Abstract
The neuropilins-1 and -2 (NRP1 and NRP2) function as receptors for both the semaphorins and vascular endothelial growth factor. In addition to their contribution to the development of the nervous system, NRP1 and NRP2 have been implicated in angiogenesis and tumor progression. Given their importance to cancer and endothelial biology and their potential as therapeutic targets, an important issue that has not been addressed is the impact of metabolic stress conditions characteristic of the tumor microenvironment on their expression and function. Here, we demonstrate that hypoxia and nutrient deprivation stimulate the rapid loss of NRP1 expression in both endothelial and carcinoma cells. NRP2 expression, in contrast, is maintained under these conditions. The lysosomal inhibitors chloroquine and bafilomycin A1 prevented the loss of NRP1 expression, but proteasomal inhibitors had no effect. The hypothesis that NRP1 is degraded by autophagy is supported by the findings that its expression is lost rapidly in response to metabolic stress, prevented with 3-methyladenine and induced by rapamycin. Targeted depletion of NRP2 using small hairpin RNA revealed that NRP2 can function in the absence of NRP1 to mediate endothelial tube formation in hypoxia. Studies aimed at assessing NRP function and targeted therapy in cancer and angiogenesis should consider the impact of metabolic stress.
The neuropilins (NRP1 and NRP2)2 were identified as receptors for the semaphorin family of axon guidance molecules, and they are critical for development of the nervous system (1, 2). A large extracellular domain and a short intracellular domain that lacks intrinsic signaling capacity characterize these receptors. The neuropilins can also bind vascular endothelial growth factor (VEGF) on endothelial and cancer cells (3) and function as coreceptors for tyrosine kinase VEGF receptors (4) and possibly other growth factor receptors (5–7). There is also evidence that the neuropilins may function independently of tyrosine kinase VEGF receptors (e.g. 8, 9). The role of the neuropilins as VEGF receptors added a new dimension to their functional importance and resulted in numerous studies that have implicated their involvement in angiogenesis and tumor progression (5, 8, 10–13). Recently, NRP-specific antibodies (Abs), either alone or in combination with anti-VEGF therapy, have been shown to impede tumor growth and spread in mice by targeting tumor cells directly or the vasculature and lymphatics (5, 11, 14, 15).
The potential contribution of the neuropilins as VEGF coreceptors or receptors that mediate key functions of tumor and endothelial cells such as migration, growth rate, and survival has been the focus of an increasing number of studies with an emphasis on NRP1 (5, 8–13, 15–17). Of note, NRP1 has been shown recently to promote the survival of endothelial cells by modulating a p53/caspase axis (9). An important area that has not been investigated, however, is the contribution of metabolic stress conditions that are characteristic of the tumor microenvironment to the regulation of NRP expression and function. Hypoxia, for example, can be postulated to be a major influence on the neuropilins because it selects for the survival of more aggressive tumor cells and enhances metastatic dissemination (18). In fact, hypoxia has been shown to regulate the expression of other receptors important for tumor progression such as c-Met (19). Hypoxia also stimulates the expression of the NRP ligand VEGF and promotes angiogenesis (18). Nutrient deprivation resulting from inadequate blood supply is another hallmark of the tumor microenvironment which also has profound effects on cellular metabolism and genomic stability (20). Given these considerations, we sought to assess the impact of metabolic stress on the expression and function of the NRPs in both tumor and endothelial cells.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions—The human breast carcinoma cell lines MDA-MB-231, MDA-MB-435, MDA-MB-453, and the human prostate carcinoma cell line PC-3 were obtained from the American Type Culture Collection (Manassas, VA). SUM159 cells were obtained from Dr. Stephen Ethier (Karmanos Cancer Institute, Detroit MI). MDA-MB-231 and MDA-MB-435 cells were maintained in Dulbecco's modified Eagle's medium (low glucose) with 5% fetal bovine serum (FBS), MDA-MB-453 cells in Dulbecco's modified Eagle's medium (high glucose) with 5% FBS, SUM159 in Ham's F-12 with 5% FBS, 5 μg/ml insulin, and 1 μg/ml hydrocortisone, and PC-3 in RPMI 1640 medium with 5% FBS. Human umbilical vein endothelial cells (HUVECs) were obtained from CAMBREX Corp. (East Rutherford, NJ) and maintained on gelatin-coated plates in medium 199 with 10% FBS, 6 μg/ml endothelial cell growth supplement, 0.5 mg/ml heparin, and 5 ng/ml basic fibroblast growth factor (Sigma-Aldrich). Unless stated otherwise, all cells were maintained at 37 °C with 5% CO2. In some experiments, cells were maintained in phosphate-buffered saline (PBS) containing 4 g/liter glucose for the indicated times or treated with either MG132 (5 nm, Calbiochem), chloroquine (100 μm, Sigma-Aldrich), bafilomycin A1 (100 nm, Sigma-Aldrich), 3-methyladenine (5 and 10 mm, Sigma-Aldrich) in PBS containing glucose, or rapamycin (5 and 500 nm, Sigma-Aldrich), as indicated in the figure legends.
