Abstract
Bit1 (Bcl-2 inhibitor of transcription) is a mitochondrial protein that induces caspase-independent apoptosis upon its release into the cytoplasm. Bit1 is primarily associated with anoikis (cell death induced by detachment from the extracellular matrix), because the apoptotic function of Bit1 is inhibited by integrin-mediated cell attachment but not by many other antiapoptotic treatments. Here, we show that protein kinase D (PKD) regulates Bit1 apoptotic function. Overexpression of constitutively active PKD or PKD activation by treatment with phorbol 12-myristate 13-acetate results in phosphorylation of two serine residues (Ser5 and Ser87) in a form of Bit1 that is confined to the cytoplasm and concomitantly increases the apoptotic activity of cytoplasmic Bit1. Conversely, suppressing PKD activity with pharmacological inhibitors or small interfering RNA approaches attenuates apoptosis induced by cytoplasmic Bit1. Furthermore, PKD regulates induction of cell death by wild-type Bit1 following loss of cell attachment to the extracellular matrix. Activation of PKD enhances Bit1 function in anoikis, whereas inhibiting PKD function with pharmacological inhibitors or small interfering RNA compromises the ability of Bit1 to induce anoikis. The induction of Bit1-mediated apoptosis by PKD is in part attributable to the release of Bit1 from mitochondria to the cytoplasm as a consequence of phosphorylation of Ser5 in the mitochondrial localization sequence of Bit1. Consistent with the regulatory role of PKD in the anoikis function of Bit1, we found that cell attachment to fibronectin inhibits PKD activity. These studies identify the PKD serine/threonine kinase as one of the signaling molecules through which integrin-mediated cell attachment controls Bit1 activity and anoikis.
The survival of adherent cells is highly dependent on substrate attachment (anchorage dependence). Loss of attachment causes cell death through an apoptosis process known as anoikis (1). Malignant cells tend to be less dependent on attachment to the extracellular matrix and more resistant to anoikis than normal cells. This anoikis resistance may enable tumor cells to survive lack of attachment during invasion and metastasis. The signals that prevent anoikis originate from integrin-mediated attachment of cells to the extracellular matrix, and some of the well known integrin signaling molecules, such as focal adhesion kinase, have been shown to regulate anoikis (2, 3). Although the same signaling molecules are also controlled by various growth factors, growth factors cannot substitute for integrin-mediated attachment, suggesting that signaling pathways specific for integrins may exist.
Bit1 (Bcl2-inhibitor of transcription 1) is a protein that appears to be part of an integrin-specific signaling pathway (4). Bit1 is a 179-amino acid mitochondrial protein with a known crystal structure (5). Upon loss of cell attachment, it is released from the mitochondria into the cytosol and promotes apoptosis. Suppression of Bit1 expression in tumor cells as well as in normal cells significantly protects cells from detachment-induced apoptosis, demonstrating a key role of Bit1 in anoikis (4, 6, 7). Unlike other apoptotic pathways, Bit1-induced apoptosis is uniquely controlled by integrin-mediated cell attachment. Only integrin-mediated cell attachment counteracts apoptosis induced by cytosolic Bit1, whereas various antiapoptotic signaling molecules, such as Bcl-2, Bcl-xL, phosphatidylinositol 3-kinase, and Akt, fail to do so. Cell attachment mediated by the α5β1 and αvβ3 integrins, which are receptors for fibronectin or vitronectin, is particularly effective in inhibiting the apoptotic activity of cytoplasmic Bit1. Interestingly, Bit1-induced cell death is independent of caspase activity but requires the presence of AES, a member of the Groucho/TLE family of transcriptional regulators (4). We have also recently shown that Bit1 is a negative regulator of Erk and provided evidence that the target of Bit1 is an Erk phosphatase (6).
The signaling mechanisms through which integrins block the apoptotic activity of Bit1 are completely unknown. In the work reported here, we set out to explore the integrin regulation of Bit1. We hypothesized that Bit1, like so many other intracellular molecules, might be regulated by phosphorylation. Indeed, we found that the atypical protein kinase C, PKC3 μ or PKD, phosphorylates Bit1 and enhances Bit1 apoptotic activity. We also provide evidence that activation of PKD is regulated by integrin-mediated cell attachment. These findings place PKD in a pathway from integrins to Bit1 and begin to delineate a novel signaling pathway that appears to be important in anoikis.
EXPERIMENTAL PROCEDURES
Chemical Reagents, Antibodies, and Plasmids—Phorbol 12-myristate 13-acetate (PMA) and the mouse monoclonal anti-green fluorescent protein (GFP) antibody (clone GFP-20) were obtained from Sigma. The kinase inhibitors Go6976 and Go6983 were purchased from Calbiochem. The anti-PKD polyclonal antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The anti-phospho-PKD (Ser744/748 and Ser916) antibodies were from Cell Signaling Technology (Beverly, MA). Both antibodies have been used to monitor PKD activation (8, 9). The mouse anti-Myc and the rabbit anti-phosphoserine antibodies were from Zymed Laboratories Inc. (South San Francisco, CA). Dr. Franz-Josef Johannes (The Fraunhofer Institute for Interfacial Engineering) provided mammalian expression vectors encoding various GFP-tagged PKD isoforms, and GST-tagged PKD vectors were obtained from Dr. Vivek Malhotra (Division of Biology, University of California San Diego). Generation of N- and C-terminally Myc-tagged Bit1 constructs was described previously (4).
Generation of Bit1 Phosphospecific Antibodies—Cocalico Biological, Inc. (Reamstown, PA) generated antibodies against the human Bit1 serine 5 or serine 87-phosphorylated motifs according to our specifications. Rabbits were immunized with keyhole limpet hemocyanin-conjugated peptides with the sequence CKVAAQCpSHAAVSAY containing the phosphorylated Ser87 residue and MPSKpSLVMEYLAHPSC containing the phosphorylated Ser5 residue. Phophospecific antibodies were purified from crude rabbit serum by sequential negative and positive affinity purification with nonphosphorylated and phosphorylated peptide, respectively. Antibody specificity was confirmed using pre-immune sera and blocking experiments with the phosphorylated peptide.
