Abstract
Autosomal dominant polycystic kidney disease (ADPKD), the most common inherited cause of kidney failure, is caused by mutations in either PKD1 (85%) or PKD2 (15%). The PKD2 protein, polycystin-2 (PC2 or TRPP2), is a member of the transient receptor potential (TRP) superfamily and functions as a non-selective calcium channel. PC2 has been found to form oligomers in native tissues suggesting that it may form functional homo- or heterotetramers with other subunits, similar to other TRP channels. Our experiments unexpectedly revealed that PC2 mutant proteins lacking the known C-terminal dimerization domain were still able to form oligomers and co-immunoprecipitate full-length PC2, implying the possible existence of a proximal dimerization domain. Using yeast two-hybrid and biochemical assays, we have mapped an alternative dimerization domain to the N terminus of PC2 (NT2-1-223, L224X). Functional characterization of this domain demonstrated that it was sufficient to induce cyst formation in zebrafish embryos and inhibit PC2 surface currents in mIMCD3 cells probably by a dominant-negative mechanism. In summary, we propose a model for PC2 assembly as a functional tetramer which depends on both C- and N-terminal dimerization domains. These results have significant implications for our understanding of PC2 function and disease pathogenesis in ADPKD and provide a new strategy for studying PC2 function.
Autosomal dominant polycystic kidney disease (ADPKD),3 the most common inherited human renal disease, has been shown to result from mutations in either PKD1 or PKD2 (1). ADPKD accounts for ∼10% of patients on renal replacement therapy and is therefore an important cause of end-stage renal failure world-wide. The cardinal feature of the ADPKD kidney is the presence of multiple fluid-filled cysts. However, cysts also arise in other epithelial structures such as the liver and pancreas. A number of non-cystic manifestations such as cardiac valve abnormalities, diverticular disease, and intracranial aneurysms have been reported (2).
Mutations in PKD2 account for 15% of all patients with ADPKD. The PKD2 protein, polycystin-2 (PC2), is a Type II membrane protein of 968 amino acids in length (3). PC2 has the properties of a high-conductance nonselective Ca2+-permeable cation channel. Because of significant homology, PC2 (or TRPP2) has been included in the TRP (transient receptor potential) superfamily of channels, which broadly function as cellular sensors for multiple stimuli (4, 5). There is evidence that PC2 may transduce a mechanosensitive Ca2+ current in primary cilia (6) although it is unclear whether the mechanosensor is PC1, PC2, or another protein. However, it has also been reported that PC2 can function downstream of G protein-coupled receptor and/or receptor-tyrosine kinase activation at the cell surface (7–9). The basolateral localization of PC2 in kidney tubules and cells has implicated a possible role in cell-cell or cell-matrix adhesion in association with PC1 (10, 11). Finally, it has been reported that PC2 can function as an endoplasmic reticulum-located Ca2+ release channel in some systems (12).
Previously we demonstrated that PC2 can exist as PC1-PC2 heterodimers as well as PC2 homodimers in native tissues (10). Interactions between PC1 and PC2 may regulate their trafficking and there is evidence for reciprocal activation or inhibition of activity in different experimental systems (13, 14). PC2 may also heterodimerize with TRPC1 through its C terminus (5, 9). PC2-TRPC1 heteromultimers have been shown to possess distinct channel properties from PC1-PC2 heterodimers, being activated in response to G protein-coupled receptor activation in the kidney epithelial cell line, mIMCD3 (9). In yeast two-hybrid assays, PC2 can homodimerize via a C-terminal domain, which is distinct from heterodimerization sequences for PC1 or TRPC1 interactions (5, 15). In this report, we describe the identification and functional characterization of a second dimerization domain for PC2 within the N terminus and propose a likely homotetrameric model for PC2 based on C- and N-terminal interactions.
EXPERIMENTAL PROCEDURES
Materials—All chemicals were purchased from Sigma unless otherwise stated. Yeast vectors pGBAD-B and pACT2-B were obtained from D. Markie (University of Otago, NZ) (16). The plasmids LDR and CF used for the FKBP-FRB dimerization system were gifts of T. Meyer (Stanford University) (17).
Generation of PKD2 Plasmids—Unless otherwise stated, the PKD2 plasmids used in this work have been previously reported (18, 19). N-terminal HA-tagged full-length and mutant (L703X) PKD2 constructs were created by replacing an XbaI and SacII fragment of a wild-type PKD2 plasmid (gift of S Somlo, Yale University) with the same fragment excised from the previously described HA-L224X plasmid (19). A C-terminal HA-tagged PKD2 mutant construct, R742X, was generated by PCR using the wild-type PKD2Pk plasmid as a template including the HA epitope tag sequence and in-frame stop codon in the reverse primer. The missense PKD2 mutation, D511V, was created by site-directed mutagenesis in the PKD2Pk plasmid template using a previously published protocol (19). The N-terminal Myc-tagged L224X plasmid was generated by PCR and subcloned into the XbaI and HindIII sites of pcDNA3.1 (-). The plasmids CFP-PKD2-(1–177) and CFP-PKD2-(1–223) were generated by fusing the N-terminal sequences of PKD2 in-frame with the CFP and FKBP cassette in the vector, CF.
Immunoblotting and Immunoprecipitation—HEK-293 and mIMCD3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transient transfection was carried out on cells cultured to 90% confluency using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Immunoblotting and immunoprecipitation (IP) was performed as previously described using epitope-specific antibodies (10). The PKD2 antibody, p30, generated to the C-terminal 258 amino acids of human PC2, has been previously reported (18).