Hypoxia Experiments—Cells were plated at a subconfluent density and incubated at 37 °C in a hypoxic work station (INVIVO2 200; Biotrace, Inc., Cincinnati, OH) for the times and [O2] indicated in the figure legends. Duplicate plates were maintained in “normoxia,” a standard tissue culture incubator maintained at 37 °C with 5% CO2.
Immunoblotting—Cells were extracted in radioimmune precipitation assay buffer (Boston Bioproducts, Worcester MA) containing protease inhibitors (Complete Mini, Roche Applied Science), cleared by centrifugation, and protein content was quantified using the Bradford assay (Bio-Rad). Samples containing equivalent amounts of protein were boiled for 5 min in sample buffer containing 62.5 mm Tris-HCl, pH 6.8, 1% SDS, 8% glycerol, 1.5% β-mercaptoethanol, and bromphenol blue. Proteins were separated by standard SDS-8% PAGE. After electrophoresis, proteins were transferred to nitrocellulose (Ready Gel Blotting Sandwiches, Bio-Rad). Nonspecific binding was prevented by incubation with 5% dried milk powder in Tris-buffered saline with 0.1% Tween 20 (TBST). Blots were incubated at 4 °C overnight with Abs specific for either NRP1 (A12 and C19, Santa Cruz Biotechnology, Santa Cruz, CA), NRP2 (H300, Santa Cruz Biotechnology; AF2215, R&D Systems), or β-actin (Sigma-Aldrich). After washing with TBST, blots were incubated with the appropriate secondary Ab conjugated to horseradish peroxidase for 1 h at room temperature. After washing with TBST, the proteins were detected using ECL (Pierce, Rockford, IL). When required, reblotting was done after standard stripping and blocking. Band intensities were quantified using Labworks 4.6 image acquisition and analysis software (UVP, Inc., Upland, CA) and normalized with the intensity of β-actin.
Expression of NRP1 and NRP2 in MDA-MB-453 Cells—NRP1 and NRP2 constructs with the same cytomegalovirus promoter, which were provided by Drs. Jonathan Raper and Francoise Bono, respectively, were expressed in MDA-MB-453 cells using Lipofectamine 2000 (Invitrogen). Briefly, 4 μg of each construct was introduced into 5 × 105 MDA-MB-453 cells, and after 1 day in fresh medium, the transfected cells were maintained in either normoxia or hypoxia (0.1% O2) for 24 h. Cell extracts were harvested for analysis of NRP1 and NRP2 expression by immunoblotting.