Cell Culture and Transfection Assays—The experiments were carried out using HEK 293T cells, unless otherwise stated. These cells and HeLa and CHO cells were cultured in Dulbecco's modified Eagle's medium with glutamine containing 10% fetal bovine serum, penicillin, and streptomycin. Human umbilical vascular endothelial cells (HUVECs) were purchased from Lonza (Walkersville, MD) and maintained using the EGM-2 bullet kit (Lonza). In some experiments, cells were plated onto non-tissue culture-treated plates precoated with 10 μg/ml fibronectin (Calbiochem) or type I collagen (Sigma) in serum-free Dulbecco's modified Eagle's medium. Suspension cultures were prepared by plating cells on dishes coated with polyHEMA (Sigma) and culturing in serum-containing medium. Transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol 18 h after plating the cells. The total amount of plasmid used per transfection was kept constant by using empty vector DNA when necessary. For apoptotic and cell viability assays, cells were harvested 48 h after transfection.
siRNA Transfection for Knockdown of PKD—The PKD siRNA SMARTpool, which consists of four individual siRNA, was used to effectively knock down PKD, whereas the siControl nontargeting siRNA pool was used as a control. Both the PKD siRNA SMARTpool and nontargeting siRNA pool were obtained from Dharmacon (Lafayette, CO). For transient transfection experiments, 293T or HUVECs (2 × 105) were transfected with 100 pmol of each siRNA pool by using the Lipofectamine 2000 transfection reagent (Invitrogen). In HUVECs, transfection mixtures were left on the cells for 4 h and then replaced with regular (EBM-2) medium. To express Bit1 in cells with down-regulated PKD, 1 day after control or PDK siRNA transfection, the cells were further transfected with 2 μg of N- or C-terminally Myc-tagged Bit1 vector or empty vector as a control. The cells were harvested 24 h later and analyzed by immunoblotting and apoptosis assay.
Subcellular Fractionation—Subcellular fractionation was performed as described previously (4). Briefly, HEK 293T cells were washed twice in PBS, resuspended in isotonic mitochondrial buffer (250 mm mannitol, 70 mm sucrose, 1 mm EDTA, 10 mm HEPES, pH 7.5, containing protease inhibitors), and homogenized in a Dounce homogenizer. The lysates were initially centrifuged at 500 × g for 10 min to remove nuclei and unbroken cells. The resulting supernatant was further centrifuged at 10,000 × g for 30 min at 4 °C to isolate the mitochondrial enriched pellet, which was resuspended in isotonic mitochondrial buffer. Both the cytosolic supernatant and mitochondrial fraction were subjected to SDS-PAGE electrophoresis and immunoblotting.
Analysis of Cell Viability and Apoptosis—Cell viability was measured by trypan blue exclusion. Apoptosis was assessed by determining the level of cytosolic nucleosomal fragments with the cell death detection enzyme-linked immunosorbent assay kit (Roche Applied Science), according to the manufacturer's instructions. To measure cell death by anoikis, cells were plated in poly-HEMA-coated 96-well plates in complete growth medium containing 0.5% methylcellulose at a density of 1.0 × 104 cells/well for 48 h. The apoptotic activity of Bit1 mutants was assessed by transfecting each mutant in CHO cells and measuring apoptosis 24 h later by annexin V staining and flow cytometry.
Protein Preparation, Immunoblotting, and Coimmunoprecipitation Assays—Protein preparation and immunoblotting were performed as described previously (4). Briefly, cells were harvested 24–48 h after transfection with various plasmids or siRNAs by adding ice-cold Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 2 mm sodium vanadate, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 5 μg/ml aprotinin) followed by a 20-min incubation at 4 °C. For immunoblot analysis, equal amounts of proteins were resolved on Tris-glycine gels (Invitrogen) and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with primary antibodies overnight at 4 °C followed by secondary antibodies conjugated to horseradish peroxidase. Membranes were developed using the ECL detection system (Amersham Biosciences).
For coimmunoprecipiation assays, GFP-tagged PKD and Myc-tagged Bit1 expression plasmids were cotransfected into 293T cells. Twenty-four hours after transfection, the cells were harvested in ice-cold Nonidet P-40 lysis buffer, and cell debris was removed by centrifugation. Myc-tagged Bit1 was immunoprecipitated with anti-Myc-agarose conjugate (Abcam) and thoroughly washed with lysis buffer. Bound proteins were resolved by SDS-PAGE, and Western blotting was performed using anti-Myc or anti-GFP antibodies.
In Vitro Kinase Assays—HEK 293T cells were transfected with Myc-tagged cytoplasmic Bit1 construct or with empty vector as a control, and Myc-Bit1 was isolated by immunoprecipitation 24 h later. The immunoprecipitates were subjected to in vitro kinase reactions in the presence or in the absence of recombinant active PKD (Calbiochem) and kinase buffer containing 0.1 mm ATP and 50 μCi of [γ-32P]ATP for 30 min at 30 °C. An equal volume of 2× SDS-PAGE loading buffer was added, and phosphorylated Bit1 was resolved by SDS-PAGE and visualized by autoradiography.
In Vivo 32P Labeling—HEK 293T cells were transiently transfected with Myc-tagged Bit1, and 18 h later, the cells were labeled with 5 mCi of [32P]orthophosphate and incubated at 37 °C for 4 h. Following incubation, the cells were washed with phosphate-buffered saline and then lysed in lysis buffer as described above. Myc-Bit1 was immunoprecipitated with anti-Myc-agarose conjugate, and immunoprecipitates were subjected to SDS-PAGE. Incorporation of 32P was visualized by autoradiography.
Statistical Analysis—Data are presented as means ± S.E. All immunoblotting, in vitro kinase activity, and apoptosis assays were performed at least twice with duplicate or triplicate samples in each experiment. Densitometric analysis was performed using Image J software. Data were analyzed for statistical significance using a paired Student's t test. A p value of <0.05 was considered significant.
