Abstract
The P2X7 receptor regulates cell growth through mediation of apoptosis. P2X7 levels are lower in cancer epithelial cells than in normal cells, and previous studies showed that expression of P2X7 was regulated post-transcriptionally. The objective of the study was to understand regulation of P2X7 mRNA stability. Overexpression of a reporter containing the full-length human P2X7 3′-untranslated region (3′-UTR) or reporters containing parts of the 3′-UTR-P2X7 were associated with increased abundance of the construct in normal cells and decreased abundance in cancer epithelial cells. Sequences within the 3′-UTR-P2X7, which are putative target sites for the microRNAs, miR-186 (middle segment) and miR-150 (distal segment), decreased the abundance of the P2X7 transcript. Overexpression in cancer cells of mutated miR-186 and miR-150 target sites was associated with lower levels of the reporter genes. In normal cells overexpression of the mutated miR-186 target site was associated with marked increased concentration, but overexpression of the miR-150 target site reporters, wild-type and mutant, did not change over time. Levels of miR-186 and miR-150 were higher in cancer than in normal cells, and treatment with miR-186 and miR-150 inhibitors increased P2X7 mRNA. In human embryonic kidney-293 cells heterologously expressing the full-length 3′-UTR-P2X7 luciferase reporter, miR-186 and miR-150 inhibitors increased luciferase activity, whereas miR-186 and miR-150 mimics decreased luciferase activity after actinomycin D treatment. These data suggest that increased expression of miR-186 and miR-150 in cancer epithelial cells decreases P2X7 mRNA by activation of miR-186 and miR-150 instability target sites located at the 3′-UTR-P2X7.
The receptor P2X7 is a membrane-bound, ligand-operated channel (1–6). ATP is the naturally occurring ligand for the P2X7, and extracellular levels of ATP may reach low micromolar levels (7–12), which are sufficient to activate the receptor (13). Activation of the receptor may induce formation of pores in the plasma membrane (14), which in epithelial cells mediate apoptosis via the caspase-9 mitochondrial pathway (15, 16). The P2X7 apoptosis effects can be regulated by receptor glycosylation (16), trafficking, plasma membrane expression (17–20), oligomerization (7, 21), and by receptor post-activation internalization, recycling, and degradation (14, 21).
In epithelial tissues the P2X7 receptor is expressed predominantly by proliferative (germinative) epithelial cells (21–23), and it controls the growth of the epithelial cells. Previous studies in human uterine epithelial cells showed that base-line and P2X7-mediated apoptosis are lower in cancer cells than in normal cells (12, 22, 23). The differences were not the result of ligand availability because steady-state levels of ATP in conditioned media of cancer epithelial cells were similar to those of normal epithelial cells (12). Similarly, there were no significant differences in P2X7 receptor activation, oligomerization, or cycling between normal and cancer cells (12). In contrast, the differences were associated with lower P2X7 mRNA and protein levels in the cancer cells than in the corresponding normal epithelial cells (17, 18). Moreover, P2X7 receptor expression was decreased in pre-cancerous epithelial tissues (22, 23). These findings are biologically and clinically important because defective apoptosis may lead to cancer (24–26), and the decreased cellular expression of P2X7 could be causally related to the development and progression of uterine cancers.
At present little is known about how epithelial cells regulate expression of the P2X7 receptor. The human P2X7 receptor gene is localized to chromosome 12q24 and includes 13 exons (27). P2X7 transcription may be regulated.3 However, a recent preliminary study showed that P2X7 expression is also regulated post-transcriptionally (23). In that preliminary experiment, normal and cancer epithelial cells were transfected with a luciferase reporter vector ligated with the 3′-UTR4 region of the human P2X7 gene. The steady-state levels of luciferase mRNA in normal cells were higher than in control cells after a 6-h incubation, but luciferase mRNA levels were lower in cancer cells (23). These data suggested that cancer epithelial cells control P2X7 mRNA expression through targets in the 3′-UTR P2X7. Moreover, because the decrease of P2X7 mRNA was quantitatively as great as that of the protein in cancer cells (22), it is possible that P2X7 mRNA is degraded at a faster rate in cancer cells than in normal epithelial cells. The objective of this study was to gain a better understanding of the mechanism by which cancer epithelial cells control the expression of P2X7 mRNA post-transcriptionally.
Degradation of mRNA can be mediated by two main mechanisms as follows: cis-dependent trans-acting regulatory proteins (28, 29) and microRNAs (miRNA) (13, 30). In this study we tested the hypothesis that post-transcriptional regulation of P2X7 mRNA expression involves miRNAs. miRNAs are small, noncoding 18–25-nucleotide RNAs that regulate mRNA targets and control critical functions across various biological processes (31). The role of miRNAs in differentiation and apoptosis was previously documented (32), and recent data underscored their role in cancer etiology (reviews in Refs. 13, 33, 34). Animal miRNAs form imperfectly base-paired duplexes with miRNA response elements in target mRNAs. This interaction can inhibit translation and lead to the degradation of the mRNA (35–37). For effective repression, base-pairing between the miRNA-response elements and the first 2–7 nt of the miRNA, the “seed” region, are important (38–41).
The specific objective of this study was to explore which miRNAs are important for P2X7 mRNA regulation. The experimental model was the well characterized system of cultured human uterine epithelial cells (14–16, 21–23). We found putative target sites for miR-186 and miR-150 within the 3′-UTR of P2X7, and both of these cis-elements conferred instability to the transcript. Mutagenesis of these sites confirmed that they are important for the maintenance of P2X7 mRNA stability. The data showed that levels of miR-186 and miR-150 were higher in cancer than in normal cells, both in vivo and in cultured cells. In addition, treatment with miR-186 and miR-150 inhibitors increased P2X7 mRNA, predominantly in the normal cells. Also, in the host human embryonic kidney-293 (HEK293) cells heterologously expressing a luciferase reporter containing the full-length human 3′-UTR-P2X7, treatment with miR-186 and miR-150 inhibitors increased luciferase activity. To determine whether a decrease in transcription or RNA stability produced the decreased luciferase activity, HEK293 cells containing the 3′-UTR-P2X7 reporter and miR-186 and miR-150 mimics were treated with actinomycin D, and the results showed a decrease in luciferase activity in the presence and absence of the drug. These data suggest that miR-186 and miR-150 stimulate a decrease in P2X7 mRNA steady-state levels by targeting instability sites within the 3′-UTR of the P2X7 gene. The data suggest that the reduced expression of P2X7 in cancer epithelial cells is the result of high steady-state levels of miR-186 and miR-150 in cancer cells, which activate the instability domains and decrease P2X7 mRNA levels possibly by inducing degradation of the transcript.
EXPERIMENTAL PROCEDURES
Human Tissues—Discarded human tissues were obtained according to IRB protocols 12-03-50 and 03-90-300 from the Human Tissue Procurement Facility of University Hospital CASE Medical Center/the Comprehensive Cancer Center Tissue Procurement Core Facility at Case Western Reserve University, Cleveland, OH. Cross-sections of uterine (ectocervical and endometrial), bladder, and breast segments were obtained from paraffin-embedded blocks that were prepared by the Department of Pathology initially to establish the diagnosis for the patient. For the purpose of this study, additional parallel 10-μm sections were cut, and slides intended for immunostaining were made by the Department of Pathology according to standard procedures. This part of the study included tissues obtained from four women with cervical cancers, three women with endometrial cancers, two women and one man with bladder cancers, and three women with breast cancers. Included for each case were cross-sections of the cancer tissue and an adjacent histologically normal tissue.
Discarded human endometrial and ectocervical tissues from women undergoing hysterectomy for indications unrelated to this study were obtained according to the above IRB protocols also from the Cooperative Human Tissue Network (NCI, National Institutes of Health) through the Human Tissue Resource Network, Department of Pathology, Ohio State University, Columbus. These tissues were used for mRNA and microRNA assays. Tissues were snap-frozen in liquid N2, shipped on dry ice, and stored at -80 °C until assayed. Included were tissues obtained from two women with endometrial adenocarcinomas, three women with cervical squamous carcinomas, and from four women with histologically normal cervix.
The histological diagnoses presented below were made by the Departments of Pathology at University Hospitals of Cleveland or at the Ohio State University. No attempt was made to categorize results by ethnic origin or race of the patients.
Image Analysis of Immunofluorescence Data—Immunostaining with the rabbit polyclonal anti-P2X7 receptor antibody (Alomone Laboratories, Jerusalem, Israel) or the anti-cytokeratin-18 (CK-18) and anti-E-cadherin antibodies (Cell Signaling Technology, Danvers, MA) and image analysis of immunofluorescence data were described (12, 21–23). Briefly, fields of interest were captured and saved in Adobe Photoshop. Pictures were scanned using UN-SCAN-IT software (Silk Scientific, Orem, UT) by choosing 3–5 representative fields for each picture. Light intensity in each field was digitized, and average pixel density for P2X7 per field was determined using the program software. Data are presented in terms of P2X7 pixel density averaged over the number of cells per field (determined by 4′,6-diamidino-2-phenylindole-stained nuclei (22)).