Yeast Two-hybrid Assays—Yeast two-hybrid assays were performed in the yeast strain AH109 containing ADE2, HIS3, and LacZ reporter genes under the control of the GAL4 upstream activating sequences (UAS) by co-transforming bait and prey constructs of the entire intracellular N terminus of human PC2 (NT2) or its truncations into yeast cells using a published protocol (20). Full-length NT2-(1–223) was subcloned into bait (BD) vector pGBKT7 and prey (AD) vector pGADT7 (Clontech), respectively, by PCR and DNA ligation. Similarly, the following NT2 truncations, NT2-(1–207), NT2-(1–198), NT2-(1–177), NT2-(1–117), NT2-(1–61), and NT2-(178–223) were constructed in both bait and prey vectors. After incubation at 30 °C for 3–4 days, positive co-transformants (containing both bait and prey plasmids) were picked from selective medium S.D./-LT (lacking tryptophan and leucine) and restreaked onto selective media S.D./-LTH (lacking tryptophan, leucine, and histidine), S.D./-LTH with 2 mm 3-amino-1,2,4-triazole (3-AT), and S.D./-LTAH (lacking tryptophan, leucine, histidine, and adenine), respectively, to activate the reporter genes HIS3 and ADE2. 2 mm 3-AT was used in the selective medium to suppress minor autoactivation of NT2-(1–198) and NT2-(1–177) in the pGBKT7 vector. The constructs pGBKT7–53 (p53) and pGBADT7-T (SV40 T-antigen) were used as a pair of positive controls. Truncations of the PC2 C terminus (CT2) and PC1 C terminus (CT1), which mimic two naturally occurring mutants lacking their respective interaction domains i.e. PKD2-R742X and PKD1-R4227X, respectively, were generated as a pair of negative controls: pGBAD-B-CT1 (4107–4227) and pACT2-B-CT2 (680–742). At least three individual colonies were chosen from each plate to quantify growth.
Live Cell Imaging—IMCD3 and HEK-293 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) with the following cDNAs: CFP-FKBP-L223 (CF-PKD2–223), CFP-FKBP-L177 (CF-PKD2–177), and Lyn11-FRB (LDR). Translocation of the fusion proteins to the cell surface was induced 24 h later by the addition of 10 μm rapamycin (Calbiochem) to the culture media. In some experiments, co-expression of a surface-localized YFP fusion protein (C1 domain of PKC-γ1, YFP-C1 (PKC)) was used to outline the plasma membrane. Live cell measurements were performed on an Olympus Imaging Systems inverted IX-71 microscope with a CFP and YFP filter set to capture cellular fluorescence images with a CCD camera (Hamamatsu), driven by Simple PCI software (C Imaging Systems). Fluorescence images of cells were taken every5sat room temperature. Changes in the mean fluorescence intensity over a given region of the cytosol and nucleus were quantified in individual cells (n = 6) using ImageJ (NIH) (21).
Electrophysiology—The whole cell voltage-clamp configuration was used in the perforated mode to measure transmembrane currents in single cells. Briefly, patch-clamp recordings were obtained from single cells at 37 °C using a Warner PC-505B amplifier (Warner Instrument Corp., Hamden, CT) and pClamp 8 software (Axon Instrument, Foster City, CA). Glass pipettes (plain, Fisher Scientific) with resistances of 5–8 MΩ were prepared with a pipette puller and polisher (PP-830 and MF-830, respectively, Narishige, Tokyo, Japan). After the whole cell configuration was achieved, cell capacitance and series resistance were compensated before each recording period. Current measurements in time course experiments were made by applying a 100-ms pulse from a holding potential -60 mV to -100 every 10 s for 5 min. Current amplitude was extracted at -100 mV and plotted as function of time (min). I-V curves were derived from a voltage step protocol as follows: from a holding potential of -60 mV, voltage steps were applied from -100 to 100 mV in 20-mV increments with 200 ms duration at 3-s intervals. Current traces were filtered at 1 kHz and analyzed off-line with pClamp 8. Statistical analysis was employed with the SigmaStat (Chicago, IL) software. Data were reported as means ± S.E. Student's t test was used for comparisons between groups. Differences were considered significant at p < 0.05. The pipette solution contained (in mm): 0.3 Amphotericin B, 110 potassium aspartate, 30 KCl, and 5 HEPES, pH 7.2. The bath solution contained (in mm): 130 KCl, 1 MgCl2, 10 HEPES, 0.1 CaCl2, and 5 glucose (pH 7.4).
Zebrafish Experiments—Wild-type zebrafish AB strains were maintained and staged according to standard protocols (22). Embryos were treated with 0.003% 1-phenyl-2-thiourea (Sigma) in ×0.5 E2 solution to reduce pigmentation. At the one-cell stage, embryos were microinjected with pkd2ATGMO or control MO (Gene Tools LLC) to block zebrafish pkd2 translation as described (23) at final MO concentrations estimated at 100 nm. In vitro-transcribed capped mRNA encoding human PKD2-L177, -L223, and D511V were synthesized using a mMessage mMachine T7 kit (Ambion), and 50 pg were injected or co-injected at the one-cell stage. Injection solution conditions have been described in a previous report (19).
RT-PCR—Total RNA was isolated from 48 hpf embryos by using the RNAqueous®-4PCR Kit (Ambion). Nested RT-PCR primers were designed from human PKD2 sequence to confirm expression in zebrafish embryos. RT-PCR was performed using the SuperScript™ III One-Step RT-PCR System with Platinum® Taq High Fidelity (Invitrogen) followed by a second PCR using Phusion™ High-Fidelity DNA Polymerase (New England BioLabs). Amplification of β-actin was performed as a positive control. The human PKD2 primers used were: 148F1: GGCCTGGAGATCGAGATG, 332R1: ACCACCATTCCGCCTTCT, 149F2: GCCTGGAGATCGAGATGC, 320R2: CCTTCTTCCCCTTCCACCT. All procedures involving kits were performed according to the manufacturers' protocols.
Immunohistochemistry—Whole zebrafish embryos were fixed in 100% MeOH at 4 °C overnight. After rehydration, embryos were washed in PBS plus 0.5% Triton X-100 and blocked in PBS-DTR (PBS plus 1% DMSO, and 0.5% Triton X-100) with 10% normal goat serum (NGS) at room temperature for 1 h. Anti-polycystin-2 (1:800; gift of I. Drummond) (24) was incubated in PBS-DTR 2% NGS for 4 °C overnight. Embryos were washed in PBTX (PBS/0.5% Triton X-100) for 2 h and then incubated in 1:1000 CY3-labeled goat anti-rabbit IgG (Jackson ImmunoResearch) in PBS-DTR 2X NGS at room temperature for 2 h. After rinsing the embryos in PBTX for 2 h, they were dehydrated, embedded in JB-4 (Polyscience), and sectioned at 10-μm thickness. Sections were stained with 1:1000 DAPI (KPL) solution for 3 min at room temperature, the slides rinsed once with PBS and then mounted with fluorescent mounting medium (KPL). Section images were taken using a Nikon E500 microscope and analyzed by the Spot v. 4.4 program.