RESULTS
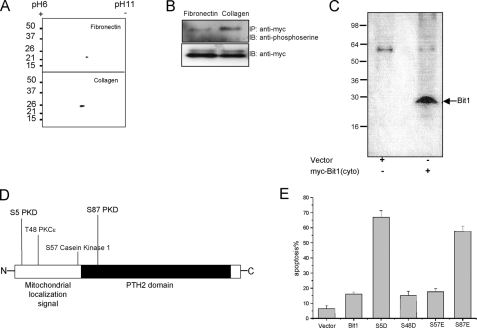
Regulation of Bit1 Function by Phosphorylation—To begin testing the hypothesis that Bit1 might be subject to phosphorylation and that phosphorylation might regulate its apoptotic potency, we made use of an earlier observation that Bit1 localized in the cytoplasm (due to an N-terminal tag that interferes with the mitochondrial localization signal of Bit1) exhibits a more potent apoptotic activity in cells plated on collagen than in cells plated on fibronectin (4). We reasoned that if phosphorylation was important for Bit1 activity, Bit1 phosphorylation should differ under these two conditions. Two-dimensional gel analysis revealed a Bit1 component with lower isoelectric point in cells plated on collagen but not in cells plated on fibronectin, suggesting that a portion of Bit1 was phosphorylated in cells plated on collagen (Fig. 1A). Consistent with this result, we found that a greater fraction of Bit1 was reactive with an anti-phosphoserine antibody when Bit1 was immunoprecipitated from HEK 293T cells plated on collagen than from cells plated on fibronectin (Fig. 1B). Moreover, Bit1 isolated from cells that had been transfected with cytosolic Bit1 and labeled with 32P produced a radioactive band by autoradiography (Fig. 1C). Collectively, these findings indicate that Bit1 can be phosphorylated in cells and that the degree of phosphorylation correlates with its apoptotic activity.
FIGURE 1.
Bit1 phosphorylation. A, HEK 293T cells plated onto dishes coated with either fibronectin or type I collagen were transfected with N-terminally Myc-tagged Bit1, which is localized in the cytoplasm. Cell lysates prepared 24 h after the transfection were separated on two-dimensional gels and immunoblotted with an anti-Myc antibody. Bit1 from collagen-plated cells included a negatively charged species, indicating possible partial phosphorylation. B, anti-Myc immunoprecipitates of lysates from cells plated on collagen show more reactivity with anti-phosphoserine antibodies than lysates from cells plated on fibronectin. C, autoradiography of N-terminally Myc-tagged Bit1 immunoprecipitated from transfected cells that were labeled with [32P]orthophosphate shows phosphorylation of Bit1. D, the Scansite 1.0 Motif Scan program (10) revealed four potential phosphorylation sites in Bit1, whereas a more recent version of the program (Scansite 2.0) (14) yields slightly different results. E, Bit1 amino acids 5, 48, 57, and 87 were each individually mutated to aspartic or glutamic acid in the N-terminally Myc-tagged cytoplasmic version of Bit1, and the apoptotic activity of the mutant proteins was assessed by annexin V staining and fluorescence-activated cell sorting analysis. IP, immunoprecipitation; IB, immunoblot.
We next searched for possible phosphorylation sites in Bit1 by employing the Scansite 1.0 Motif Scan program to predict phosphorylation sites for various kinases (10). As shown in Fig. 1D, Bit1 contains three serine residues and one threonine residue that are predicted phosphorylation sites. The serine residues at positions 5 and 87 are putative PKD phosphorylation sites, whereas serine 57 is a potential phosphorylation site for casein kinase 1, and threonine 48 is a potential site for PKCε. To address the importance of these phosphorylation sites in the apoptotic function of Bit1, we mutated each of them to negatively charged phosphomimetic amino acids (aspartic acid or glutamic acid) and determined the effect on Bit1 activity (Fig. 1E). Introducing a negatively charged amino acid at positions Ser5 and Ser87 greatly increased the apoptotic activity of Bit1, whereas mutating these two putative PKD phosphorylation sites to a neutrally charged alanine did not alter Bit1 apoptotic function. Furthermore, mutation of serine 57 to aspartic acid or threonine 48 to glutamic acid also did not change Bit1 apoptotic activity. These results suggest that the serine residues at positions 5 and 87 are functional phosphorylation sites in Bit1 and that PKD may be the kinase that phosphorylates these sites.