Cell Cultures, Transfections, Immunoprecipitation, Western Blots, and Apoptosis Assays—Primary cultures of human ectocervical-vaginal epithelial cells (hEVEC), a well characterized model of the normal human ectocervical epithelium, were generated from discarded normal ectocervical-vaginal tissues as described (42). Cultures of normal human keratinocytes were generated from discarded foreskins that were obtained following nonritual circumcision of newborn males at the nursery of the Department of Obstetrics and Gynecology, University Hospital CASE Medical Center. Tissues were obtained according to approved IRB protocols 12-03-50, 03-90-300, and 08-06-28. The human cervical epithelial cancer HeLa cells, HEK293 cells, and Madin-Darby canine kidney cells (MDCK, strain II) were obtained from the ATCC. HEK293 and MDCK cells lack endogenous expression of the P2X7 receptor. MDCK cells expressing tetracycline-regulated repressor (21) were used for heterologous transfection expression experiments. For inducible expression of P2X7 receptors in MDCK cells, the human P2X7 gene (NM_002562), including a Myc tag attached to the N terminus, was subcloned into pcDNA4/TO vector (Invitrogen) with HindIII and NotI sites. Primers, the method of transfections, and the generation of stable MDCK clones were described (21). Stable clones were maintained in tetracycline-free medium and 100 μg/ml Zeocin. Expression of Myc-P2X7 genes was induced by treatment with 100 ng/ml doxycycline, and expression of Myc-P2X7 protein in stable MDCK cells was confirmed by RT-PCR (21). Methods of cell cultures were described (12).
Immunoprecipitation using the anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was described (21). Western blots on cell lysates using the anti-Myc or anti-P2X7 antibody or anti-tubulin antibody (from the Developmental Studies Hybridoma Bank, University of Iowa, Iowa City) were described (16). Apoptosis was quantified using the cell death detection ELISA kit (Roche Applied Science) (16).
P2X7-Antisense Oligonucleotides—P2X7-specific antisense oligonucleotides (ASO) and random control oligonucleotides (RCO) were designed from the published sequence of the human P2X7 gene (NM_002562). The sequences of the 20-mer ASO that would hybridize to the coding region of exon 7 (nt 724–741) and the RCO are shown in Table 1. In the RCO the antisense sequence was randomly replaced with adenine and thymine residues so that the oligonucleotides had the same length (20-mer) and GC content (33%) as the ASO. The RCO was designed such that no cross-hybridization against the P2X7 gene would occur. To assess the effects of the ASO and RCO on P2X7 mRNA expression, cultured cells were treated with or without ASO or RCO at concentrations of 10 μm. qPCR assays were carried out after 2 days, and Western blots and apoptosis determinations were made after 3 days.
TABLE 1.
Sequences of P2X7 oligonucleotides, miR-186 and miR-150, and primers for P2X7 3′-UTR and luciferase The underlined segments within the miR-186 and miR-150 sequences are the putative seed sequences that base pair with their respective targets.
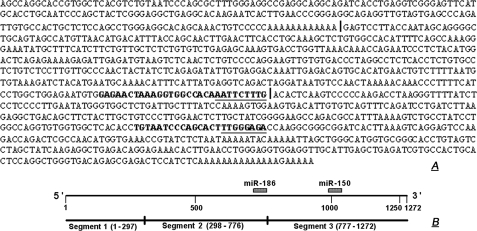
3′-UTR-P2X7 Assays—Fig. 1, top, shows the sequence of the 3′-untranslated region (3′-UTR, 1272 nt) of the human P2X7 gene (NM_002562). Vertical lines in Fig. 1, top, within the 3′-UTR P2X7 sequence indicate subregions of the 3′-UTR (Segment-1, nt 1–297; Segment-2, nt 298–776; Segment-3, nt 777–1272; Fig. 1, bottom). Primers for amplification of the segments are shown in Table 1. The corresponding cDNAs were synthesized by reverse transcription using human keratinocyte RNA as the template and oligo(dT) (16) as the primer. The full-length P2X7 3′-UTR was cloned by digestion of the two PCR fragments with XbaI and StuI and ligation of the products into pGL3 promoter luciferase reporter vector (SV40 promoter, Promega, Madison, WI), containing XbaI-5′ and XbaI-3′ sites, using the rapid ligation kit (Roche Applied Science). P2X7 3′-UTR Segments 1–3 were cloned by digestion with XbaI and ligation into the same vector.
FIGURE 1.
A, sequence of the human P2X7 3′-UTR. Vertical lines within the sequence delineate subregions (segments) of the 3′-UTR that were subcloned into reporter plasmids to test RNA stability. Sequences in bold are putative target sites for miR-186 (end of Segment-2) and miR-150 (middle of Segment-3). Underlined are putative sites within the target that base pair with the seed sequence of the respective miRNAs. B, schema of the full-length human P2X7 3′-UTR with its division into Segments 1–3, and the putative target sites for miR-186 and miR-150.
Primers for experiments testing putative miR-186 and miR-150 target regions within the human P2X7 3′-UTR are shown in Table 1. Each complementary pair of primers was ligated into the pGL3 promoter vector XbaI site and cloned as described above. All constructs were confirmed by sequencing.
Control plasmids or test plasmids (P2X7-3′-UTR full-length luciferase, P2X7-3′-UTR-Segment-1-luciferase, P2X7-3′-UTR-Segment-2-luciferase, or P2X7-3′-UTR-Segment-3-luciferase) were transfected into hEVEC or HeLa cells. Cells were plated in 6-well plates at a density of 2·105 cells per well, resulting in subconfluent cultures. After 14 h of incubation the culture medium was replaced with fresh medium plus 100 μl of serum-free medium (per well) containing 2.5 μl of FuGENE 6 (Roche Applied Science) and 750 ng of DNA of the control or test vectors. After 48 h cells were shifted to fresh medium lacking plasmids, and at times 0, 3, and 6 h, cells were harvested in RNA lysis buffer (Qiagen, Valencia, CA; RNeasy mini kit). Total RNA was extracted, and luciferase mRNA levels were determined by the one-step real time RT-PCR using the primers described in Table 1. qPCR assays utilized 600 ng of RNA/sample as described (22, 23).
Real Time PCR—Total RNA from tissues or from cultured cells was extracted by RNeasy mini kit (Qiagen, Valencia, CA), and one-step real time qPCR was carried out as described (22, 23). Primers for the P2X7, E-cadherin, and cytokeratin-18 (CK-18) (used for data normalization) and PCR conditions were described (22, 23). Relative quantification (RQ) was calculated using Applied Biosystems SDS software (Foster City, CA) based on the equation RQ = 2-ΔΔCt, where Ct is the threshold cycle to detect fluorescence.
Micro-RNA in Situ Hybridization—Pre-designed miRCURY™ in situ detection probes for miRNA-186, miRNA-150, and U6 snRNA were obtained from Exiqon Inc. (Woburn, MA). Cells (2·105) were plated on Lab-Tek chamber slides (Nalge Nunc International, Rochester, NY) and cultured at 37 °C overnight. Cells were rinsed with PBS (pH 7.4) and fixed for 10 min in paraformaldehyde (4% in PBS). For deproteinization, cells were washed twice for 5 min at 37 °C in PBS, incubated for 10 min at 37 °C with 10 μg/ml proteinase K (in 5 mm Tris, 13.8 mm NaCl, 0.27 mm KCl), washed for 30 s with 0.2% glycine in PBS, and twice (30 s) with PBS. Cells were fixed again for 10 min in 4% paraformaldehyde and rinsed twice for 5 min in PBS. For prehybridization, cells were incubated for 2 h at 55°C in a humidified chamber in 0.5 ml of hybridization buffer (composed of 50% formamide, 5× SSC (150 mm NaCl plus 15 mm sodium citrate), 0.1% Tween 20, 9.2 mm citric acid (pH 6), 50 μg/ml heparin, 500 μg/ml yeast RNA). For hybridizations, probes (miR-186, miR-150, and U6 [Applied Biosystems, Foster City, CA]) were diluted to 200 nm in hybridization buffer. Slides containing cells were incubated in 500 μl of hybridization mix per slide, with a set of a negative control (blank, no probe added) and a positive control (U6 snRNA). Cells were hybridized overnight at 55 °C in a humidified chamber, rinsed in 2× SSC at 55 °C and three times for 30 min in 2× SSC plus 50% formamide at 55 °C, followed by 5-min washes at room temperature in PBST (PBS plus 0.1% Tween 20). For immunological detection, hybridized cells were blocked for 1 h at 23 °C in blocking buffer (10 mg/ml bovine serum albumin in PBST), followed by incubation overnight at 23 °C in a humidified chamber at 4 °C with 1:200 anti-digoxigenin-AP antibody (Fab fragments, Roche Applied Science). Slides were washed five times at 23 °C for 5 min in PBST; 0.5 ml of AP-substrate (Roche Applied Science, catalog number11442074001) was added in a humidified chamber for 4 h of incubation at 23 °C in the dark. Slides were washed three times for 5 min in PBST and mounted in aqueous mounting medium. Staining was evaluated using Nikon Eclipse 80i microscope, and image analysis of the staining was done as described above.