Histochemistry—48-days postfertilization (dpf) embryos were fixed in 1.5% glutaraldehyde (Electron Microscopy Sciences), 1% paraformaldehyde (Electron Microscopy Sciences), 70 mm NaPO4 pH 7.4, 3% sucrose at 4 °C overnight, dehydrated, embedded in glycolmethacrylate (JB4; Polyscience), and sectioned at 4-μm thickness using HM325 (Richard-Allan Scientific). Slides were stained with hematoxylin and eosin (BBC Biochemical) according to the protocol from Jennifer R. Panizzi and Lilianna Solnica-Krezel.4
RESULTS
Evidence for a Second Dimerization Domain in PKD2—We initially generated three naturally occurring PKD2 mutants, one with a single missense change in the third transmembrane domain (D511V) and two others lacking the C-terminal homodimerization domain, L703X and R742X (Fig. 1). The D511V mutation has previously been shown to completely abolish PC2 channel activity whereas L703X and R742X have altered channel properties in some expression systems (12, 25). Relevant to the present study, D511V has been demonstrated to function as a dominant negative interfering allele in mIMCD3 (9) and LLC-PK1 cells (26). As previously shown for endogenous PC2 (10), full-length PKD2 could be detected mainly as monomers under normal reducing conditions on SDS-PAGE but as oligomers under non-reducing conditions (Fig. 2A). Apart from prominent dimers, several other high molecular weight bands were detected under non-reducing conditions. These could represent homophilic binding between PC2 subunits or heterophilic interactions with other proteins. PKD2-D511V, showed a similar pattern of oligomerization under these conditions (data not shown). Unexpectedly we found that PKD2-L703X, could still form dimers under non-reducing conditions (Fig. 2A). Similar results were found for PKD2-R742X (not shown). As predicted, PKD2-D511V bound to wild-type PKD2, but unexpectedly, so did PKD2-R742X in co-immunoprecipitation (co-IP) assays (Fig. 2, B and C). To exclude the effect of the epitope tags in the interaction, we repeated these experiments with different PKD2 full-length (Myc-tagged) and mutant (C-terminal HA-tagged) constructs and obtained similar results (data not shown). We therefore hypothesized that a proximal interacting domain could lie upstream of the previously described distal C-terminal domain, possibly in the N terminus. Indeed, the isolated PC2 N-terminal domain (NT2-(1–223)) could interact with itself in HEK293 cells (Fig. 2D).
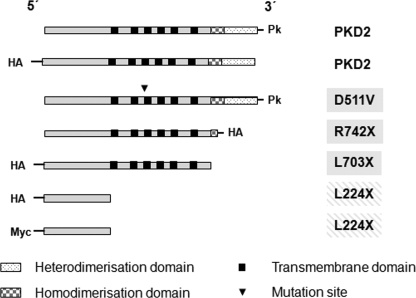
FIGURE 1.
Schematic diagram of epitope-tagged PKD2 constructs used in the study with their predicted domains. Apart from wild-type PKD2, a site-specific mutant (D511V), two mutants with truncations of the C terminus (R742X and L703X), and the isolated N-terminal domain (L224X) were generated for study.
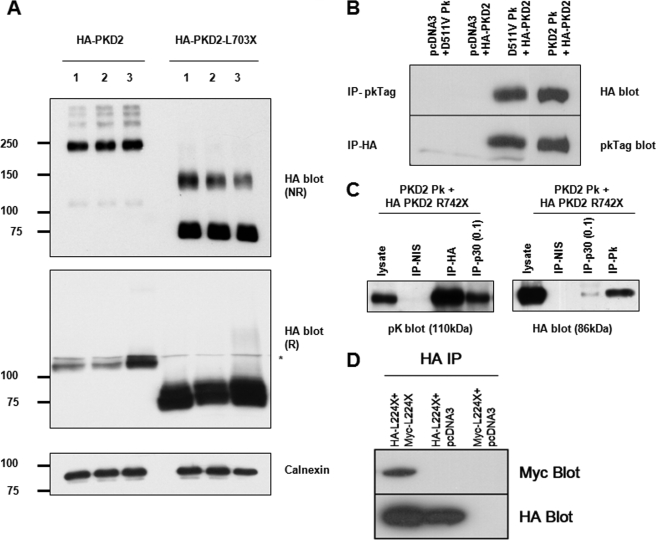
FIGURE 2.
Biochemical evidence for a proximal dimerization sequence in the N terminus of PC2. A, detection of epitope-tagged wild-type and mutant PC2 expressed in HEK293 cells by immunoblotting under reducing (R) and non-reducing (NR) conditions in three individual samples. Note that both forms of PC2 are detected predominantly as monomers under reducing SDS-PAGE but that there are prominent slower migrating species visible under non-reducing states. For full-length PC2, apart from prominent dimeric species (∼220 kDa), there are also higher bands suggestive of more complex oligomeric structures (e.g. tetramers). For L703X, the monomeric species are more notable than for wild-type PC2 under NR conditions but there is also detectable dimer formation (∼150kDa). Calnexin was used as an endogenous control for loading. * indicates a minor nonspecific band detected by the HA antibody. B, coimmunoprecipitation of epitope-tagged PC2-D511V (D511VPk) and wild-type PC2 (HA-PKD2). PC2-D511V is predicted to retain both hetero- and homodimerization C-terminal domains. C, PC2 mutant lacking both C-terminal dimerization domains (HA PKD2 R742X) associates with wild-type PC2 (PKD2Pk). The p30 and pK antibodies only recognize C-terminal epitope in full-length PC2. However, the HA antibody, which only recognizes the mutant R742X was able to co-IP wild-type PC2 (PKD2Pk) suggesting an interacting sequence proximal to the truncation. 1 mg of total cell lysate was used for IP with HA or NIS (non-immune serum) and 0.1 mg (1/10) for IP with p30 as indicated. 30 μg of lysate were loaded as a positive control. The converse experiment showed that p30 or pK antibodies could pull-down R742X. D, co-immunoprecipitation of HA-tagged N-terminal PC2 protein (NT2) containing the first 223 amino acids (L224X) with co-expressed Myc-tagged L224X. These results implied the existence of an N-terminal dimerization domain.