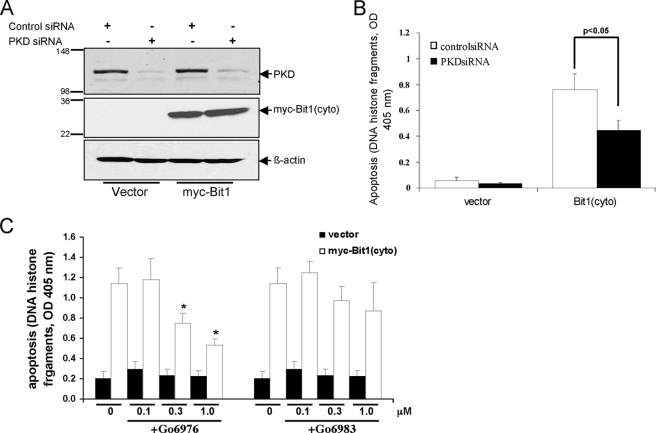
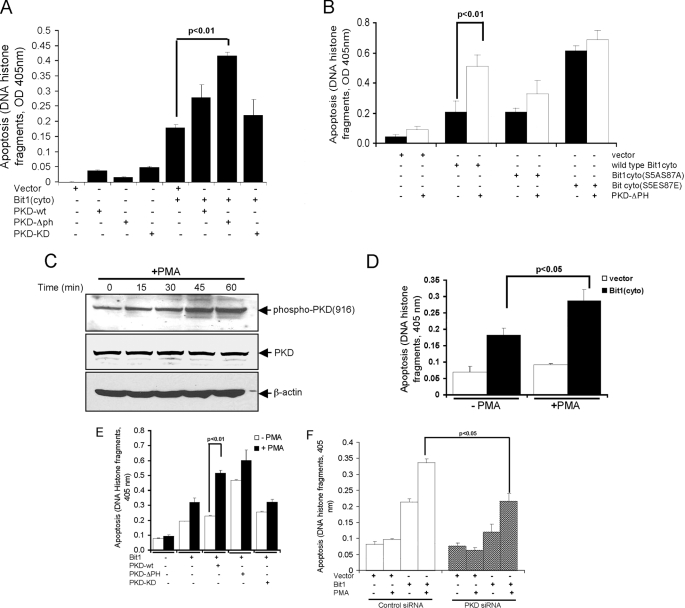
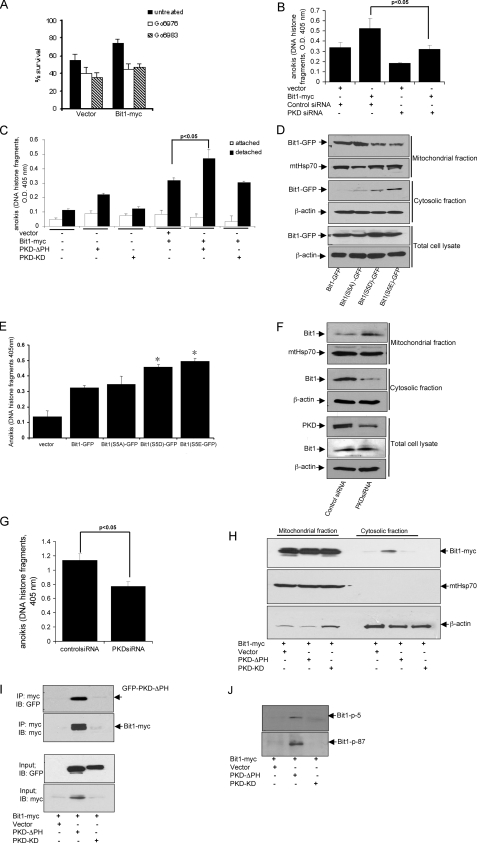
PKD Knockdown Attenuates and PKD Activation Enhances Bit1 Apoptotic Activity—We knocked down PKD expression in HEK 293T cells by transfecting a mixture of siRNAs (Fig. 2A). Down-regulation of PKD expression significantly decreased Bit1-induced apoptosis (Fig. 2B). Consistent with this result, the PKC inhibitor Go6976, which has a preferential inhibitory activity against PKD, significantly attenuated Bit1-induced apoptosis in a dose-dependent manner in HeLa cells (Fig. 2C). The more general PKC inhibitor Go6983, which lacks direct PKD inhibitory function, failed to significantly alter Bit apoptosis. These data indicate that PKD potentiates Bit1 apoptotic activity. Indeed, transient cotransfection of cells with the cytosolic form of Bit1 together with constitutively active PKD greatly enhanced Bit1 apoptotic activity compared with empty vector control (p < 0.01). Wild type and catalytically inactive PKD did not significantly alter Bit1 apoptotic function (Fig. 3A). The effect of constitutively active PKD on Bit1 appeared to require the presence of Bit1 serine residue 5 and/or 87, since mutating both of these residues to alanine reduced the effect of cotransfected, constitutively active PKD (Fig. 3B). Furthermore, the high apoptotic activity of Bit1 containing glutamic acid at positions 5 and 87 was not further enhanced by active PKD. These results indicate that PKD kinase activity is required in order to regulate Bit1 apoptotic function and that serine residue 5 and/or 87 are targets for phosphorylation by PKD.
FIGURE 2.
PKD knockdown attenuates Bit1 apoptotic activity. HEK 293T cells were transfected with control siRNA or with a mixture of PKD siRNAs and, 24 h later, with N-terminally Myc-tagged Bit1 or the empty vector. Twenty-four hours after the second transfection, cells were harvested and subjected to immunoblotting with antibodies to PKD and Myc to confirm down-regulation of PKD and overexpression of Bit1 (A) or assayed for apoptosis using a cell death enzyme-linked immunosorbent assay kit (B). The histogram shows means ± S.E. from three independent experiments. C, HeLa cells were pretreated with various doses of PKC inhibitor Go6983 or Go6976 for 6 h and then transfected with either N-terminally Myc-tagged Bit1 or empty vector. At 24 h postransfection, apoptotic cells were quantified by cell death enzyme-linked immunosorbent assay. Three independent experiments were performed in triplicate, and the data are expressed as means ± S.E.; *, p < 0.05 versus untreated cells.
FIGURE 3.
PKD kinase activity augments Bit1-induced apoptosis in 293T cells. A, cells were cotransfected with expression vectors encoding N-terminally Myc-tagged Bit1 and GFP-tagged versions of wild type (wt), constitutively active (ΔPH), or catalytically inactive (KD) PKD. The extent of apoptosis was determined 24 h later. B, constitutively active PKD-ΔPH and various mutant forms of N-terminally Myc-tagged Bit1 (wild type, S5A/S87A, and S5E/S87E) were cotransfected into cells, and apoptosis was measured 24 h later. C, cells were cultured with or without 10 ng/ml PMA, and activation of PKD was assessed at various time points by immunoblotting with a phosphospecific antibody recognizing the phosphorylated serine 916 motif in the activation loop. The same membrane was reprobed separately with an antibody against total PKD and β-actin. D, cells transfected with N-terminally Myc-tagged Bit1 or empty vector were left untreated or treated with 10 ng/ml PMA for 20 h before measuring apoptosis. E, cells cotransfected with N-terminally Myc-tagged Bit1, and various forms of PKD were left untreated or treated with 10 ng/ml PMA for 20 h before measuring apoptosis. F, cells were transfected with control or mixed PKD siRNAs and, 24 h later, with N-terminally Myc-tagged Bit1 or empty vector. The cells were then incubated with or without 10 ng/ml PMA for 24 h. In A, B, D, E, and F, experiments were performed in triplicate, and the histograms show means ± S.E.