Micro-RNA Real Time RT-PCR—Human endometrial tissues were homogenized by grinding with mortar and pestle under liquid N2. Cultured cells were harvested off the plates by pipetting. Total RNA from the lysates of the endometrial tissues or the cultured cells was isolated using the mirVana miRNA isolation kit (Ambion, supplied by Applied Biosystems, Foster City, CA) according to the supplier's instructions. Purity and concentration of the samples were assessed by spectrophotometry. Real time PCR quantification of miRNA-186, miRNA-150, and U6 snRNA (endogenous control) was performed following the protocol described for the mirVana™ qRT-PCR miRNA detection kit from Ambion, using 7500 real time PCR system instrument (Applied Biosystems, Foster City, CA). PCRs were performed in triplicate using specific mirVana™ qRT-PCR primer sets for miRNA-150, miRNA-186, or U6. The assay includes two steps as follows: RT to synthesize cDNA from miRNA and PCR amplification. For RT, the reaction master mixture containing 5× RT Buffer, 1× RT Primers, 5× Array-Script Enzyme Mix, and nuclease-free water were mixed with 2.5 μg of total RNA in a 50-μl volume. Mixtures were incubated for 30 min at 37 °C in a thermal cycler and then for 10 min at 95 °C to inactivate the ArrayScript and cooled to 4 °C to stop the reaction. Real time PCR was done using QuantiTect SYBR Green RT-PCR kit (Qiagen). The reaction containing 2× QuantiTect SYBR Green RT-PCR Master Mix, 10× primer mix, 100 ng of RT product, and nuclease-free water in a 20-μl volume was processed as follows: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 60 s (repeated three times). Relative quantification was calculated as described above, and miRNAs data were calculated in terms of Ct and normalized to U6 RNA.
miR-186 and miR-150 Inhibition and Mimics Assays—Assays utilized Ambion AM4510 kit and miR-186 and miR-150 inhibitors and mimics (Ambion, Austin TX) according to the supplier's instructions. Immediately prior to performing the assays, two solutions were prepared as follows: a 1:50 mixture of siPORT NeoFX transfection agent in Opti-MEM medium and a mixture of 6.25 nm miR-186 or miR-150 inhibitors or mimics in 300 μl of Opti-MEM medium (per well). The two solutions were mixed; 600 μl of the combined mixture were dispensed into each well of an empty 6-well culture plate, and cells were added into each well for assays.
For effects of miRNAs inhibitors on P2X7 mRNA stability, hEVEC or HeLa cells (2.5·105 in 2 ml culture medium) were added into each well, and cultures were incubated at 37 °C for 48 h. Cells were harvest in RNA lysis buffer for total RNA and qPCR determinations of P2X7 mRNA (normalized to CK-18). The efficiency of miR-186 and miR-150 inhibitors transfections was tested using PTK9 mRNA assays (supplied with the kit) (not shown).
Effects of mimics and inhibitors of miR-186 and/or miR-150 on 3′-UTR-P2X7 stability were determined in HEK293 cells heterologously expressing the full-length 3′-UTR-P2X7. HEK293 cells (1·105/ml) were co-transfected in 12-well plates using the siPORT neoFX protocol (Ambion). For each well, 400 ng of the pGL3/P2X7-3′-UTR report vector, 12 ng of a control vector containing Renilla luciferase and pRL-CMV (Promega), and 10 nm mimics or inhibitors of miR-186 and/or miR-150 (Ambion) were used. Firefly and Renilla luciferase activities were measured consecutively by using dual-luciferase reporter assay system (Promega) 48 h after transfection. 3′-UTR-P2X7-dependent luciferase activity was determined in terms of Fluc/Rluc.
Data Analysis—Data were analyzed using GraphPad Instat, GraphPad Software Inc., San Diego, CA. Significance of differences among groups was estimated by t test, or by one-way or two-way analysis of variance with Tukey-Kramer multiple comparisons post test analysis or the Dunnett's test.
Supplies—All chemicals, unless specified otherwise, were obtained from Sigma.
RESULTS
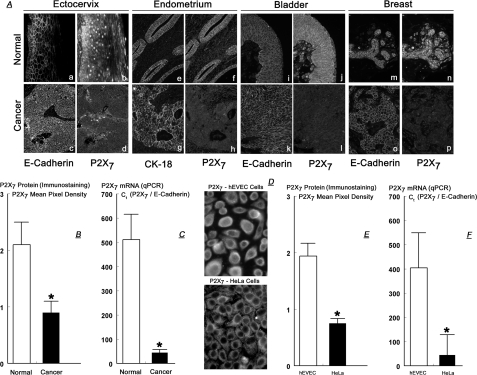
P2X7 Protein and mRNA Levels in Normal and Cancer Epithelial Cells—Fig. 2A shows immunoreactivity to anti-P2X7 antibody of tissue cross-sections obtained from four patients with ectocervical (panels a–d), endometrial (panels e–h), bladder (panels i–l), and breast cancers (panels m–p). In each case tissues were obtained from a site that included the cancerous tissue and from an adjacent histologically normal site. Tissues were co-immunostained with anti-P2X7, plus anti-E-cadherin or anti-CK-18 antibodies. Immunostaining with anti-E-cadherin and anti-CK-18 antibodies was used to identify the epithelial component of the tissue, because the P2X7 receptor is expressed predominantly by epithelial cells (22, 23). The ectocervical and endometrial data (Fig. 2A, panels a–h) confirmed previous reports (22, 23), whereas the bladder and breast tissue data are novel. In the normal tissues P2X7 immunoreactivity was localized mainly within the epithelial component (Fig. 2A, panels a and b, e and f, i and j, and m and n), and in the case of the ectocervix predominantly in the basal-parabasal layers (Fig. 2A, panels a and b).
FIGURE 2.
A, tissue cross-sections from four patients with ectocervical squamous carcinoma (panels a–d), endometrial adenocarcinoma (panels e–h), bladder transitional cell carcinoma (panels i–l), and breast ductal adenocarcinoma (panels m–p). Tissues were co-immunostained with anti-P2X7 plus anti-E-cadherin or with anti-P2X7 plus anti-CK-18 antibodies (10 times). Data are representative of a total of 12 similar cases. B, semiquantitative analysis of P2X7 immunostaining in four cases of ectocervical cancers. C, qPCR data of P2X7 mRNA (normalized to E-cadherin mRNA) in normal (n = 4) and cancer (n = 3) ectocervical tissues. D, immunostaining with anti-P2X7 antibody of cultured hEVEC and HeLa cells (20 times). E, semi-quantitative analysis of P2X7 immunostaining in hEVEC and HeLa cells. F, qPCR data of P2X7 mRNA (normalized to E-cadherin mRNA) in hEVEC and HeLa cells. Experiments D–F were repeated five times. B, C, E, and F, *, = p < 0.001 compared with normal tissues or hEVEC cells.
In all cases P2X7 immunoreactivity was reduced in the cancer tissues (Fig. 2A, panels d, h, l, and p) compared with the normal tissues (Fig. 2A, panels b, f, j, and n). Semi-quantitative analysis of P2X7 immunostaining data in four cases of ectocervical cancers showed a 2.3-fold lower P2X7 immunostaining density in the cancer tissues than in the normal tissues from the same patients (Fig. 2B).
Fig. 2C shows qPCR data of P2X7 mRNA in normal and cancer ectocervical tissues. When normalized to E-cadherin mRNA, P2X7 mRNA in cancer ectocervical tissues was 10-fold lower than P2X7 mRNA levels in normal and ectocervical tissues.
Similar data were obtained in cultures of human normal (hEVEC) and cancer (HeLa) cervical epithelial cells. P2X7 immunoreactivity was lower in HeLa than in hEVEC cells (Fig. 2D) by 2.2-fold (Fig. 2E), and P2X7 mRNA (normalized to E-cadherin mRNA) was 10-fold lower in HeLa than in hEVEC cells (Fig. 2F). These data confirm (22, 23) that P2X7 protein and mRNA levels are lower in cancer epithelial tissues compared with normal epithelial cells.
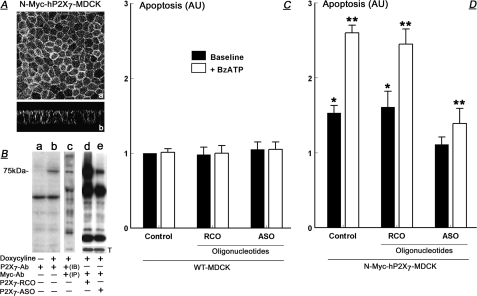
Requirement and Specificity of the P2X7 Receptor for BzATP-related Apoptosis—It was previously suggested that the P2X7 mechanism controls growth of epithelial cells through activation and modulation of apoptosis (12, 15). To test this hypothesis more directly, experiments utilized MDCK cells, which lack endogenous expression of the P2X7 receptor (21). MDCK cells were transfected with doxycycline-inducible reporter containing the human full-length P2X7 receptor tagged with N-Myc at the N terminus (N-Myc-hP2X7-MDCK), and stable transfectants were used for experiments. Laser-confocal microscopy of doxycycline-treated N-Myc-hP2X7-MDCK cells showed P2X7 immunoreactivity decorating the plasma membrane (Fig. 3A). Western blots of doxycyline-treated N-Myc-hP2X7-MDCK cell lysates revealed immunoreactivity of a 75-kDa form with anti-P2X7 antibody (Fig. 3B, lanes a and b). In Fig. 3B, lane b, immunoblotting in the presence of the P2X7 antigen (21) blocked the 75-kDa immunoreactivity (data not shown; see Ref. 21), indicating the specificity of the immunoreactivity. Specificity of the 75-kDa immunoreactivity was further demonstrated by immunoprecipitation of lysates of doxycyline-treated N-Myc-hP2X7-MDCK cells with the anti-Myc antibody followed by immunoblotting with the anti-P2X7 antibody, demonstrating again the 75-kDa immunoreactivity (Fig. 3B, lane c). Treatment of N-Myc-hP2X7-MDCK cells with antisense P2X7 oligonucleotides inhibited expression of the 75-kDa P2X7 receptor (Fig. 3B, lanes d and e). These data suggest that the transfected N-Myc-hP2X7 receptor is properly inserted into the plasma membrane of MDCK cells and is expressed in the form of the functional 75-kDa receptor (21).