Defining the Dimerization Domain using Yeast Two-hybrid Assays—To characterize and refine this domain further, yeast two-hybrid assays were performed. As shown in Fig. 3, yeast co-transformants containing NT2-(1–223)-pGBKT7 (bait) and NT2-(1–223)-pGADT7 (prey) were able to grow on selective media requiring the activation of two reporters i.e. HIS3 and ADE2. NT2 did not interact with the C terminus of PC2 (CT2, data not shown). Further studies were carried out to refine the minimal interacting region using deletion constructs. As showed in Fig. 3, the truncation NT2-(1–207) could still interact with NT2-(1–223), while truncations NT2-(1–177) and NT2-(1–198) lost the ability to interact with NT2-(1–223). Shorter sequences of NT2 such as NT2-(1–61) and NT2-(1–117) were unable to interact with full-length NT2-(1–223) (not shown). These results indicate that the region from codons 199–207 is an essential part of the N-terminal interacting domain. The sequence NT2-(178–223) showed a weaker interaction than NT2-(1–223) in our assay (data not shown) suggesting that flanking sequences might be important in determining binding affinity. Fig. 3C shows the high sequence conservation of this region between human, mouse, and zebrafish PC2. Between codons 190 and 207, 12 of 17 amino acids (70.6%) are identical or similar compared with human/mouse PC2. This contrasts with the minimal sequence conservation between human and zebrafish PC2 in the preceding sequence of NT2 (codons 119–189).
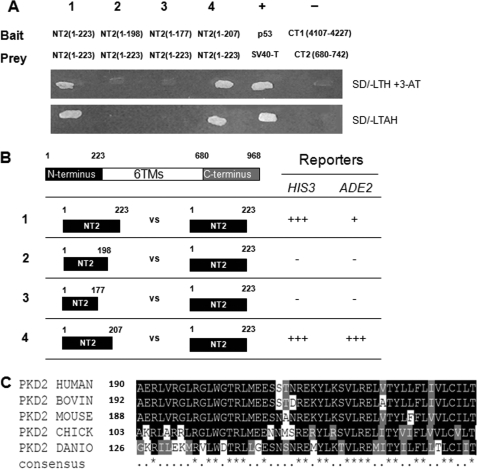
FIGURE 3.
Dimerization of the polycystin-2 N terminus (NT2) detected by yeast two-hybrid assays. A, growth of yeast co-transformants cultured on selective media S.D./-LTH with 2 mm 3-AT and S.D./-LTAH to activate HIS3 and ADE2 selection markers, respectively. The pairs of NT2 sequences tested are numbered from 1–4 and their sequences indicated. Constructs pGBKT7–53 (p53) and pGBADT7-T (SV40-T antigen) were used as a pair of positive controls while pGBAD-B-CT1 (4107–4227) and pACT2-B-CT2 (680–742) were used as a pair of negative controls. B, truncations of the N terminus of polycystin-2 (NT2) marked by the number of starting and ending amino acid residue and their interaction with NT2. The fragments on the left column are in bait (BD) constructs, while the fragments in the right column are in prey (AD) constructs. The numbers in the left column indicate the pairs of NT2 constructs tested and correspond to those shown in A. Positive (interaction) and negative results are indicated by +++, ++, +, and - for the appearance of positive growth on selective medium within 24 h, 48 h, and 3–7 days or no positive growth in 2 weeks, respectively. C, multiple sequence alignment of PC2 orthologues from different species overlapping with the sequence of human NT2-(190–238). Amino acids which show perfect conservation down to zebrafish are indicated by an asterisk (*) in the bottom line. In the region between amino acids 190 and 223, 23 of 33 amino acids (70%) are identical or similar. This contrasts with the region between amino acids 119 and 189 where only 1 of 70 amino acids (1.4%) show identity from human to Danio. The accession numbers for each sequence are as follows: Q13563 (Homo sapiens), O35245 (Mus musculus), Q4GZT3 (Bos taurus), Q5ZM00 (Gallus gallus), and Q6IVV8 (Danio rerio).
Induction of Zebrafish Pronephric Cyst Formation by Co-injection of PKD2-L223 mRNA—We have previously established the zebrafish as a relevant model system to study human ADPKD (19, 24). Disruption of zebrafish pkd2 expression with morpholinos (MO) results in cyst formation in the glomerulus and pronephric tubules in conjunction with changes in body axis curvature and hydrocephalus. All of these features were rescued by co-injection of human PKD2 mRNA (19, 24). Because of sequence conservation between humans and zebrafish, we reasoned that the zebrafish model could be used to test the functionality of the N-terminal domain of PC2 by a dominant negative mechanism. These results are summarized in Table 1. To establish if a dominant negative effect could be observed in this system, we initially tested the effect of injecting human PKD2-D511V synthetic mRNA into one-cell stage zebrafish embryos. Injection of human PKD2-D511V synthetic mRNA alone (Fig. 4E) induced the same phenotypic changes seen in pkd2ATGMO-injected embryos (Fig. 4D). Moreover, as shown in Fig. 4, co-injection of PKD2-D511V mRNA with pkd2ATGMO could not rescue the pkd2ATGMO induced phenotype (Fig. 4F) unlike wild-type PKD2 mRNA. Therefore, these results established a a dominant negative mechanism for PKD2-D511V in zebrafish and fully supported previous data using the same construct in mIMCD3 cells (9).
TABLE 1.