As an alternative approach to activate PKD, we treated cells with the PKC activator PMA (8, 11) and monitored PKD activation by measuring autophosphorylation at serine 916 (9). PMA induced a marked increase in PKD activation (Fig. 3C) and augmented Bit1-induced apoptosis (Fig. 3D). To determine whether the PMA-mediated induction of Bit1 apoptotic activity was indeed mediated by PKD, cells were cotransfected with cytosolic Bit1 and various versions of PKD and subsequently treated with PMA. PMA caused a significant increase in Bit1 apoptotic activity in the presence of transfected wild type PKD but had much less effect on cells transfected with the constitutively active or catalytically inactive variants (Fig. 3E). Thus, PMA-mediated enhancement of Bit1-induced apoptosis is mostly dependent on PKD catalytic activity. Indeed, down-regulating PKD with siRNAs significantly reduced PMA stimulation of Bit1-dependent apoptosis (Fig. 3F). Taken together, these results suggest a role for PKD in PMA-induced Bit1 apoptosis and further indicate that PKD kinase activity is critical for the ability of this kinase to regulate apoptosis induced by Bit1.
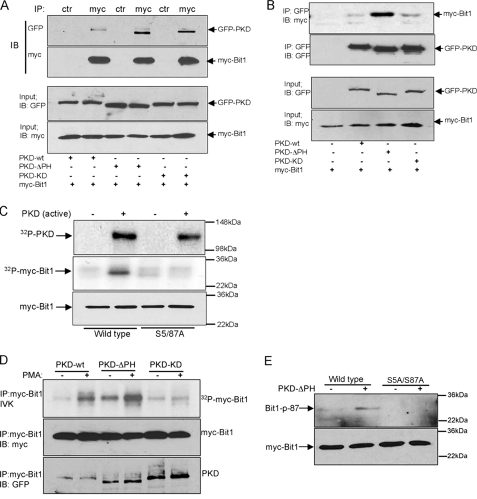
PKD Interacts with and Phosphorylates Bit1—To further explore the potential involvement of PKD in Bit1 phosphorylation, we examined whether PKD associates with Bit1 in cells. Cytoplasmic Bit1 was cotransfected with wild type, constitutively active, or kinase-inactive PKD into HEK 293T cells, and the association of PKD with Bit1 was determined by co-immunoprecipitation. All three PKD variants were detected in Bit1 immunoprecipitates, with constitutively active PKD showing the strongest association (Fig. 4A). The preferential binding of constitutively active PKD to Bit1 was confirmed by the reciprocal immunoprecipitation of PKD, followed by immunoblotting for Bit1 (Fig. 4B). The binding of kinase-inactive PKD to a PKD substrate (histone deacetylase 5) has been observed before (12).
FIGURE 4.
PKD associates with and phosphorylates Bit1. A, cells were cotransfected with expression vectors encoding N-terminally Myc-tagged Bit1 and GFP-tagged PKD variants. Cell lysates were prepared 24 h later and immunoprecipitated with anti-Myc or control rabbit IgG, and the immunoprecipitates were immunoblotted with anti-GFP and anti-Myc antibodies. The input panels show the levels of the transfected proteins in the original lysates. B, cells were transfected as in A, but immunoprecipitates were with anti-GFP antibodies and immunoblots with anti-Myc antibodies. C, N-terminally Myc-tagged wild type Bit1 or Bit1S5A/S87A expressed in HEK 293T cells were immunoprecipitated with an anti-Myc antibody. The immunoprecipitates were subjected to an in vitro kinase assay in the presence or absence of recombinant active PKD, followed by SDS-PAGE and autoradiography (top two panels) or immunoblotting with anti-Myc antibody (bottom panel). Constitutively active PKD phosphorylates both itself (top panel) and wild type Bit1 (middle panel) but not Bit1S5A/S87A. D, HEK 293T cells cotransfected with N-terminally c-Myc-tagged Bit1 and GFP-tagged PKD variants were treated with 10 ng/ml PMA for 30 min or left untreated. Kinase assays (top panel) show that Myc-Bit1 immunoprecipitated from the lysates is phosphorylated by wild type and constitutively active, but not kinase-dead, PKD and that the activity of wild type PKD is PMA-dependent. Total Bit1 and PKD were detected by immunoblotting with anti-Myc and anti-GFP (bottom two panels). E, a phosphospecific antibody against the Ser87-phosphorylated motif recognizes wild type Bit1, but not the Bit1S5A/S87A mutant, in HEK 293T cells cotransfected with the Bit1 variants and constitutively active PKD. IP, immunoprecipitation; IB, immunoblot.
To determine whether PKD can directly phosphorylate Bit1, in vitro kinase assays were performed with Bit1 isolated from transfected cells in the presence of recombinant active PKD. Constitutively active PKD caused phosphorylation of wild type Bit1 but not of Bit1 in which serine residues 5 and 87 had been mutated to alanine (Fig. 4C). Wild type and catalytically inactive PKD showed little activity toward wild type Bit1 (Fig. 4D), but treating the cells with PMA enhanced Bit1 phosphorylation by wild type PKD. PMA treatment further enhanced Bit1 phosphorylation by constitutively activated PKD, possibly due to the presence of endogenous PKD in the immune complexes. In contrast, catalytically inactive PKD failed to phosphorylate Bit1 even in the presence of PMA. We also assessed the ability of PKD to phosphorylate Bit1 by using antibodies that specifically recognize Bit1 phosphorylated at serine 5 or 87. The antibodies against the Ser87 site, but not those against the Ser5 site, recognized Bit1 that had been coexpressed with constitutively active PKD (Fig. 4E) (data not shown). The N-terminal tag on Bit1 may have prevented Ser5 phosphorylation or recognition of Ser5-phosphorylated Bit1 by the anti-phospho-Ser5 antibody, since this antibody did recognize C-terminally tagged Bit1 (see Fig. 5J). Neither antibody recognized Bit1 S5A/S87A. Collectively, these data show that PKD is capable of phosphorylating Bit1 in vitro as well as in the cellular environment and that at least Ser87 is a PKD phosphorylation site.
FIGURE 5.