Flow Cytometric Analysis—HUVECs were detached with 5 mm EDTA in PBS, blocked with 3% FBS in PBS, incubated at 4 °C for 30 min with anti-BDCA-4 (NRP1) monoclonal Ab (1 μg/ml; AD5–17F6, Miltenyi Biotec, Auburn, CA) and anti-NRP2 (H300; 5 μg/ml) in 1% bovine serum albumin in PBS, and stained at 4 °C for 30 min with fluorescein isothiocyanate (FITC)-labeled anti-mouse IgG for NRP1 and FITC-labeled anti-rabbit IgG for NRP2 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Data were collected using a BD LSRII flow cytometer (BD Biosciences, San Jose, CA), and a live (DAPI-negative) single-cell population was analyzed for examining the cell surface expression of NRPs.
Generation and Use of AP-Sema3A and AP-Sema3F—Human Sema3A and Sema3F ligated to the 3′ end of the alkaline phosphatase gene (AP-Sema3A and AP-Sema3F) constructs were generously provided by Dr. Alex Kolodkin. To generate AP-Sema protein, 25 μg of each construct was introduced into human embryonic kidney 293 T cells by Lipofectamine 2000. Conditioned medium containing secreted recombinant proteins was harvested 72 h after transfection. The identity of each recombinant protein was verified by immunoblotting using an alkaline phosphatase Ab (GenHunter, Nashville, TN). The concentration of each recombinant protein was determined using an AP enzymatic assay kit (GenHunter) based on the specific activity of alkaline phosphatase (units/ml). HUVECs were plated on chamber slides and incubated with AP, AP-Sema3A, or AP-Sema3F conditioned medium containing equivalent amounts of AP activity for 1.5 h at 4 °C. The cells were then fixed with formalin solution for 15 min at room temperature after five washes with PBS. Endogenous phosphatase activity was inactivated by incubation at 65 °C for 20 min in AP buffer (150 mm NaCl/20 mm Hepes, pH 7.5). All assays were done in duplicate. Cell surface expression of AP-Sema was visualized by incubation with AP assay reagent S (GenHunter) for 16 h at room temperature, and representative photomicrographs were taken under the microscope.
NRP2 shRNA—Lentiviruses containing the following NRP2 shRNAs were generated and titrated according to the manufacturer's instructions: Oligonucleotide ID TRCN0000063308 and TRCN0000063312 for 8 and 12 of shNRPZ (shRNA for NRP2), respectively, and green fluorescent protein control in pLKO.1 (Open Biosystems, Huntsville, AL) and used to infect HUVECs following standard protocols.
Endothelial Tube Formation Assay—HUVECs (5 × 105 cells) were infected with shRNA-expressing lentivirus (5 × 106 cfu) for green fluorescent protein and NRP2 in the presence of 6 μg/ml Polybrene (Sigma-Aldrich). At 72 h after infection, cells were plated on Matrigel as described (21) with modifications. Growth factor-reduced Matrigel (1 mg/ml, BD Biosciences, Franklin Lakes, NJ) was added to each well of a 96-well plate on ice, allowed to polymerize for 15 min at room temperature, and then placed in a humidified incubator at 37 °C. HUVECs (104 cells/well) were added to the Matrigel-coated plates. Cells were then incubated in hypoxia. After 24 h, photomicrographs of each well (magnification, ×40) were taken using an Olympus IX71 inverted microscope (Optical Analysis Corp., Nashua, NH) equipped with a Retiga 1300 CCD digital camera (QImaging, Burnaby, BC Canada), and they were processed using IPLab software (Scanalytics, inc., Fairfax, VA). Tube formation was quantified by determining the number of branch points/field. The effect of the shRNAs on NRP expression before and after the Matrigel assays was assessed by immunoblotting and flow cytometry.
Statistical Analysis—Values of results are expressed as means and S.D., and significance was established by Student's t test. In all analyses, the level of statistical significance was more than the 95% confidence level (p < 0.05).