Zebrafish phenotypes after injection of capped PKD2 mRNA and/or pkd2 morpholino at the one-cell embryo stage
| Total embryos injected | Normal embryos | Cysts and body axis curvature | |
|---|---|---|---|
| A. Control MO | 303 | 303 (100%) | 0 (0%) |
| B. Human PKD2-L177 | 254 | 250 (98%) | 4 (2%) |
| C. Human PKD2-L223 | 252 | 20 (8%) | 232 (92%) |
| D. pkd2ATGMO | 222 | 12 (5%) | 210 (95%) |
| E. Human PKD2-D511V | 296 | 6 (5%) | 281 (95%) |
| F. pkd2ATGMO + human PKD2-D511V | 103 | 2 (2%) | 101 (98%) |
FIGURE 4.
Human PKD2-L223 and D511V induce pronephric cysts in the zebrafish and downregulate zebrafish polycystin-2 expression. A, 48 hpf zebrafish injected with a control MO have a normal body; histology section of 48 hpf embryos showing a glomerulus (glm) in the midline and pronephric tubules connected to bilateral pronephric ducts. Endogenous zebrafish PC2, detected with a specific antibody that does not cross-react with human PC2, is distributed in the basolateral membranes and apical cilia in the anterior pronephric ducts (see also H). B, 48-hpf human PKD2-L177 mRNA-injected embryos show normal whole mount histology cross-section and zebrafish PC2 expression. C, human PKD2-L223 mRNA-injected embryos showing pronephric cysts, body axis curvature, and reduced zebrafish PC2 expression. D, pkd2ATGMO-injected embryos showing pronephric cysts, body axis curvature, and hydrocephalus. pkd2ATGMO blocked endogenous zebrafish pkd2 translation leading to a reduction in PC2 expression. E, human PKD2-D511V mRNA-injected embryos also developed body axis curvature, cyst, and hydrocephalus. F, co-injection of 50 pg of human PKD2-D511V was unable to rescue the pkd2ATGMO phenotype and induced more severe body axis curvature, cysts, and hydrocephalus than pkd2ATGMO alone. G, RT-PCR analysis for human PKD2 (upper panel) and β-actin (lower panel) mRNA expression. Endogenous zebrafish PC2 expression is clearly down-regulated by co-injection of PKD2-L223 (C) and PKD2-D511V (E) mRNA to a similar level as pkd2ATGMO (D).
Next, we injected PKD2-L223 in zebrafish embryos and tested whether it could result in a phenotype similar to the phenotype obtained by injection of pkd2ATGMO or PKD2-D511V. Fig. 4C shows that PKD2-L223 induced body axis curvature, pronephric cyst formation and hydrocephalus whereas injection of human PKD2-L177 mRNA lacking the dimerization domain did not (Fig. 4B). All injected embryos with PKD2-L177 exhibited normal histology as compared with embryos injected with control MO (Fig. 4A). Expression levels of PKD2-D511V, PKD2-L223 and PKD2-L177 were shown to be equivalent in injected embryos by nested RT-PCR using human-specific primers (Fig. 4G). In summary, these data suggest that PKD2-L223 must have interfered with wild-type PC2 and/or its interacting partners. However, because the N-terminal domain of PC2 has been shown not to interact with known C-terminal binding partners such as PKD1 or TRPC1, it is highly likely that PKD2-L223 mediated its effect by direct binding to wild-type PKD2.
Embryos injected with either PKD2-D511V (Fig. 4E) or PKD2-L223 (Fig. 4C) mRNA all showed a reduction of endogenous zebrafish PC2 expression similar to that seen with pkd2ATGMO (Fig. 4D). These results raised the possibility that PKD2-D511V or PKD2-L223 may have bound to wild-type PKD2 and/or marked it for degradation. An alternative possibility was that the acute binding of the mutant protein could have directly inhibited surface channel activity resulting in cysts with degradation occurring as a later event.
Inducible Translocation of PKD2-(223) to the Plasma Membrane Inhibits Endogenous and Transfected PKD2 Surface Currents—To address whether PKD2-1–223 could acutely inhibit surface PC2 channels, we exploited a novel ligand-induced (rapamycin) chemical dimerization system (summarized in Fig. 5F) based on the rapamycin-induced dimerization between FKBP and FRB (17). The FRB (FKBP-rapamycin binding) domain was fused to a plasma membrane targeting sequence of the Rho GTPase Lyn (LDR) while CFP-tagged FKBP (FK506- and rapamycin-binding protein) was fused to the N terminus of PKD2-(1–223) to generate CF-PKD2-(223). As a control, we utilized PKD2-(1–177) to generate CF-PKD2-(177). In the absence of rapamycin, both CF-PKD2 fusions are primarily expressed in the cytoplasm. On addition of rapamycin in the presence of the FRB domain (LDR plasmid), there was a rapid translocation of both fusion proteins to the plasma membrane in HEK293 (not shown) and mIMCD3 cells (Fig. 5) as shown by the loss of cytoplasmic CFP fluorescence in individual cells over time. Moreover, the decrease in fluorescent intensity (ΔF) over baseline intensity (F0) triggered by rapamycin was significantly altered for cytosolic CFP compared with the nuclear signal in the presence of LDR (n = 6) (Fig. 5E). The translocated CFP signal clearly overlapped with the YFP signal of a co-expressed surface-localized protein, YFP-C1 (PKC). The original CF (CFP-FKBP-Inp54p) construct served as a positive control (data not shown) (21).
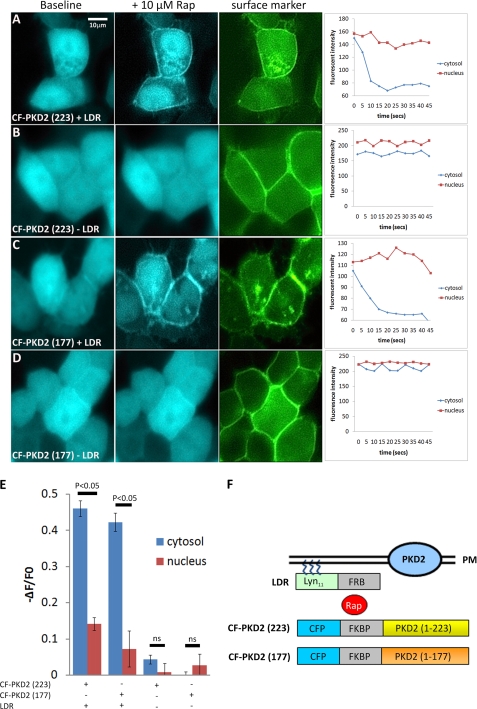
FIGURE 5.