FIGURE 3.
Heterologous expression in MDCK cells of the human full-length P2X7 and effects of treatments with BzATP on apoptosis. Cells were transfected with the human full-length P2X7 receptor Myc-tagged at the N terminus (N-Myc-hP2X7-MDCK), and expression of the receptor was induced by treatment with doxycycline. Cells were also co-treated with the vehicle (Control); with antisense P2X7 oligonucleotides (ASO), or with random-control P2X7 oligonucleotides (RCO). A, P2X7 immunoreactivity in control cells captured by laser confocal microscopy at the planar (panel a) and vertical (panel b) axes. B, Western immunoblots of lysates of N-Myc-hP2X7-MDCK cells using the indicated antibodies. Expression of the P2X7 receptor was induced by treatment with doxycycline, and cells were treated with P2X7 RCO or ASO as indicated. In lane c lysates were immunoprecipitated (IP) with the anti-Myc antibody and immunoblotted (IB) with the anti-P2X7-antobody. In lanes d and e, membranes were reprobed and immunoblotted with anti-tubulin antibody (T), showing equal protein gel loading. Experiments repeated three times. C and D, levels of apoptosis (arbitrary units (AU)) in wild-type (WT) MDCK cells or in N-Myc-hP2X7-MDCK cells. Treatments included P2X7 RCO or ASO and BzATP (100 μm, 8 h) as indicated. Shown are means ± S.D. of three experiments repeated in triplicates. * = p < 0.05 compared with Control WT-MDCK; ** = p < 0.01 compared with control N-Myc-hP2X7-MDCK.
To test the effect of heterologous expression of the human full-length P2X7 receptor in MDCK cells on base-line and agonist-induced apoptosis, N-Myc-hP2X7-MDCK cells were treated with the P2X7-specific agonist 2′,3′-0-(4-benzoylbenzoyl)-adenosine 5′-triphosphate (BzATP). In wild-type MDCK cells (WT-MDCK), treatment with BzATP did not affect baseline steady-state levels of apoptosis (Fig. 3C). In contrast, baseline apoptosis of N-Myc-hP2X7-MDCK cells was 1.5-fold greater than of WT-MDCK cells (probably induced by ATP secreted by the cells (1, 5)), and treatment with BzATP augmented the apoptosis 2.5-fold (Fig. 3D). Treatments with antisense P2X7 oligonucleotides or with random control P2X7 oligonucleotides in WT-MDCK cells had no effect on apoptosis (Fig. 3C). In contrast, treatment of N-Myc-hP2X7-MDCK cells with antisense P2X7 oligonucleotides (but not with random-control P2X7 oligonucleotides) inhibited both the base-line apoptosis and the BzATP-augmented apoptosis (Fig. 3D). These data indicate that the P2X7 receptor is an important mediator of BzATP-dependent apoptosis.
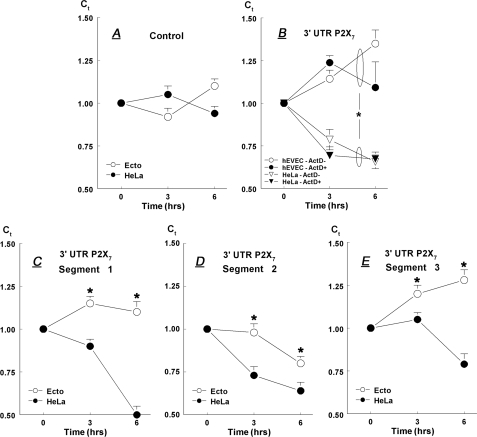
Stability of a P2X7-3′-UTR-Luciferase Construct—To test the hypothesis that regions in the 3′-UTR of the P2X7 gene confer instability to the transcript, we examined a series of reporter constructs in transient transfection assays containing different regions of the P2X7 3′-UTR. In the first experiment a cDNA stretch corresponding to the entire 3′-UTR region of the human P2X7 gene (nt 1–1272; Fig. 1A) was ligated to the SV40 promoter-luciferase reporter vector, and the plasmid was transfected into hEVEC or HeLa cervical epithelial cells. Control experiments included cells transfected with the luciferase reporter vector only. After 48 h of transfection cells were shifted to fresh medium lacking added plasmids, and at times 0, 3, and 6 h cells were harvested; total RNA was extracted, and luciferase mRNA levels were determined by one-step real time RT-PCR. qPCR data of luciferase mRNA (in terms of Ct, relative to CK-18 mRNA) were normalized to 0-h results.
In cells transfected with the control luciferase reporter vector, steady-state levels of luciferase mRNA changed relatively little (Fig. 4A). In hEVEC cells transfected with the entire length of 3′-UTR-P2X7-luciferase vector, steady-state levels of luciferase mRNA increased significantly over the 6-h incubation period. In contrast, in HeLa cells transfected with that vector luciferase mRNA decreased significantly (Fig. 4B).
FIGURE 4.
Steady-state levels of luciferase mRNA (relative to CK-18 mRNA) in hEVEC and HeLa cells. Cells were transfected with either the control luciferase reporter vector (A); the full-length 3′-UTR-P2X7--luciferase vector (B); or Segment-1 (C), Segment-2 (D), or Segment-3 (E) of the 3′-UTR-P2X7--luciferase vector. B, cells in some cultures were treated for 0, 3, and 6 h with 10 μm actinomycin-D. Shown are means ± S.D. of 2–3 experiments repeated in triplicate. * = p < 0.05–0.01.
To test whether the differences in steady-state levels of the P2X7-3′-UTR-luciferase construct in HeLa versus the hEVEC cells involve modulation of transcription of the luciferase reporter vector, the experiments were repeated with the addition of the transcription inhibitor actinomycin-D. Treatment with 10 μm actinomycin-D did not show significant additional effects (Fig. 4B), suggesting that the lower steady-state levels of the P2X7-3′-UTR-luciferase construct in HeLa cells compared with the hEVEC cells are not the result of decreased transcription but rather increased instability of the construct.
To better understand the instability of the 3′-UTR-P2X7-luciferase construct in HeLa cells, the 3′-UTR of the human P2X7 gene was divided into three segments (Segment-1, nt 1–297; Segment-2, nt 298–776; and Segment-3, nt 777–1272) (Fig. 1, A and B). cDNA corresponding to the three segments was generated, and luciferase mRNA levels were determined at 0, 3, and 6 h.
In hEVEC cells transfected with vectors containing Segment-1 or Segment-3, luciferase mRNA increased, whereas in HeLa cells it decreased (Fig. 4, C and E) similar to changes in cells transfected with the vector containing the full-length 3′-UTR-P2X7 (Fig. 4B). In cells transfected with the vector containing Segment-2, luciferase mRNA decreased both in hEVEC and HeLa cells, but the decrease in HeLa cells occurred earlier and was greater than in hEVEC cells (Fig. 4D). Collectively, the data in Fig. 4 show greater instability of the overexpressed full-length P2X7 3′-UTR reporter and each of the Segments 1–3 reporters in the cancer epithelial cells compared with the normal epithelial cells.
Stability of Constructs with Sequences Matching miR-186 and miR-150—Bioinformatic analysis identified putative miRNA target sites within the human P2X7 3′-UTR. In this study we examined the role of miR-186 and miR-150 on P2X7 mRNA stability (sequences shown in Table 1). A putative target of miR-186 was found at the end of Segment-2 of the human P2X7 3′-UTR, and a putative target of miR-150 was found in the middle of Segment-3 (Fig. 1, A, bold letters, and B).
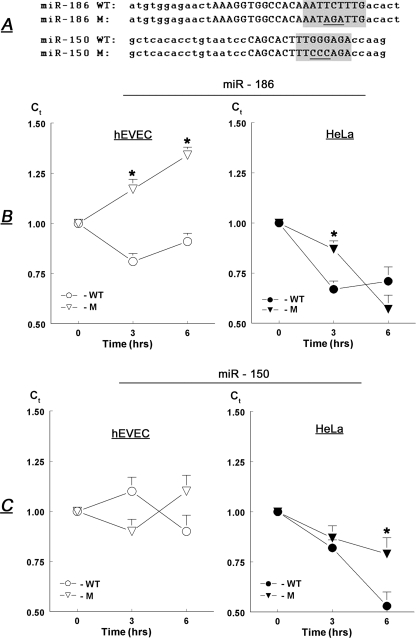
To test the potential role of miR-186 and miR-150 in the regulation of P2X7 mRNA, we studied the stability of luciferase constructs attached to short segments of the P2X7 3′-UTR containing the putative target sites. In addition, short stretches within the putative target sites of each of the two miRNAs were mutated to disrupt the region of base pairing between the miRNA and target, and the stability of the reporters carrying the mutated 3′-UTRs was tested. For miR-186, the TCT was changed to AGA, and for miR-150 the GGG was changed to CCC in the putative target sites (Fig. 5A, underlined).
FIGURE 5.