RESULTS
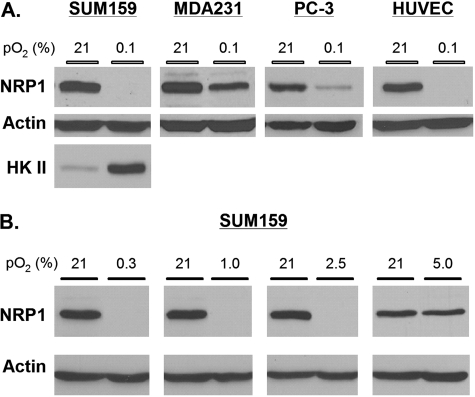
Hypoxia Suppresses NRP1 but Not NRP2 Expression—To assess the impact of hypoxia on NRP1 expression, MDA-MB-231 and SUM159 breast carcinoma cells, PC-3 prostate carcinomas, and HUVECs were maintained in either hypoxic (0.1% O2) or normoxic conditions (21% O2) for 24 h. NRP1 expression was assayed in extracts from these cells by immunoblotting. Compared with normoxia, hypoxia suppressed NRP1 expression in all of these cells (Fig. 1A). In contrast, hypoxia increased the expression of hexokinase II in SUM159 cells, a hypoxiainducible glycolytic enzyme (22), providing a positive control for the hypoxic environment (Fig. 1A). Subsequently, we examined the impact of a range of [O2] on NRP1 expression. As shown in Fig. 1B, NRP1 expression was suppressed up to 2.5% O2 but not at 5% O2.
FIGURE 1.
Hypoxia suppresses NRP1 expression in carcinoma and endothelial cells. A, MDA-MB-231 and SUM159 breast carcinoma cells, PC-3 prostate carcinoma cells, and HUVECs were maintained at 37 °C in either 21% O2 or 0.1% O2 for 24 h. Protein extracts (50 μg) obtained from these cells were immunoblotted with the indicated antibodies (mouse anti-NRP1 and goat anti-hexokinase II (HK II; Santa Cruz Biotechnology) and rabbitβ-actin (Sigma-Aldrich). Similar results were observed in at least three independent experiments. B, SUM159 breast cancer cells were maintained in a range of oxygen tensions (0.3–5.0% O2) for 1 day, and the relative expression of NRP1 was compared with that in normoxia for each of these oxygen tensions.
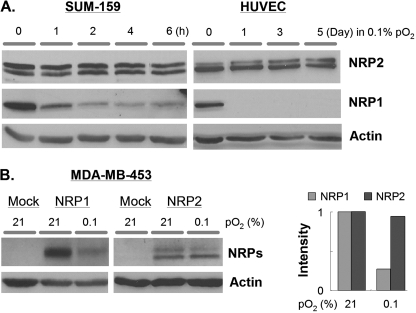
Given the striking suppression of NRP1 expression by hypoxia, we sought to determine the impact of hypoxia on the expression of NRP2, which is also expressed by endothelial cells and some breast carcinoma cells (12, 23). Exposure of SUM159 cells to hypoxia (0.1%) for time periods that ranged from 0 to 6 h repressed NRP1 expression but had no effect on NRP2 expression (Fig. 2A). Exposure of HUVECs to hypoxia resulted in significant repression of NRP1 expression within 3–6 h, similar to SUM159 cells (data not shown). This repression was sustained for at least 5 days in hypoxia (Fig. 2A). In marked contrast, NRP2 expression in HUVECs was not diminished significantly even after 5 days in hypoxia (Fig. 2A). Long term hypoxic stimulation did not induce a significant increase in apoptosis in these cells (data not shown).
FIGURE 2.
NRP2 expression is not suppressed by hypoxia. A, protein extracts (50 μg) obtained from either SUM159 cells or HUVECs that had been maintained in normoxia or hypoxia (0.1% O2) for the indicated times were immunoblotted with a NRP2 Ab (goat anti-NRP2, R&D Systems). The blots were also probed with a β-actin Ab to assess protein loading. B, MDA-MB-453 breast carcinoma cells were transfected with either a mock, NRP1, or NRP2 expression plasmid. The cells were maintained in either normoxia or hypoxia (0.1% O2) for 24 h, and the expression of each NRP was assessed by immunoblotting. The bar graph depicts the relative expression of each NRP as determined by densitometry using β-actin as a control.