Rapamycin-induced translocation of CFP-PKD2 fusions to the plasma membrane. A–D, CFP fluorescent images in live mIMCD3 cells co-transfected with the plasmids CF-PKD2-(177) or CF-PKD2-(223) in the presence or absence of LDR. The left hand panels represent baseline CFP (blue), the middle panels are CFP signals (blue) 5–45 s following the addition of rapamycin (Rap, 10 μm) to the medium and the right panels, YFP fluorescence (green) of the fusion protein, YFP-C1-(PKC), which is constitutively localized at the plasma membrane. The translocation of both CFP-PKD2 fusion proteins induced by Rap in the presence of LDR can be seen graphically by the rapid reduction in the cytoplasmic CFP signal within the time frame shown (5–45 s). In contrast, nuclear expression of both fusion proteins is present at baseline but does not change following Rap. E, change in cytosolic CFP fluorescence intensity (ΔF) expressed as a ratio of baseline CFP fluorescence (F0) was significantly altered compared with nuclear CFP fluorescence following Rap in the presence of LDR (n = 6). F, schematic diagram of the rapamycin-induced chemical dimerization strategy used to translocate CFP-PKD2 fusions to the plasma membrane (PM). The FRB (FKBP-rapamycin binding) domain was fused to a plasma membrane targeting sequence of the Rho GTPase Lyn (LDR), while CFP-tagged FKBP (FK506- and rapamycin-binding protein) was fused to the N terminus of PKD2 (1–177 or 1–223) to generate CF-PKD2-(177) and CF-PKD2-(223), respectively. Addition of Rap induces rapid heterodimerization between the PM-anchored FRB and FKBP fusion proteins, thus bringing the CF-PKD2 fusions into close proximity of PM-located PKD2 channels.
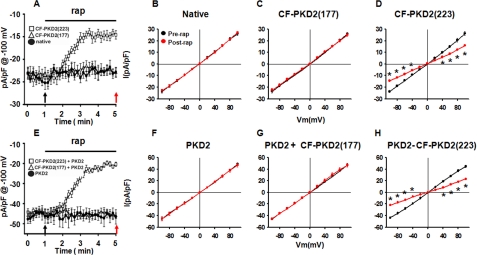
Fig. 6 shows that addition of rapamycin induced a time-dependent reduction in whole cell current amplitude of mIMCD3 cells transiently transfected with LDR+CF-PKD2-(223) cells from -23.7 ± 0.9 pA/pF to -14 ± 0.6 pA/pF at -100 mV (Fig. 6, A and D). The reduction in current amplitude was significant up to -40 mV (Fig. 6D). Rapamycin did not have an effect in untransfected (from -23.6 ± 1.2 pA/pF to -22.7 ± 1.2 pA/pF at -100 mV) (Fig. 6B) or LDR+CF-PKD2 (177)-transfected cells (from -23.5 ± 1.4 pA/pF to -22.3 ± 1.4 pA/pF at -100 mV) (Fig. 6C). It should be noted that constitutive overexpression of PKD2-D511V in these cells suppressed whole cell currents from -24.3 ± 3.4 pA/pF (n = 9) to -10.3 ± 2.28 pA/pF (n = 7) under identical recording conditions (9) suggesting a similar mechanism of whole cell current density inhibition by PKD2-D511V or PKD2-L223. The small difference in whole cell current density (∼4 pA/pF) between PKD2-D511V and PKD2-L223 may be explained by the higher affinity of PKD2-D511V than PKD2-L223 for binding to wild-type PKD2 or binding of PKD2-D511V to other interacting partners of PKD2 such as TRPC1, which was shown to be required for basal activity of native PKD2 in these cells (9). In contrast, PKD2-L223 should not associate with PKD1 or TRPC1 (5, 15) and therefore its effect on whole cell current density should be specific to wild-type PKD2, at least based on existing data.
FIGURE 6.
Inhibition of plasma membrane PKD2 channel activity by CF-PKD2-(223). Time-dependent inhibition of native (A) or transfected PKD2 (E) channel activity by rapamycin (rap)-induced translocation of a CFP fusion of the PC2 N terminus (NT2, 1–223) to the plasma membrane. mIMCD3 cells were transiently transfected with LDR plus CF-PKD2-(177) or CF-PKD2-(223) in the absence (A) or presence (E) of transfected wild-type mouse PKD2. Translocation of CF-PKD2-(177) or CF-PKD2-(223) to the plasma membrane was induced by the addition of 10 μm rapamycin to the bath solution. Current densities at -100 mV were obtained by 100-ms pulses from -60 mV to -100 mV applied every 10 s. Arrows indicate time points at which voltage steps were applied to derive I-V curves shown in B, C, D, F, G, and H. I-V curves derived from native (B), LDR plus CF-PKD2-(177) (C), or LDR plus CF-PKD2-(223) (D)-transfected mIMCD3 cells before (black) or after (red) the addition of rapamycin in the bath solution are shown. I-V curves derived from PKD2 (F), PKD2, LDR, and CF-PKD2-(177) (G), or PKD2, LDR, and CF-PKD2-(223) (H)-co-transfected mIMCD3 cells before (black) or after (red) the addition of rapamycin to the bath solution are shown. *, p < 0.05.