A, sequences within the putative target sites for miR-186 and miR-150 of the human P2X7 3′-UTR (uppercase letters) that were chosen for the experiments in B and C. Shaded areas are putative site of base pairing between the target and the seed sequence of the miRNAs. Short regions within the target were mutated (underlined). WT, wild type; M, mutated. B and C, steady-state levels of luciferase mRNA (relative to CK-18 mRNA) in hEVEC and HeLa cells transfected with luciferase reporter vectors containing wild type or mutated putative target sites for miR-186 (B) or miR-150 (C). Shown are means ± S.D. of two experiments repeated in triplicate. * = p < 0.05–0.01.
Luciferase mRNA levels in hEVEC cells transfected with reporter containing the wild-type miR-186 target site decreased slightly (Fig. 5B, ○). In contrast, luciferase mRNA increased in hEVEC cells transfected with reporter containing the mutated miR-186 target site (Fig. 5B, Δ). Luciferase mRNA levels decreased in HeLa cells transfected with reporter containing either the wild-type miR-186 target site (Fig. 5B, •) or the mutated miR-186 target site (Fig. 5B, ▴), but at 3 h the 3′-UTR containing the mutated 186 target site was significantly more abundant than the wild type. Luciferase mRNA levels did not change significantly in hEVEC cells transfected with reporter containing either the wild-type miR-150 target site (Fig. 5C, ○) or the mutated miR-186 target site (Fig. 5C, Δ). Luciferase mRNA levels decreased in HeLa cells transfected with reporter containing either the wild-type miR-150 target site (Fig. 5C, •) or the mutated miR-150 target site (Fig. 5C, ▴), but at 6 h the 3′-UTR containing the mutated miR-150 target site was more abundant than the wild type. Collectively, the data in Fig. 5 indicate that regions within Segment-2 and Segment-3 of the human P2X7 3′-UTR, which contain the miR-186 and miR-150 target sites, respectively, confer instability to the P2X7 transcript.
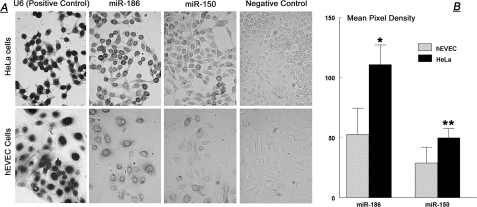
Cellular Levels of miR-186 and miR-150 Are Higher in Cancer than in Normal Cultured Cervical Cells—In situ hybridization assays revealed a significantly stronger perinuclear-cytoplasmic staining for both miR-186 and miR-150 in HeLa cells than in hEVEC cells (Fig. 6A). Semiquantification of the in situ staining density showed 2.2-fold greater pixel density per cell for miR-186 in HeLa than in hEVEC cells and a 1.7-fold greater pixel density per cell for miR-150 in HeLa than in hEVEC cells (Fig. 6B).
FIGURE 6.
A, in situ hybridization of miR-186 and miR-150 in hEVEC and HeLa cells (10 times). The experiment was repeated twice. B, semi-quantitative analysis of the miRNA in situ hybridization data. Pictures were scanned using UN-SCAN-IT; regions encircling cells of interest (n = 10 for each slide) were chosen, and mean (±S.D.) pixel density was determined. *, p < 0.001 compared with hEVEC; **, p = 0.003 compared with hEVEC.
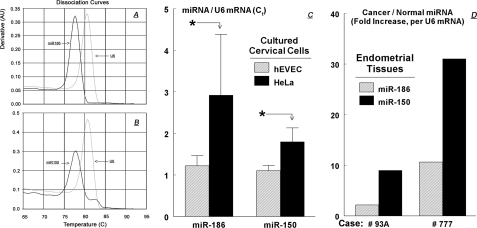
The in situ hybridization data for miRNAs were confirmed using miRNA qPCR assays. The dissociation-temperature curves for miR-150 (Fig. 7A), miR-186 (Fig. 7B), and U6 RNA yielded monophasic single peaks at 77.5 °C for miR-150 and miR-186, and at 81 °C for U6, indicating the generation of single PCR products. The miRNAs qPCR data (Fig. 7C) showed a 1.7-fold higher miRNA-150/U6 ratio in HeLa cells compared with hEVEC cells and a 2.9-fold higher miRNA-186/U6 ratio in HeLa cells compared with hEVEC cells. Collectively, the data in Fig. 6 and Fig. 7, A–C, indicate that steady-state cellular levels of miR-186 and miR-150 are higher in the cancer HeLa cells than in the normal hEVEC cells.
FIGURE 7.
miRNA qPCRs. A and B show the dissociation temperature curves for miR-186 (A), miR-150 (B), and the positive control U6 snRNA (lysates from hEVEC cells). C, levels of miR-186 and miR-150 in hEVEC and HeLa cells (qPCR, means ± S.D., two experiments in triplicate, normalized to U6 RNA). *, p < 0.001 compared with hEVEC. D, levels of miR-186 and miR-150 in normal and cancerous human endometrial tissues. Discarded surgical uterine tissues were obtained from two patients with endometrial endometrioid (type I) adenocarcinomas. In each case fresh samples were collected from a site that included the cancerous tissue and from an adjacent macroscopic (and histologically confirmed) normal endometrial site. For each case, qPCR Ct data of miR-186 and miR-150 levels in cancer tissues were normalized to those in the normal tissue of the same case.
miR-186 and miR-150 Levels Are Higher in Endometrial Cancer Tissue than in Normal Endometrium—Levels of miR-186 and miR-150 were also determined in human endometrial cancer and normal tissues. The endometrium was chosen as a prototype example because it is embryologically related to the cervix, and because it is relatively easy to dissect and collect the epithelial components of normal and cancerous endometrial tissues. Discarded surgical uterine tissues were obtained from two patients with endometrial endometrioid (type I) adenocarcinomas. In each case fresh samples were collected from a site that included the cancerous tissue and from an adjacent macroscopic (and histologically confirmed) normal endometrial site. The miRNAs qPCR data showed that in both cases miR-186 and miR-150 levels in the cancer tissues were higher than those in the normal tissues. Thus, miR-186 levels in the cancer tissues were 2.2- and 10.6-fold higher than in the corresponding normal tissues, and miR-150 levels in the cancer tissues were 9.0- and 31.1-fold higher than in the corresponding normal tissues (Fig. 7D). These data suggest that steady-state cellular levels of miR-186 and miR-150 are higher also in the endometrial cancer cells than in the normal endometrial cells in vivo.
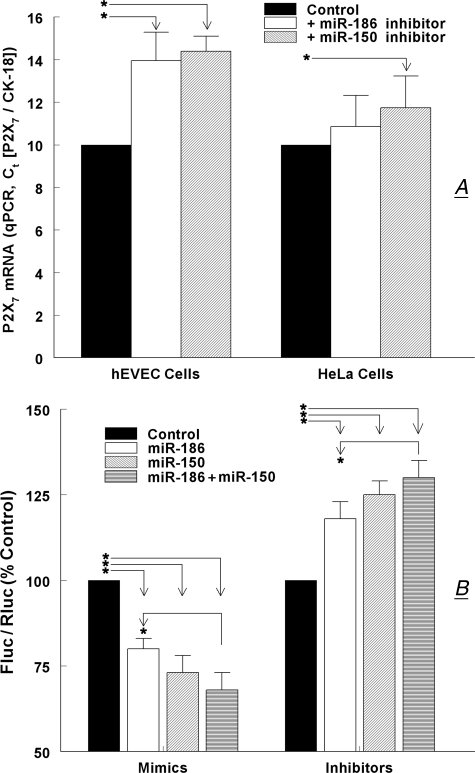
miR-186 and miR-150 Modulation of P2X7 mRNA and 3′-UTR-P2X7 Stability—To test more directly the hypothesis that miR-186 and miR-150 confer instability to the P2X7 transcript, hEVEC and HeLa cells were transfected with specific inhibitors of the miRNAs. In hEVEC cells, transfections with the miR-186 inhibitor increased P2X7 mRNA levels by 40% and transfections with the miR-150 inhibitor increased P2X7 mRNA levels by 44% (Fig. 8A). In HeLa cells, transfections with the miR-186 inhibitor had no significant effect on P2X7 mRNA levels, whereas transfections with the miR-150 inhibitor increased P2X7 mRNA levels by 17% (Fig. 8A).
FIGURE 8.
Effects of miR-186 and miR-150 mimics and inhibitors on P2X7 mRNA. A, effects of miR-186 and miR-150 inhibitors on steady-state levels of P2X7 mRNA in hEVEC and HeLa cells. Cells were transfected with specific inhibitors of the miRNAs, and P2X7 mRNA levels (normalized to CK-18) were determined by qPCR. Shown are means ± S.D. of three experiments in triplicate. *, p < 0.01 compared with controls. B, effects of miR-186 and miR-150 mimics and inhibitors on luciferase activity in HEK293 cells transfected with the full-length 3′-UTR-P2X7--luciferase vector. HEK293 were also transfected with the Renilla luciferase and pRL-CMV vectors, and co-transfected with mimics and/or inhibitors of the miR-150 and miR-186. Changes in luciferase activity were determined in terms of Fluc/Rluc 48 h after transfection, and data were normalized to steady-state Fluc/Rluc activity in nontreated cells. Shown are means ± S.D. of two experiments in triplicate. *, p < 0.01.