Additional evidence to support the differential regulation of NRP1 and NRP2 expression by hypoxia was obtained using MDA-MB-453 breast carcinoma cells. These cells lack expression of both NRP1 and NRP2 (Fig. 2B). For this reason, we transfected cDNAs for either NRP1 or NRP2 into these cells and assessed the impact of hypoxia (24 h) on the expression of these exogenous proteins. As shown in Fig. 2B, hypoxia stimulated a substantial reduction in NRP1 but not NRP2 expression.
Analysis of NRP surface expression by flow cytometry revealed that exposure of HUVECs to hypoxia for 24 h stimulated a substantial reduction (∼70%) in NRP1 surface expression compared with cells maintained in normoxia (Fig. 3A). In contrast, the surface expression of NRP2 was not diminished significantly by exposure to hypoxia (Fig. 3A). To assess the impact of hypoxia on the NRPs further, we took advantage of the fact that Semaphorin 3A binds to NRP1 (24) and Semaphorin 3F binds to NRP2 (25). HUVECs were maintained in normoxia or hypoxia for 24 h and then incubated at 4 °C in the presence of either an alkaline phosphatase-tagged Semaphorin 3A (AP-Sema3A) or alkaline phosphatase-tagged Semaphorin 3F (AP-Sema3F) ligand. Subsequently, the cells were fixed, and in situ alkaline phosphatase reaction was carried out to visualize semaphorin binding. Both AP-Sema3A and AP-Sema3F bound robustly to HUVECs maintained in normoxia (Fig. 3B). No significant change in the binding of AP-Sema3F was detected in HUVECs maintained in hypoxia (Fig. 3B). In contrast, however, a marked decrease in AP-Sema3A binding was observed in hypoxia compared with normoxia (Fig. 3B).
FIGURE 3.
Cell surface expression of NRP1 is diminished in hypoxia. A, cell surface expression of the NRPs was assessed on HUVECs by flow cytometry using anti-BDCA-4 (NRP1) or anti-NRP2 (H300) Abs with fluorescein isothiocyanate (FITC)-labeled secondary Abs. Only live cells (DAPI– population) were analyzed. The data from multiple experiments were quantified and are shown in the bar graph. B, AP-conjugated, recombinant human Sema3F (for NRP2) and Sema3A (NRP1) proteins (provided by Dr. Alex Kolodkin (Johns Hopkins University) were allowed to interact with HUVECs in either normoxia (21% O2) or hypoxia (0.1% O2) for 24 h at 4 °C. AP alone was used for negative control. Bound semaphorins were visualized using an AP enzymatic assay (GenHunter) following the manufacturer's protocol. Inset, the expression of the NRPs under each of the above experimental conditions was assessed by immunoblotting as described in Figs. 1 and 2.
Hypoxia and Other Stress Conditions Induce the Degradation of NRP1 by a Mechanism Characteristic of Autophagy—One hypothesis to explain the rapid loss of NRP1 expression in hypoxia is protein degradation. Initially, we assessed the possibility that hypoxia induced the proteasomal-mediated degradation of NRP1 using MG132, a proteasomal inhibitor (26). This inhibitor did not impede the loss of NRP1 expression in hypoxia (data not shown). Subsequently, we investigated whether hypoxia stimulates the lysosomal degradation of NRP1. As shown in Fig. 4A, treatment of HUVECs with the lysosomal inhibitor chloroquine (27) prevented the loss of NRP1 expression that occurs in hypoxia. In contrast, chloroquine had no significant effect on NRP2 expression, nor did it affect the expression of NRP1 in normoxia (Fig. 4A). These data were confirmed using another lysosomal inhibitor, bafilomycin A1 (28, 29). Similar to chloroquine, bafilomycin A1 prevented the loss of NRP1 expression in hypoxia, but it did not impact NRP2 expression significantly (Fig. 4B). However, chloroquine did not inhibit the loss of NRP1 surface expression as assessed by flow cytometry (Fig. 4C). These data suggest that loss of NRP1 under hypoxia is caused by internalization and degradation of NRP1.