To further confirm the specificity of CF-PKD2-(223) on PKD2, we overexpressed full-length PKD2 and tested the effect of CF-PKD2-(177) or CF-PKD2-(223) on transfected PKD2. Overexpression of PKD2 resulted in an increase in overall whole cell current density from -23.6 ± 1.2 pA/pF to -45.4 ± 1.8 pA/pF at -100 mV (Fig. 6, B and F, black plot), consistent with the formation of active channels at the plasma membrane (9). Addition of rapamycin to the bath induced a time-dependent reduction in whole cell currents in PKD2-, LDR-, and CF-PKD2-(223)-cotransfected cells from -43.5 ± 1 pA/pF to -21.8 ± 1 pA/pF at -100 mV (Fig. 6H). However, in PKD2-transfected alone (Fig. 6F) or PKD2-, LDR-, and CF-PKD2-(177)-cotransfected cells, rapamycin did not affect whole cell currents (Fig. 6G). These data provide direct evidence for a dominant negative effect of CF-PKD2-(223) on native or transfected PKD2 surface channel activity. In this system, binding of PKD2-L223 resulted in acute inhibition of channel activity because the effect was observed almost immediately following induced translocation of PKD2-L223 to the plasma membrane.
DISCUSSION
In the present study, we have identified and functionally characterized a new dimerization domain in the N-terminal cytosolic region of PC2. This domain is shown to have a physiologically relevant role in zebrafish development as it phenocopied known loss-of-function constructs of PC2. We propose that the identification of this domain has important implications in type 2 ADPKD pathophysiology.
The tendency of native PC2 to oligomerize led us initially to investigate how PC2 homodimerization could be regulated. Unexpectedly, we found that two naturally occurring PC2 mutants lacking the C-terminal homodimerization domain (L703X, R742X) could still form oligomers and bind to full-length PC2 in mammalian cells. These findings led us to demonstrate the existence of a more proximal dimerization domain within the N-terminal domain and its functionality in two assays of PC2 activity i.e. nephrogenesis in zebrafish embryos and channel activity in mIMCD3 cells. These findings are compatible with a likely dominant negative effect in both models. Overall, our data would support a direct acute inhibitory effect of the mutant protein (PKD2-L223) on the PC2 channel itself, which also leads to subsequent degradation of PC2. Recently, it was reported that the transgenic expression of PKD2-L703X in rats gave rise to a cystic phenotype by an undetermined mechanism (27). We believe that our findings of an N-terminal dimerization domain support a dominant negative mechanism as a plausible explanation of the phenotype in this model.
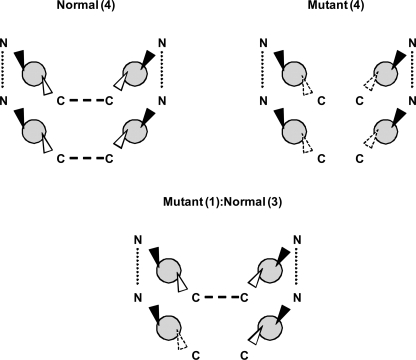
The existence of both N- and C-terminal dimerization domains in PC2 provide supportive evidence that PC2 is likely to form functional homotetramers, a possible model is shown in Fig. 7. This model does not require the binding of PC1 or that of other TRP subunits (such as TRPC1) both of which have been shown to interact with PC2 via distinct sequences or amino acids in the C-terminal domain (5). The evidence that PC2 homodimers can function independently in vivo is indirect: PC2 can function independently of PC1 at the embryonic node in the determination of LR asymmetry in the axial body plan (28). Nevertheless, an important question is what regulates the assembly of PC2 monomeric subunits into homotetramers or alternatively heterotetramers with PC1 or other TRP subunits. In addition, we do not know if PC2 truncation mutants (e.g. L703X), which retain the putative pore region and which can still dimerize via the N-terminal domain are still functional. In some assays, there is evidence for altered PC2 localization (e.g. increased cell surface expression) and for altered channel properties (e.g. loss of calcium responsiveness, voltage-dependence) of this mutant (12, 29).
FIGURE 7.
A tetrameric model of PC2 channel assembly. The existence of N- and C-terminal dimerization domains would facilitate the assembly of four PC2 monomers into a homotetramer. By contrast, a mutant PC2 monomer lacking the distal C-terminal dimerization domain (such as L703X) would still be capable of forming dimers via the proximal N-terminal domain: these mutant dimers could be functional. The incorporation of one mutant PC2 monomer with 3 normal PC2 monomers would permit the formation of a tetrameric structure but is likely to be non-functional.
Our results also raise the possibility as to whether cyst formation in PKD2 patients could arise by a dominant-negative mechanism as shown for the D511V mutant in addition to two-hit and haploinsufficiency models (30). If PC2 forms an oligomeric structure, the association of a mutant protein with wild-type subunits would result in the generation of non-functional multimeric complexes (Fig. 7). For a tetrameric model, potentially 15 of 16 possible combinations between mutant and wild-type subunits could be affected. Therefore, although there will be 50% reduction in the wild-type protein, the reduction in function could approximate 100%. This mechanism would only be effective assuming that a stable mutant mRNA transcript and protein are produced in patients at levels that would bind in a stoichiometric fashion to wild-type monomers. Nonetheless, given the lack of specific inhibitors to PC2, the dominant negative strategy we have described could be a useful way to study the function of the endogenous protein in different systems.
Acknowledgments
We thank D. J. Moon and Y. Xu for technical assistance, and T. Meyer, I. Drummond, S. Somlo, and D. Markie for kind gifts of reagents. No conflict of interest is declared.
This work was supported, in whole or in part, by National Institutes of Health Grants R21-DK069604, RO1-DK078209 (to T. O.), and R01-DK59599 (to L. T.). This work was also funded by grants from the PKD Foundation (69a2r and 119a2r), John S. Gammill Endowed Chair in Polycystic Kidney Disease, Research Councils UK (RA108836) (to A. J. S.), and the Wellcome Trust (GR071201) (to A. C. M. O.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ADPKD, autosomal dominant polycystic kidney disease; PKC, protein kinase C; PBS, phosphate-buffered saline; TRP, transient receptor potential; HA, hemagglutinin; IP, immunoprecipitation; CFP, cyan fluorescent protein; NT, N terminus; MO, morpholino.
Personal communication.