To better understand how miR-186 and miR-150 regulate stability of the P2X7 transcript, HEK293 cells transfected with the full-length 3′-UTR-P2X7--luciferase vector plus the Renilla luciferase and pRL-CMV vectors were also co-transfected with mimics or inhibitors of the miR-186 or/and miR-150. Effects of the mimics and inhibitors on 3′-UTR-P2X7-dependent luciferase activity were determined in terms of Firefly/Renilla luciferase activity (Fluc/Rluc) 48 h after transfection, and data were normalized to steady-state Fluc/Rluc activity in nontreated cells (vehicle only).
Transfections with mimics for miR-186 and for miR-150 decreased Fluc/Rluc activity by 20 and 27%, respectively, whereas transfections with mimics for miR-186 plus miR-150 decreased Fluc/Rluc activity by 32% (p < 0.01 compared with control, Fig. 8B). The combined effect of the miR-186 plus miR-150 mimics was greater than the effect of the miR-186 mimic alone (Fig. 8B, p < 0.01).
Transfections with inhibitors for miR-186 and for miR-150 increased Fluc/Rluc activity by 11 and 21%, respectively, whereas transfections with inhibitors for miR-186 plus miR-150 increased Fluc/Rluc activity by 30% (p < 0.01 compared with control, Fig. 8B). The combined effect of miR-186 plus miR-150 inhibitors was greater than the effect of the miR-186 inhibitor alone (Fig. 8B, p < 0.01).
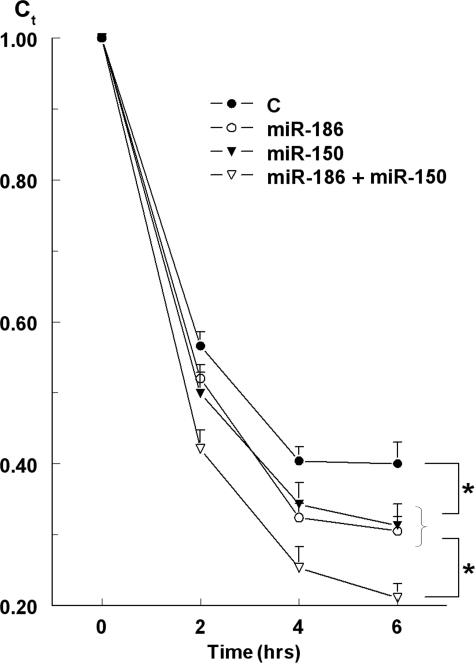
A detailed time course analysis of the effects of miR-186 and miR-150 mimics on the stability of the 3′-UTR-P2X7 is shown in Fig. 9. HEK293 cells transfected with the full-length 3′-UTR-P2X7-luciferase vector were co-transfected with mimics of the miR-150 and miR-186 alone or together. After 48 h cells were treated with 5 μm actinomycin-D for 6 h, a treatment that in preliminary experiments was shown not to affect the viability of the cells or the steady-state levels of GAPDH. At times t = 0, 2, 4, and 6 h post-treatment, luciferase mRNA (relative to GAPDH) was determined by qPCR. In the presence of actinomycin-D, luciferase mRNA levels in cells transfected only with the 3′-UTR-P2X7-luciferase reporter decreased in a time-related manner over the 6-h incubation period by about 60% (t½ ∼82 min). Co-transfections with either miR-186 or miR-150 mimics resulted in greater (about 70%, p < 0.05) and faster (t½ ∼75 min, p < 0.05) decreases in luciferase mRNA levels (Fig. 9). Co-transfections with both miR-186 plus miR-150 mimics decreased luciferase mRNA levels by about 80% (t½ ∼71 min; p < 0.05 for both compared with co-transfections with miR-186 or miR-150 alone) (Fig. 9). The combined effect of miR-186 plus miR-150 mimics suggests an additive effect of the two mimics (Fig. 9).
FIGURE 9.
Effects of miR-186 and miR-150 mimics on the stability of heterologously expressed full-length 3′-UTR-P2X7 in HEK293 cells. HEK293 cells transfected with the full-length 3′-UTR-P2X7--luciferase vector were co-transfected with mimics of the miR-150 and miR-186. After 48 h cells were treated with 5 μm actinomycin-D, and at times t = 0, 2, 4, and 6 h after actinomycin D treatment, luciferase mRNA (relative to GAPDH) was determined by qPCR. Shown are means ± S.D. of one experiment in triplicate; data for each category were normalized to luciferase/GAPDH mRNA (Ct = 1att = 0). *, p < 0.01. All four curves could be fitted into simple exponentials, and the t½ of the declines were calculated from the curves.
DISCUSSION
The objective of this study was to gain a better understanding of the regulation of P2X7 mRNA expression. This question is biologically and clinically important because P2X7-mediated apoptosis controls the growth of epithelial cells (12, 15), and expression of P2X7 mRNA and protein are lower in uterine endometrial and cervical precancerous and cancer epithelial cells compared with normal epithelial cells (22, 23). In this study we made the novel observation that the P2X7 receptor is also reduced in epithelial cancers of the bladder and breast. We found that sequences within the 3′-UTR-P2X7 were putative target sites for miR-186 and miR-150, and our data show for the first time that both elements conferred instability to the P2X7 transcript. The data showed more abundant expression of miR-186 and miR-150 in cancer than in normal cells, both in vivo and in cultured cells in vitro, and transfection with miR-186 and miR-150 inhibitors increased P2X7 mRNA, predominantly in the normal cells. In the host HEK293 cells, which lack endogenous expression of the P2X7 and which were transfected with the luciferase full-length 3′-UTR-P2X7 reporter, co-transfection with miR-186 and miR-150 mimics decreased, whereas co-transfection with inhibitors of the miRNAs increased the luciferase activity. Also, co-treatment with actinomycin-D did not have a significant effect in that system, suggesting that the changes in the P2X7-3′-UTR-luciferase mRNA expression are not the result of decreased transcription but rather increased instability of the construct. These data indicate that miR-186 and miR-150 confer instability to P2X7 mRNA by activation of miR-186 and miR-150 instability target sites located at the 3′-UTR-P2X7, and suggest that activation of the miRNAs sites decrease P2X7 mRNA levels possibly by stimulating degradation of the transcript. Based on these findings we suggest that the low steady-state levels of P2X7 mRNA in cancer epithelial cells, which can contribute to cancer growth through abrogation of P2X7-dependent apoptosis, are the result of increased expression of miR-186 and miR-150 that stimulate degradation of the P2X7 transcript.
The data showed that regions within the P2X7 3′-UTR determine the stability of the transcript, but the degree of stability of the P2X7 transcript depended on the cell type. Thus, overexpression of a full-length 3′-UTR-P2X7 reporter, or reporters containing the proximal (Segment-1) or distal (Segment-3) parts of the 3′-UTR-P2X7 were associated with increased stability of the construct in normal cells and decreased stability in cancer cells. In contrast, overexpression of a reporter containing the middle (Segment-2) part of the 3′-UTR-P2X7 was associated with decreased stability both in normal and in cancer cells. In both cell types we used the same reporter genes, and given the fact that the structure of the P2X7 3′-UTR is identical in hEVEC (normal) and HeLa (cancer) cells,3 the hypothesis is that the differences in P2X7 3′-UTR stability depend on differences in the intracellular activity of trans-acting factors in normal and cancer cells that target and regulate the cis-acting sites.
This hypothesis is supported by the present findings that cellular levels of miR-186 and miR-150 were higher in the cancer cells than in the normal cells. Human miRNAs reside in genomic regions that are involved in the regulation of cell growth and are prone to alterations in cancer cells, and abnormally expressed miRNAs in human cancers target transcripts of protein-coding genes involved in tumorigenesis or apoptosis (33, 34, 43). To date, most types of tumors analyzed by miRNA profiling have shown significantly different miRNA profiles compared with normal cells from the same tissue (43). Martinez et al. (44) reported that miR-218 was underexpressed in human papillomavirus-positive cervical cancer cells compared with normal cells, and Lui et al. (45) found that miR-21 was overexpressed, whereas miR-let-7b, miR-let-7c, miR-23b, miR-196b, and miR-143 were underexpressed in cervical cancer cells. This study was the first to focus on miRNAs that regulate expression of the human P2X7 receptor, and the present data show a correlation between the miR-186 and miR-150 profile in cancer/normal cells with regulation of the P2X7 mechanism that could be related to cancer development.
The experiments in this study focused on the role that miR-186 and miR-150 play in regulating the expression of P2X7. The rationale for choosing these miRNAs was severalfold. First is the near perfect match between the seed sequences of the miRNAs and their putative target sites within the human P2X7 3′-UTR, 7- and 8-mer for miR-186 and miR-150, respectively, along with additional putative base pairing (32, 40, 46, 47). Second is the relatively high 94 and 59% context scores for miR-150 and miR-186, respectively (46). This score takes into account the seed pairing with target, additional 3′ end of the miRNA pairing with the target, local AU contribution, site type contribution, and the position of the target within the 3′-UTR, all factors identified empirically as being important for identifying true miRNA targets (46, 47). Third is the evolutionary conservation of the putative miR-186 target site between human, mouse, rat, and dog. Fourth is the fact that the miR-186 putative target site on the P2X7 3′-UTR is not shared by other known miRNAs, whereas the miR150 target site is a region that is shared by other miRNAs and it may be a signature site for regulation. Finally, is the very intriguing observation that the two miRNAs appear to show different responses in normal and cancer cells. The miR-186 target site is located in the middle part (Segment-2) of the 3′-UTR-P2X7, which demonstrated decreased stability when expressed in either normal or cancer cells. In contrast, the miR-150 putative target site is located in the distal part (Segment-3) of the 3′-UTR-P2X7, which demonstrated increased stability in normal cells and decreased stability in cancer cells.
