (1995) Prog. Biophys. Mol. Biol. 63 31-59 [DOI] [PubMed] [Google Scholar]
- 36.Lewis, H. A., Musunuru, K., Jensen, K. B., Edo, C., Chen, H., Darnell, R. B., and Burley, S. K. (2000) Cell 100 323-332 [DOI] [PubMed] [Google Scholar]
- 37.Liu, Z. H., Luyten, I., Bottomley, M. J., Messias, A. C., Houngninou-Molango, S., Sprangers, R., Zanier, K., Kramer, A., and Sattler, M. (2001) Science 294 1098-1102 [DOI] [PubMed] [Google Scholar]
- 38.Braddock, D. T., Louis, J. M., Baber, J. L., Levens, D., and Clore, G. M. (2002) Nature 415 1051-1056 [DOI] [PubMed] [Google Scholar]
- 39.Braddock, D. T., Baber, J. L., Levens, D., and Clore, G. M. (2002) EMBO J. 21 3476-3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du, Z., Yu, J., Chen, Y., Andino, R., and James, T. L. (2004) J. Biol. Chem. 279 48126-48134 [DOI] [PubMed] [Google Scholar]
- 41.Mulder, F. A., Schipper, D., Bott, R., and Boelens, R. (1999) J. Mol. Biol. 292 111-123 [DOI] [PubMed] [Google Scholar]
- 42.Valverde, R., Pozdnyakova, I., Kajander, T., Venkatraman, J., and Regan, L. (2007) Structure 15 1090-1098 [DOI] [PubMed] [Google Scholar]
- 43.Musco, G., Stier, G., Joseph, C., Morelli, M. A. C., Nilges, M., Gibson, T. J., and Pastore, A. (1996) Cell 85 237-245 [DOI] [PubMed] [Google Scholar]
- 44.Lewis, H. A., Chen, H., Edo, C., Buckanovich, R. J., Yang, Y. Y., Musunuru, K., Zhong, R., Darnell, R. B., and Burley, S. K. (1999) Structure Fold Des. 7 191-203 [DOI] [PubMed] [Google Scholar]
- 45.Baber, J. L., Libutti, D., Levens, D., and Tjandra, N. (1999) J. Mol. Biol. 289 949-962 [DOI] [PubMed] [Google Scholar]
- 46.Maguire, M. L., Guler-Gane, G., Nietlispach, D., Raine, A. R., Zorn, A. M., Standart, N., and Broadhurst, R. W. (2005) J. Mol. Biol. 348 265-279 [DOI] [PubMed] [Google Scholar]
- 47.Sidiqi, M., Wilce, J. A., Vivian, J. P., Porter, C. J., Barker, A., Leedman, P. J., and Wilce, M. C. J. (2005) Nucleic Acids Res. 33 1213-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García-Mayoral, M. F., Hollingworth, D., Masino, L., Díaz-Moreno, I., Kelly, G., Gherzi, R., Chou, C.-F., Chen, C.-Y., and Ramos, A. (2007) Structure 15 485-498 [DOI] [PubMed] [Google Scholar]
- 49.Dejgaard, K., and Leffers, H. (1996) Eur. J. Biochem. 241 425-431 [DOI] [PubMed] [Google Scholar]
- 50.Chmiel, N. H., Rio, D. C., and Doudna, J. A. (2006) RNA 12 283-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Z., Day, N., Trifillis, P., and Kiledjian, M. (1999) Mol. Cell. Biol. 19 4552-4560 [DOI] [PMC free article] [PubMed] [Google Scholar]