PKD regulates Bit1 anoikis function and the release of Bit1 into the cytosol 293T cells. A, cells transfected with C-terminally Myc-tagged Bit1 (Bit-Myc, which can localize to mitochondria) or with empty vector, were placed 24 h later onto polyHEMA-coated wells with or without the indicated PKC inhibitors. Live cells were quantified 48 h later. B, cells were dually transfected with a mixture of PKD siRNAs or with control siRNA and 24 h later with C-terminally Myc-tagged Bit1 or empty vector. The cells were then placed on polyHEMA 48 h later, and the extent of anoikis was determined another 48 h later. C, cells were cotransfected with expression vectors encoding C-terminally Myc-tagged Bit1 and GFP-tagged PKD mutants. The cells were replated 24 h later onto tissue culture plates (attached) or polyHEMA-coated plates to induce anoikis (detached), and cell death was measured 48 h later. D, HEK 293T cells were transfected with various forms of C-terminally GFP-tagged Bit1 (Bit1-GFP). Cells were harvested 24 h later and subjected to subcellular fractionation, and the resulting cytosolic and mitochondrial fractions were probed by immunoblotting with an anti-GFP antibody. The mitochondrial protein HSP 70 (mtHsp70) and β-actin were used as mitochondrial and cytosolic marker, respectively. E, cells transfected with various forms of C-terminally GFP-tagged Bit1 were cultured on polyHEMA-coated plates for 48 h, and cell death was quantified. *, p < 0.05, compared with cells transfected with wild type Bit1-GFP. F and G, HUVECs were transfected with control or PKD siRNAs, and 24 h later, the cells were placed on polyHema, incubated for 12 h, and subjected to either subcellular fractionation (F) or cell death enzyme-linked immunosorbent assay (G). In F, the mitochondrial, cytosolic, and total cell lysate fractions were separated by SDS-PAGE and probed by immunoblotting against the indicated proteins. H, cells cotransfected with C-terminally Myc-tagged Bit1- and GFP-tagged PKD mutants were cultured and analyzed as in D. I and J, the cytosolic fraction derived from H was subjected to immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-Myc, anti-GFP, or phosphospecific antibodies to the Bit1 Ser5- or Ser87-phosphorylated motifs (p-S5 and p-S87). All experiments were performed at least three times in triplicate, and the histograms show means ± S.E. IP, immunoprecipitation; IB, immunoblot.
PKD Enhances the Cytosolic Localization and Anoikis Function of Bit1—We also examined whether PKD may regulate Bit1 activity in anoikis. HEK 293T cells were transfected with C-terminally Myc-tagged Bit1 (Bit-Myc), which has a functional mitochondrial localization signal and can therefore associate with mitochondria (4), and placed it onto polyHEMA-coated plates to induce anoikis. The PKC inhibitors Go6976 and Go6983 both significantly inhibited anoikis in Bit1-transfected as well as control-transfected HEK 293T cells (Fig. 5A). Furthermore, PKD siRNAs impaired and constitutively active PKD enhanced Bit1-dependent anoikis, whereas catalytically inactive PKD had no effect (Fig. 5, B and C). One might have expected inactive PKD to have a dominant negative effect on anoikis, which was not the case. It may be that the amount of Bit1 available to phosphorylation by PKD was not a limiting factor, since Bit1 was overexpressed. Alternatively or in addition, the results shown in Fig. 4 suggest that inactive PKD binds Bit1 less avidly than activated PKD and thus may be a poor competitor for endogenous active PKD. These findings indicate that PKD regulates Bit1 function in anoikis through phosphorylation.
Negative charges in a mitochondrial localization signal can reduce mitochondrial import efficiency (13). We speculated that phosphorylation at the Ser5 residue, which is within the Bit1 mitochondrial localization signal, may enhance the release of Bit1 into the cytosol and subsequent anoikis function of Bit1. To address the potential role of the Ser5 residue in Bit1 localization, we mutated the Ser5 residue of C-terminally GFP-tagged Bit1 (Bit1-GFP) to the negatively charged phosphomimetic aspartic acid or glutamic acid (Bit1S5D-GFP and Bit1S5E-GFP) or to alanine (Bit1S5A-GFP). When transfected into HEK 293T cells, both wild type Bit1-GFP and Bit1S5A-GFP were significantly enriched in the mitochondrial fraction and barely detectable in the cytosolic fraction (Fig. 5D). In contrast, Bit1S5D-GFP and Bit1S5E-GFP were increased in the cytosolic fraction, whereas their mitochondrial levels were decreased. Consistent with their increased cytosolic localization, both Bit1S5D-GFP and Bit1S5E-GFP were more effective at promoting anoikis than wild type Bit1-GFP or Bit1S5A-GFP (Fig. 5E).
We also studied the effect of PKD on the accumulation of cytosolic Bit1 following loss of cell attachment. We used HUVECs in this experiment, because normal cells are more sensitive to anoikis than tumor cells (1), and knockdown of Bit1 expression in HUVECs significantly protected cells from anoikis (data not shown), as previously demonstrated in other normal cell lines (4, 6, 7). Down-regulating PKD expression by siRNA greatly reduced cytosolic concentration of endogenous Bit1 (Fig. 5F) and significantly increased resistance to anoikis (Fig. 5G). No cytosolic Bit1 was detected in attached HUVECs (result not shown).
Cotransfecting catalytically active PKD with wild type Bit1-GFP increased the cytosolic levels of Bit1 in HEK 293 cells (Fig. 5H). Furthermore, cytosolic Bit1 and active PKD immunoprecipitated as a complex (Fig. 5I). In contrast, Bit1 remained primarily in the mitochondrial fraction following cotransfection with vector or inactive PKD (Fig. 5H), and a complex of Bit1 with constitutively active PKD was not detected in the mitochondrial fraction (data not shown). Bit1 in the cytosolic Bit1-PKD complex was recognized by phosphospecific antibodies against the phosphorylated Ser5 and Ser87 sites (Fig. 5J). These findings indicate that Bit1 phosphorylation at serine 5 and 87 by PKD can lead to increased cytosolic sequestration and enhanced activity of Bit1 in anoikis. Phosphorylation of Ser5 may play a role in the release of Bit1 into the cytosol.