Both in normal and in cancer cells overexpression of a reporter containing the wild type miR-186 target site was associated with instability of the reporter, and the effect was greater in the cancer cells than in the normal cells, possibly because of the higher levels of miR-186 in the former (present data). Overexpression in cancer cells of the mutated miR-186 target site was associated with lesser decrease in the reporter levels. In contrast, in normal cells overexpression of a reporter containing the mutated miR-186 target site was associated with marked stability. The sequence contained in the wild-type miR-186 target site reporter (Fig. 5A) did not match with other known miRNAs, so the reversal of instability is most likely not because of an effect induced by another miRNA. The most likely explanation for the reversal of instability is that the stability of the sequence critically depends on cellular levels of miR-186 within a relatively narrow range. Thus, a 2–3-fold increase in miR-186 levels in cancer cells over those normally present in wild-type cells (Fig. 7B) sufficed to induce instability of the wild-type miR-186 reporter. This conclusion is supported by the inhibition assays, where transfections with an miR-186 inhibitor increased P2X7 mRNA by 40% in normal cells but had no significant effect in cancer cells. In this case, the miR-186 inhibitor was more effective in cells expressing lower levels of miR-186 (normal cells) than in cells expressing higher levels of the miR-186 (cancer cells). The conclusion is supported by a recent study in HeLa cells where inhibition of miR-186 decreased cell growth (48). Our data provide a cellular mechanistic explanation for the effect, namely through up-regulation of P2X7 mRNA and receptor protein, and augmentation of P2X7-mediated apoptosis.
The miR-186 target site is present at the 3′ end of the middle segment of the 3′-UTR-P2X7 (Segment-2). The RNA stability data of Segment-2 and those of the miR-186 target site were similar in normal and in cancer cells (Figs. 4D and 5B). Interestingly, bioinformatic data suggested that in addition to miR-186, Segment-2 may contain putative target sites for other miRNAs as well. These include putative target sites for miR-138; a signature cluster of miR-588, miR-204, and miR-211; miR-198; a second signature cluster of miR-198 and miR-214; miR-768–3p; miR-323; miR-203; and miR-766. At present, little is known about the role of those miRNAs in the regulation of stability of Segment-2 3′-UTR-P2X7. One possibility is that one or more of the above miRNAs also regulate stability of the middle portion, Segment-2, of the 3′-UTR of P2X7. If this is the case and the different miRNAs have opposing effects on the same transcript, the effects could cancel each other. This speculation is supported by findings in HeLa and A549 cells (human lung epithelial carcinoma cells) in which inhibition of miRNA-214 decreased apoptosis (48) and overexpression of miR-214 increased caspase-3 activity (49), suggesting that miR-214 is a pro-apoptotic factor. In contrast, inhibition of miR-204 increased apoptosis (48), suggesting that miR-214 would tend to inhibit apoptosis. Because Segment-2 of the 3′-UTR-P2X7 contains putative target sites for miR-204 and miR-214, it is possible that their pro/contra-apoptotic effects would cancel, and the apoptosis-related direction of the cell, as determined by Segment-2 of the 3′-UTR-P2X7 will be controlled mainly by miR-186.
In cancer cells, overexpression of the putative wild-type miR-150 target site was associated with instability of the reporter, whereas overexpression of the mutated miR-150 target site was associated with attenuated decrease in reporter levels after 6 h. In contrast, in normal cells expression of the miR-150 target site reporters, wild type and mutant, did not change over time. Previous studies reported potential growth- and differentiation-dependent regulatory roles for miR-150 in cells of the red and white lineages (50) and in immune cells (51–53). In addition, miR-150 is differentially expressed in related cancers (50, 54, 55). Overexpression of miR-150 in human skin BJ fibroblasts decreased caspase-3 activity (49), suggesting an anti-apoptotic role. In contrast, in the cancer HeLa and A549 cells, inhibition of miR-150 decreased cell growth (48), suggesting a pro-apoptotic role in these cells. The present data are in agreement with those of Cheng et al. (48) showing a pro-apoptotic effect in cancer epithelial cells only. A possible explanation for these observations is that the stability of the sequence depends on cellular levels of miR-150, which are higher in cancer cells than in the normal cells (present data), similar to the case with the miR-186. This hypothesis is also supported by the miR-150 inhibition assays, where transfections with an miR-150 inhibitor increased P2X7 mRNA by 44% in normal cells and by only 17% in cancer cells. Accordingly, an equimolar dose of the miR-150 inhibitor is more effective in normal cells, which express low amounts of miR-150, than in cancer cells, which express high levels of the miR-150.
The miR-150 target site is present in the middle part of the 3′-UTR-P2X7 Segment-3. The stability data of Segment-3 and those of the miR-150 target site were similar in the cancer cells (Figs. 4E and 5C). However, the data in normal cells were different because overexpression of Segment-3 3′-UTR-P2X7 was associated with increased stability of the construct (Fig. 4E), whereas in cells overexpressing the wild-type miR-150 target site, the stability of the reporter did not change over time (Fig. 5C). Segment-3 of the 3′-UTR of P2X7 also contains additional putative miRNAs target sites, including target sites for miR-765; miR-28; a presumed signature cluster of miR-214, miR-522, miR-17, miR-512–3p, miR-302c, miR-20b, and miR-106a (plus miR-150); miR-197; miR-378; a presumed second signature cluster of miR-340, miR-999a, and miR-370; and miR-564. At present, little is known about the role of those miRNAs in the regulation of stability of Segment-3 3′-UTR-P2X7, although some have been associated previously with carcinogenesis (33, 34).
Our data also show that overexpression of a reporter containing the 3′-UTR-P2X7 Segment-1 was associated with increased stability of the construct in normal cells and with decreased stability in cancer cervical cells. This sequence contains putative miRNAs target sites for several miRNAs, but presently there is little known about the role of these miRNAs in cancer. Further studies are needed to determine whether these additional putative miRNAs target sequences within Segment-1 of the 3′-UTR are important for expression of P2X7.
An additional question that the present data raise is about the cumulative contributions of Segments 1–3 to the stability of the full-length 3′-UTR of P2X7. The data in cancer cells were uniform, namely decreased stability of Segments 1–3 reporters, which was similar to that observed in cells overexpressing the full-length 3′-UTR-P2X7 reporter. In normal cells the increased stability of Segments 1 and 3 reporters was similar to that observed in cells overexpressing full-length 3′-UTR-P2X7 reporter. However, experiments using Segment 2 reporter in normal cells were associated with decreased stability of the reporter. Thus, although the effect on the full-length 3′-UTR-P2X7 reporter could be influenced by the contributions made by each of Segments 1–3, little is known about the relative contributions to the stability of the full-length 3′-UTR-P2X7. Accordingly, the relative impact of Segments-1 and -3, alone or in combination, could be greater than that of Segment-2. Similarly, it was recently shown that miRNA targets near both ends of a long 3′-UTR are more effective than targets more centrally located (46).
Interestingly, the effects of miR-186 and miR-150 mimics (as well as inhibitors) were only partially additive, suggesting interactive regulation. Future studies will evaluate this hypothesis in greater detail.
In summary, we show for the first time that the stability of the human 3′-UTR of P2X7 depends both on miRNAs target sites and on the intracellular activity of miRNAs. We identified miR-150 and miR-186 as two factors that play a role in the regulation of P2X7 3′-UTR. The data also show differences in the regulation of the P2X7 3′-UTR in cancer versus normal cervical epithelial cells that could play a role in the different profile of expression of P2X7 mRNA and protein receptor in cancer versus normal cells. These data may be important for our understanding of regulation of P2X7-mediated apoptosis and its role in epithelial cell cancer development.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15955. This work was also supported by an unrestricted grant by CytoCore Inc. (to G. I. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
G. I. Gorodeski, unpublished data.
The abbreviations used are: UTR, untranslated region; qPCR, quantitative PCR; miRNA, microRNA; RT, reverse transcription; MDCK, Madin-Darby canine kidney cells; Bz, 2′,3′-O-(4-benzoylbenzoyl); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PBS, phosphate-buffered saline; hEVEC, human ectocervical-vaginal epithelial cell; snRNA, small nuclear RNA; nt, nucleotide; WT, wild type.