References
- 1.Calvet, J. P., and Grantham, J. J. (2001) Semin. Nephrol. 21 107-123 [DOI] [PubMed] [Google Scholar]
- 2.Wilson, P. D. (2004) N. Engl. J. Med. 350 151-164 [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki, T., Wu, G., Hayashi, T., Xenophontos, S. L., Veldhuisen, B., Saris, J. J., Reynolds, D. M., Cai, Y., Gabow, P. A., Pierides, A., Kimberling, W. J., Breuning, M. H., Deltas, C. C., Peters, D. J., and Somlo, S. (1996) Science 272 1339-1342 [DOI] [PubMed] [Google Scholar]
- 4.Montell, C., Birnbaumer, L., Flockerzi, V., Bindels, R. J., Bruford, E. A., Caterina, M. J., Clapham, D. E., Harteneck, C., Heller, S., Julius, D., Kojima, I., Mori, Y., Penner, R., Prawitt, D., Scharenberg, A. M., Schultz, G., Shimizu, N., and Zhu, M. X. (2002) Mol. Cell 9 229-231 [DOI] [PubMed] [Google Scholar]
- 5.Tsiokas, L., Arnould, T., Zhu, C., Kim, E., Walz, G., and Sukhatme, V. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3934-3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nauli, S. M., Alenghat, F. J., Luo, Y., Williams, E., Vassilev, P., Li, X., Elia, A. E., Lu, W., Brown, E. M., Quinn, S. J., Ingber, D. E., and Zhou, J. (2003) Nat. Genet. 33 129-137 [DOI] [PubMed] [Google Scholar]
- 7.Ma, R., Rundle, D., Jacks, J., Koch, M., Downs, T., and Tsiokas, L. (2003) J. Biol. Chem. 278 52763-52772 [DOI] [PubMed] [Google Scholar]
- 8.Bai, C. X., Kim, S., Li, W. P., Streets, A. J., Ong, A. C., and Tsiokas, L. (2008) EMBO J. 27 1345-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai, C. X., Giamarchi, A., Rodat-Despoix, L., Padilla, F., Downs, T., Tsiokas, L., and Delmas, P. (2008) EMBO Rep. 9 472-479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newby, L. J., Streets, A. J., Zhao, Y., Harris, P. C., Ward, C. J., and Ong, A. C. (2002) J. Biol. Chem. 277 20763-20773 [DOI] [PubMed] [Google Scholar]
- 11.Streets, A. J., Newby, L. J., O'Hare, M. J., Bukanov, N. O., Ibraghimov-Beskrovnaya, O., and Ong, A. C. (2003) J. Am. Soc. Nephrol. 14 1804-1815 [DOI] [PubMed] [Google Scholar]
- 12.Koulen, P., Cai, Y., Geng, L., Maeda, Y., Nishimura, S., Witzgall, R., Ehrlich, B. E., and Somlo, S. (2002) Nat Cell Biol. 4 191-197 [DOI] [PubMed] [Google Scholar]
- 13.Hanaoka, K., Qian, F., Boletta, A., Bhunia, A. K., Piontek, K., Tsiokas, L., Sukhatme, V. P., Guggino, W. B., and Germino, G. G. (2000) Nature 408 990-994 [DOI] [PubMed] [Google Scholar]
- 14.Delmas, P., Nauli, S. M., Li, X., Coste, B., Osorio, N., Crest, M., Brown, D. A., and Zhou, J. (2004) FASEB J. 18 740-742 [DOI] [PubMed] [Google Scholar]
- 15.Tsiokas, L., Kim, E., Arnould, T., Sukhatme, V. P., and Walz, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6965-6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semple, J. I., Prime, G., Wallis, L. J., Sanderson, C. M., and Markie, D. (2005) BioTechniques 38 927-934 [DOI] [PubMed] [Google Scholar]
- 17.Inoue, T., Heo, W. D., Grimley, J. S., Wandless, T. J., and Meyer, T. (2005) Nat. Methods 2 415-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong, A. C., Ward, C. J., Butler, R. J., Biddolph, S., Bowker, C., Torra, R., Pei, Y., and Harris, P. C. (1999) Am. J. Pathol. 154 1721-1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streets, A. J., Moon, D. J., Kane, M. E., Obara, T., and Ong, A. C. (2006) Hum. Mol. Genet. 15 1465-1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and Woods, R. A. (2002) Methods Enzymol. 350 87-96 [DOI] [PubMed] [Google Scholar]
- 21.Suh, B. C., Inoue, T., Meyer, T., and Hille, B. (2006) Science 314 1454-1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westerfield, M. (1995) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Brachydanio), University of Oregon Press, Eugene, OR
- 23.Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S., and Hopkins, N. (2004) Development 131 4085-4093 [DOI] [PubMed] [Google Scholar]
- 24.Obara, T., Mangos, S., Liu, Y., Zhao, J., Wiessner, S., Kramer-Zucker, A. G., Olale, F., Schier, A. F., and Drummond, I. A. (2006) J. Am. Soc. Nephrol. 17 2706-2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, X. Z., Segal, Y., Basora, N., Guo, L., Peng, J. B., Babakhanlou, H., Vassilev, P. M., Brown, E. M., Hediger, M. A., and Zhou, J. (2001) Biochem. Biophys. Res. Commun. 282 1251-1256 [DOI] [PubMed] [Google Scholar]
- 26.Ma, R., Li, W. P., Rundle, D., Kong, J., Akbarali, H. I., and Tsiokas, L. (2005) Mol. Cell. Biol. 25 8285-8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallagher, A. R., Hoffmann, S., Brown, N., Cedzich, A., Meruvu, S., Podlich, D., Feng, Y., Konecke, V., de Vries, U., Hammes, H. P., Gretz, N., and Witzgall, R. (2006) J. Am. Soc. Nephrol. 17 2719-2730 [DOI] [PubMed] [Google Scholar]
- 28.McGrath, J., Somlo, S., Makova, S., Tian, X., and Brueckner, M. (2003) Cell 114 61-73 [DOI] [PubMed] [Google Scholar]
- 29.Cai, Y., Maeda, Y., Cedzich, A., Torres, V. E., Wu, G., Hayashi, T., Mochizuki, T., Park, J. H., Witzgall, R., and Somlo, S. (1999) J. Biol. Chem. 274 28557-28565 [DOI] [PubMed] [Google Scholar]
- 30.Ong, A. C., and Harris, P. C. (2005) Kidney Int. 67 1234-1247 [DOI] [PubMed] [Google Scholar]