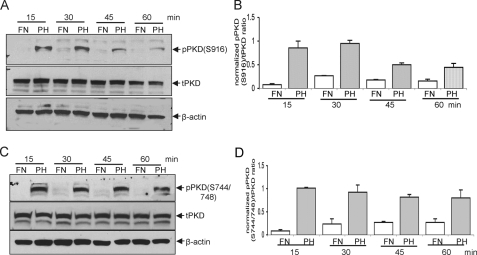
Cell Attachment to Fibronectin Suppresses PKD Activation—Cell attachment, particularly to fibronectin or vitronectin, suppresses Bit1-induced apoptosis (4). To investigate the role of PKD in this regulatory event, we examined the level of PKD Ser916 and Ser744/748 phosphorylation (activation) in HEK 293 cells plated on various substrates. Endogenous PKD in cells adhering to fibronectin remained unphosphorylated. In contrast, phosphorylation of PKD at both Ser916 (Fig. 6, A and B) and Ser744/748 sites (Fig. 6, C and D) was markedly increased as early as 15 min after the cells were suspended on polyHEMA and was sustained for up to 1 h. These results indicate that PKD is activated upon loss of cell attachment to fibronectin, placing PKD in a potentially important position in the regulation of anoikis.
FIGURE 6.
HEK 293 cells in suspension have enhanced PKD kinase activity. A, cells were allowed to remain attached to fibronectin (FN) or resuspended and placed on polyHEMA (PH). PKD activation was determined by immunoblotting with antibodies recognizing PKD phosphorylated at Ser916 (pPKD)(A) and Ser744/748 (C). Total PKD (tPKD) was determined from immunoblots of cell lysates, and the phospho-PKD/total PKD ratios were then quantified by densitometry analysis of immunoblots (B and D). Data are expressed as means ± S.E. from three independent experiments.
DISCUSSION
We have previously identified Bit1 as a mitochondrial protein that is released into the cytosol during anoikis and that is uniquely regulated by integrin-mediated attachment (4). Unlike other known anoikis effectors, Bit1 induces a form of caspase-independent apoptotic cell death that is uniquely counteracted by integrin-mediated cell attachment. We have recently shown that one of the downstream effectors of Bit1 is the Erk mitogen-activated protein kinase pathway. Mouse embryo fibroblasts from Bit1 knock-out mice and cultured cells in which Bit1 had been down-regulated by siRNA interference exhibited enhanced Erk activation. These findings indicate that Bit1 negatively regulates the Erk mitogen-activated protein kinase survival pathway (6). Intriguingly, the Scansite 2.0 Motif Scan program (14) identifies an Erk binding site in Bit1. Although the increased resistance to anoikis observed in mouse embryo fibroblasts lacking Bit1 and in Bit1 knockdown cells has been in part attributed to increased Erk activation (6), the upstream regulators of Bit1 apoptotic function have not been identified.
In this study, we show that the serine/threonine kinase PKD acts as an upstream activating kinase that regulates the function of Bit1 in anoikis by phosphorylating the Ser5 and Ser87 residues. Consistent with PKD acting upstream of Bit1, we found that overexpression of constitutively active PKD promotes Bit1 phosphorylation and enhances Bit1 apoptotic function, whereas Bit1 lacking the Ser5 and Ser87 phosphorylation sites (Bit1S5A/S87A) is unresponsive to the stimulatory effect of active PKD. Furthermore, inhibition of PKD with pharmacological inhibitors or by siRNA interference attenuated the apoptotic activity of Bit1. Taken together, our findings indicate a critical role for PKD in regulating Bit1 apoptotic function.
Based on our current data, PKD may enhance Bit1 apoptotic function via two mechanisms. First, phosphorylation of Bit1 following its release from mitochondria upon loss of substrate attachment may increase the apoptotic function of Bit1. This mechanism is supported by our findings demonstrating that active PKD phosphorylates N-terminally tagged Bit1 (which is exclusively cytosolic) and enhances its apoptotic function. Second, PKD may also enhance Bit1 anoikis activity by reducing the mitochondrial localization of Bit1 and increasing its release into the cytosol. Indeed, knocking down PKD in detached HUVECs decreased cytosolic Bit1 concentration and increased resistance to cell death.
One of the two PKD phosphorylation sites in Bit1 is the Ser5 residue, which is located within the mitochondrial localization sequence. Consistent with the notion that negative charges in a mitochondrial localization signal impair mitochondrial association, mutating the Ser5 residue to phosphomimetic amino acids enhanced the cytosolic localization and apoptotic potential of Bit1. Furthermore, coexpression of active PKD with C-terminally tagged Bit1 (whose mitochondrial localization is normally regulated) resulted in enhanced cytosolic localization of Bit1. Both of these mechanisms may contribute to the PKD-mediated induction of Bit1 apoptotic function.
In various in vitro systems, PKD has been shown to be activated through PKC-dependent phosphorylation of its activation loop (8, 9). Here, we report that stimulation of PKC signaling by treatment of cells with PMA, a general PKC activator, results in an increased phosphorylation of PKD that coincides with robust Bit1 apoptotic activity. The PMA-stimulated Bit1 activity is in part mediated through PKD, as shown by overexpressing as well as inhibiting PKD. These data indicate that PKC-dependent regulation of PKD activation results in Bit1 phosphorylation and induction of apoptosis. Consistent with a possible role of PKC upstream of PKD, PKCα activation has been previously shown to play a critical role in promoting anoikis in gastric cancer cells, and inhibition of PKCα by integrin-dependent pathways may be critical in suppressing anoikis (15). It is also noteworthy that the phosphorylation status of PKCs, including the novel PKC δ and ε, has been shown to be regulated by integrins (16).
Interestingly, Bit1 function was dramatically inhibited by the direct PKD inhibitor Go6976 but was not significantly attenuated by the general PKC inhibitor Go6983. This suggests that PKC-independent pathways may also contribute to PKD activation upstream of Bit1. Indeed, PKD can be activated by direct binding of the βγ subunits of heterotrimeric G proteins to its pleckstrin homology domain (17). Furthermore, tyrosine phosphorylation of the pleckstrin homology domain of PKD by the Abl kinase precedes PKD phosphorylation by PKCδ in response to oxidative stress (18). PKD activation in cellular processes, such as endothelin-induced HDAC5 nuclear export (12) and osteoblastic cell differentiation (19), has also been attributed to PKC-independent mechanisms.