References
- 1.Dubyak, G. R., and el-Moatassim, C. (1993) Am. J. Physiol. 265 C577-C606 [DOI] [PubMed] [Google Scholar]
- 2.Buell, G., Collo, G., and Rassendren, F. (1996) Eur. J. Neurosci. 8 2221-2228 [DOI] [PubMed] [Google Scholar]
- 3.Surprenant, A., Rassendren, F., Kawashima, E., North, R. A., and Buell, G. (1996) Science 272 735-738 [DOI] [PubMed] [Google Scholar]
- 4.Soto, F., Garcia-Guzman, M., and Stuhmer, W. (1997) J. Membr. Biol. 160 91-100 [DOI] [PubMed] [Google Scholar]
- 5.Ralevic, V., Martinez, I., Gardiner, A. S., Board, K. F., and Burnstock, G. (1998) Pharmacol. Rev. 50 413-492 [PubMed] [Google Scholar]
- 6.Virginio, C., MacKenzie, A., North, R. A., and Surprenant, A. (1999) J. Physiol. (Lond.) 519 335-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khakh, B. S., Burnstock, G., Kennedy, C., King, B. F., North, R. A., Seguela, P., Voigt, M., and Humphrey, P. P. A. (2001) Pharmacol. Rev. 53 107-118 [PubMed] [Google Scholar]
- 8.Sperlágh, B., Haskó, G., Németh, Z., and Vizi, E. S. (1998) Neurochem. Int. 33 209-215 [DOI] [PubMed] [Google Scholar]
- 9.Grahames, C. B. A., Michel, A. D., Chessell, I. P., and Humphrey, D. P. A. (1999) Br. J. Pharmacol. 127 1915-1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen, K. L., and Novak, I. (2003) Cell. Physiol. Biochem. 13 93-102 [DOI] [PubMed] [Google Scholar]
- 11.Loomis, W. H., Namiki, S., Ostrom, R. S., Insel, P. A., and Junger, W. G. (2003) J. Biol. Chem. 278 4590-4596 [DOI] [PubMed] [Google Scholar]
- 12.Wang, Q., Wang, L., Feng, Y. H., Li, X., Zeng, R., and Gorodeski, G. I. (2004) Am. J. Physiol. 287 C1349-C1358 [DOI] [PubMed] [Google Scholar]
- 13.Wang, Y., Stricker, H. M., Gou, D., and Liu, L. (2007) Front. Biosci. 12 2316-2329 [DOI] [PubMed] [Google Scholar]
- 14.Wang, L., Feng, Y. H., and Gorodeski, G. I. (2005) Endocrinology 146 164-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Q., Li, X., Wang, L., Feng, Y. H., Zeng, R., and Gorodeski, G. I. (2004) Endocrinology 145 5568-5579 [DOI] [PubMed] [Google Scholar]
- 16.Feng, Y. H., Wang, L., Wang, Q., Li, X., Zeng, R., and Gorodeski, G. I. (2005) Am. J. Physiol. 288 C1342-C1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, B. J., Zhang, W. Y., Bendall, L. J., Chessell, I. P., Buell, G. N., and Wiley, J. S. (2000) Am. J. Physiol. 279 C1189-C1197 [DOI] [PubMed] [Google Scholar]
- 18.Li, G. H., Lee, E. M., Blair, D., Holding, C., Poronnik, P., Cook, D. I., Barden, J. A., and Bennett, M. R. (2000) J. Biol. Chem. 275 29107-29112 [DOI] [PubMed] [Google Scholar]
- 19.Bobanovic, L. K., Royle, S. J., and Murrell-Lagnado, R. D. (2002) J. Neurosci. 22 4814-4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerra, A. N., Fisette, P. L., Pfeiffer, Z. A., Quinchia-Rios, B. H., Prabhu, U., Aga, M., Denlinger, L. C., Guadarrama, A. G., Abozeid, S., Sommer, J. A., Proctor, R. A., and Bertics, P. J. (2003) J. Endotoxin Res. 9 256-263 [DOI] [PubMed] [Google Scholar]
- 21.Feng, Y. H., Li, X., Wang, L., Zhou, L., and Gorodeski, G. I. (2006) J. Biol. Chem. 281 17228-17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., Zhou, L., Feng, Y. H., Abdul-Karim, F., and Gorodeski, G. I. (2006) Cancer Epidemiol. Biomark. Prev. 15 1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X., Qi, X., Zhou, L., Catera, D., Rote, N. S., Potashkin, J., Abdul-Karim, F. W., and Gorodeski, G. I. (2007) Gynecol. Oncol. 106 233-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasser, S., and Raulet, D. (2006) Semin. Cancer Biol. 16 344-347 [DOI] [PubMed] [Google Scholar]
- 25.Kujoth, G. C., Leeuwenburgh, C., and Prolla, T. A. (2006) Cancer Res. 66 7386-7389 [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Nieto, S., and Zhivotovsky, B. (2006) Curr. Pharm. Des. 12 4411-4425 [DOI] [PubMed] [Google Scholar]
- 27.Buell, G. N., Talabot, F., Gos, A., Lorenz, J., Lai, E., Morris, M. A., and Antonarakis, S. E. (1998) Receptors Channels 5 347-354 [PubMed] [Google Scholar]
- 28.Guhaniyogi, J., and Brewer, G. (2001) Gene (Amst.) 265 11-23 [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, P., and Tollervey, D. (2001) Curr. Opin. Biol. 13 32320-32325 [DOI] [PubMed] [Google Scholar]
- 30.Ambros, V. (2004) Nature 431 350-355 [DOI] [PubMed] [Google Scholar]
- 31.Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H., Ferrando, A. A., Downing, J. R., Jacks, T., Horvitz, H. R., and Golub, T. R. (2005) Nature 435 834-838 [DOI] [PubMed] [Google Scholar]
- 32.Farh, K. K., Grimson, A., Jan, C., Lewis, B. P., Johnston, W. K., Lim, L. P., Burge, C. B., and Bartel, D. P. (2005) Science 310 1817-1821 [DOI] [PubMed] [Google Scholar]
- 33.Calin, G. A., and Croce, C. M. (2006) Cancer Res. 66 7390-7394 [DOI] [PubMed] [Google Scholar]
- 34.Calin, G. A., and Croce, C. M. (2006) Nat. Rev. 6 857-866 [DOI] [PubMed] [Google Scholar]
- 35.Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., and Pasquinelli, A. E. (2005) Cell 122 553-563 [DOI] [PubMed] [Google Scholar]
- 36.Olsen, P. H., and Ambros, V. (1999) Dev. Biol. 216 671-680 [DOI] [PubMed] [Google Scholar]
- 37.Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M. (2005) Nature 433 769-773 [DOI] [PubMed] [Google Scholar]
- 38.Lai, E. C. (2004) Genome Biol. 5 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennecke, J., Stark, A., Russell, R. B., and Cohen, S. M. (2005) PLoS Biol. 3 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005) Cell 120 15-20 [DOI] [PubMed] [Google Scholar]
- 41.Xie, X., Lu, J., Kulbokas, E. J., Golub, T. R., Mootha, V., Lindblad-Toh, K., Lander, E. S., and Kellis, M. (2005) Nature 434 338-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorodeski, G. I., Romero, M. F., Hopfer, U., Rorke, E., Utian, W. H., and Eckert, R. L. (1994) Differentiation 56 107-118 [DOI] [PubMed] [Google Scholar]
- 43.Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M., and Croce, C. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2999-3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez, I., Gardiner, A. S., Board, K. F., Monzon, F. A., Edwards, R. P., and Khan, S. A. (2008) Oncogene 27 2575-2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lui, W. O., Pourmand, N., Patterson, B. K., and Fire, A. (2007) Cancer Res. 67 6031-6043 [DOI] [PubMed] [Google Scholar]
- 46.Grimson, A., Farh, K. K., Johnston, K., Garrett-Engele, P., Lim, L. P., and Bartel, D. P. (2007) Mol. Cell 27 91-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen, C. B., Shomron, N., Sandberg, R., Hornstein, E., Kitzman, J., and Burge, C. B. (2007) RNA (N. Y.) 13 1894-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng, A. M., Byrom, M. W., Shelton, J., and Ford, L. P. (2005) Nucleic Acids Res. 33 1290-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovcharenko, D., Kelnar, K., Johnson, C., Leng, N., and Brown, D. (2007) Cancer Res. 67 10782-10788 [DOI] [PubMed] [Google Scholar]
- 50.Bruchova, H., Yoon, D., Agarwal, A. M., Mendell, J., and Prchal, J. T. (2007) Exp. Hematol. 35 1657-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, H., Neilson, J. R., Kumar, P., Manocha, M., Shankar, P., Sharp, P. A., and Manjunath, N. (2007) PLoS ONE 2 e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, C., Pedro-Calado, D., Galler, G., Thai, T. H., Patterson, H. C., Wang, J., Rajewsky, N., Bender, T. P., and Rajewsky, K. (2007) Cell 131 146-159 [DOI] [PubMed] [Google Scholar]
- 53.Zhou, B., Wang, S., Mayr, C., Bartel, D. P., and Lodish, H. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7080-7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulci, V., Chiaretti, S., Goldoni, M., Azzalin, G., Carucci, N., Tavolaro, S., Castellano, L., Magrelli, A., Citarella, F., Messina, M., Maggio, R., Peragine, N., Santangelo, S., Romana-Mauro, F., Landgraf, P., Tuschl, T., Weir, D. B., Chien, M., Russo, J. J., Ju, J., Sheridan, R., Sander, C., Zavolan, M., Guarini, A., Foa, R., and Macino, G. (2007) Blood 109 4944-4951 [DOI] [PubMed] [Google Scholar]
- 55.Guglielmelli, P., Tozzi, L., Pancrazzi, A., Bogani, C., Antonioli, E., Ponziani, V., Poli, G., Zini, R., Ferrari, S., Manfredini, R., Bosi, A., and Vannucchi, A. M. (2007) Exp. Hematol. 35 1708-1718 [DOI] [PubMed] [Google Scholar]