A unique characteristic of Bit1 is that attachment of cells to fibronectin or vitronectin through the α5β1 and αvβ3 integrins, but not many other potent antiapoptotic factors, can effectively counteract apoptosis induced by cytoplasmic Bit1 (4). The data presented here indicate that attachment to fibronectin may inhibit Bit1 apoptotic function at least in part by attenuating PKD activation. We found that cells attached to fibronectin exhibit low basal PKD phosphorylation and are less responsive to PMA-induced PKD phosphorylation (data not shown). Moreover, loss of attachment to fibronectin leads to enhanced PKD phosphorylation. The suppression of PKD activation by cell attachment to fibronectin raises the possibility that integrin-mediated signals may confer anchorage-dependent survival at least in part by inhibiting PKD activation. Although the precise mechanisms used by integrins to regulate PKD activation are not known, it is conceivable that integrin-mediated attachment may activate a phosphatase, contributing to the accumulation of dephosphorylated PKD species. Indeed, cell matrix regulation of the phosphorylation status of PKCε has been attributed in part to a dephosphorylation process (20).
Anoikis is a form of apoptosis induced in epithelial cells following disruption of cellular interaction with extracellular matrix proteins. Integrin-mediated attachment abrogates anoikis primarily by generating antiapoptotic signals in host cells. In particular, integrin-mediated signals may lead to activation of focal adhesion kinase (2) and phosphatidylinositol 3-kinase (21), both of which confer survival signals from the extracellular matrix. In the present study, we show that loss of anchorage increases PKD activation and that activated PKD phosphorylates Bit1 and potentiates Bit1 apoptotic function. These findings reveal a novel function of PKD as a modulator of anoikis.
Acknowledgments
We thank Drs. Eva Engvall for comments on the manuscript, Grace Wood for the isolation of mouse embryo fibroblasts, the Burnham Institute Animal Facility for assistance with mouse work, and Albert Wahbe for assistance with photography.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants CA102583 and CA098162. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKC, protein kinase C; PKD, protein kinase D; PMA, phorbol 12-myristate 13-acetate; GFP, green fluorescent protein; siRNA, small interfering RNA; HUVEC, human umbilical vascular endothelial cell.
References
- 1.Frisch, S. M., and Ruoslahti, E. (1997) Curr. Opin. Cell Biol. 9 701–706 [DOI] [PubMed] [Google Scholar]
- 2.Frisch, S. M., Vuori, K., Ruoslahti, E., and Chan-Hui, P. V. (1996) J. Cell Biol. 134 793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz, M. (2001) Trends Cell Biol. 11 466–470 [DOI] [PubMed] [Google Scholar]
- 4.Jan, Y., Matter, M., Pai, J.-T., Chen, Y. L., Pilch, J., Komatsu, M., Ong, E., Fukuda, M., and Ruoslahti, E. (2004) Cell 116 751–762 [DOI] [PubMed] [Google Scholar]
- 5.de Pereda, J. M., Wass, W. F., Jan, Y., Rouslahti, E., Schimmel, P., and Pascual, J. (2004) J. Biol. Chem. 279 8111–8115 [DOI] [PubMed] [Google Scholar]
- 6.Kairouz-Wahbe, R., Biliran, H., Xiuquan, L., Khor, I.-W., Wankell, M., Besch-Williford, C., Pascual, J., Oshima, R., and Ruoslahti, E. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1528–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchentouf, M., Benabdallah, B. F., Rousseau, J., Schwartz, L. M., and Tremblay, J. P. (2007) Am. J. Transplant. 7 1491–1505 [DOI] [PubMed] [Google Scholar]
- 8.Iglesias, T., Waldron, R. T., and Rozengurt, E. (1998) J. Biol. Chem. 273 27662–27667 [DOI] [PubMed] [Google Scholar]
- 9.Vertommen, D., Rider, M., Ni, Y., Waelkens, E., Merlevede, W., Vandenheede, J. R., and Van Lint, J. (2000) J. Biol. Chem. 275 19567–19576 [DOI] [PubMed] [Google Scholar]
- 10.Yaffe, M. B., Leparc, G. G., Lai, J., Obata, T., Volinia, S., and Cantley, L. C. (2001) Nat. Biotechnol. 19 348–353 [DOI] [PubMed] [Google Scholar]
- 11.Rosengurt, E., Rodriguez-Pena, A., Coombs, M., and Sinnet-Smith, J. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 5748–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., and McKinsey, T. A. (2004) Mol. Cell Biol. 24 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claros, S. G., and Vincens, P. (1996) Eur. J. Biochem. 241 779–786 [DOI] [PubMed] [Google Scholar]
- 14.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda, H., Adachi, M., Miyazawa, M., Hinoda, Y., and Imai, K. (1999) Oncogene 18 5604–5609 [DOI] [PubMed] [Google Scholar]
- 16.Ivaska, J., Kermorgant, S., Whelan, R., Parsons, M., and Parker, P. J. (2003) Biochem. Soc. Trans. 31 90–93 [DOI] [PubMed] [Google Scholar]
- 17.Jamora, C., Yamanouye, N., Van Lint, J., Laudenslager, J., Vandenheede, J. R., Faulkner, D. J., and Malhotra, V. (1999) Cell 98 59–68 [DOI] [PubMed] [Google Scholar]
- 18.Storz, P., Doppler, H., and Toker, E. (2004) Mol. Cell. Biol. 24 2614–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemonnier, J. C., Ghayor, C., Guicheux, J., and Caverzasio, J. (2004) J. Biol. Chem. 279 259–264 [DOI] [PubMed] [Google Scholar]
- 20.England, K., Watson, J., Beale, G., Warner, M., Cross, J., and Rumsby, M. (2001) J. Biol. Chem. 276 10437–10442 [DOI] [PubMed] [Google Scholar]
- 21.Khwaja, A., Rodrigues-Viciana, P., Wennstrom, S., and Warne, P. H. (1997) EMBO J. 16 2783–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]