FIGURE 4.
Hypoxia stimulates the lysosomal degradation of NRP1. A, HUVECs were maintained in either normoxia (21% O2) or hypoxia (0.1% O2) in the presence or absence of chloroquine (100 μm, Sigma-Aldrich) for the indicated times. The relative expression of NRP1, NRP2, and actin was assessed by immunoblotting and densitometry as described in Figs. 1 and 2. B, a similar experiment was performed using bafilomycin A1 (100 nm, Sigma-Aldrich). The data shown in the bar graph is one representative result from three independent experiments (±S.D.). C, cell surface expression of the NRPs was assessed on a live single-cell suspension of HUVECs in the presence or absence of chloroquine by flow cytometry as described in Fig. 3. The data from two experiments were quantified and are shown in the bar graph.
Autophagy is one form of lysosomal degradation that is induced by stress conditions, including hypoxia and nutrient deprivation (30). To evaluate the possibility that NRP1 is degraded by an autophagic mechanism, we maintained HUVECs in PBS supplemented only with glucose. As shown in Fig. 5A, NRP1 expression was depleted within 40 min in this medium, but no change was evident in the expression of NRP2. Analysis of NRP surface expression by flow cytometry revealed that nutrient deprivation for 2 h stimulated a significant reduction (∼40%) in NRP1, but not in NRP2, surface expression compared with unstarved cells (Fig. 5B). We also treated HUVECs in PBS/glucose with 3-methyladenine, an inhibitor of phosphatidylinositol trisphosphate kinase III that is reported to be an autophagy inhibitor in mammalian cells (31). This inhibitor prevented the loss of NRP1 expression that was induced by nutrient deprivation (Fig. 5C). Given that TORC1 has been shown to inhibit autophagy through inactivation of Atg13, one of the early autophagy genes, by phosphorylation (30), we treated HUVECs with rapamycin, a TORC1 inhibitor, to induce autophagy. At concentrations as low as 5 nm, a significant decrease in NRP1 but not NRP2 expression was observed (Fig. 5D). Taken together, the data in Fig. 5 support the hypothesis that NRP1 is degraded by an autophagic mechanism in response to stress conditions.
FIGURE 5.
NRP1 is degraded by a mechanism characteristic of autophagy. A and B, the expression of NRP1 and NRP2 in HUVECs that had been maintained in PBS containing 4 g/liter glucose for the indicated times was assessed by immunoblotting (A) and by flow cytometry (B) as described in Fig. 3. C, HUVECs were maintained in either normal medium or PBS/glucose for 2 h in either the presence or absence of 3-methyladenine (3MA)(5 and 10 mm), and NRP expression was assessed by immunoblotting. D, HUVECs maintained in normal culture medium were treated with rapamycin (5 and 500 nm) for 24 h in normoxia, and NRP expression was assessed by immunoblotting.
NRP2 Mediates Endothelial Cell Tube Formation in Hypoxia—One implication of our data is that NRP2 mediates distinct cellular functions in hypoxia which are independent of NRP1. HUVECs exhibit the ability to form tubes in hypoxia when plated on Matrigel (21), a form of branching morphogenesis (Fig. 6). To assess the involvement of NRP2 in this process, we generated NRP2-specific shRNAs and expressed them in HUVECs. These shRNAs depleted NRP2 expression but had no effect on NRP1 (Fig. 6A). Subsequently, we evaluated the impact of these shRNAs on tube formation in hypoxia. As shown in Fig. 6B, loss of NRP2 abrogated the ability of HUVECs to form tubes in hypoxia. Quantification of branch point formation, a measure of branching morphogenesis, substantiated this conclusion (Fig. 6C). Analysis of NRP expression by flow cytometry in these cultures revealed that the tube-forming process itself did not affect their surface expression (data not shown), that NRP2 expression was diminished in cells that expressed the NRP2 shRNAs, and that NRP1 expression was lost in all cell populations in hypoxia (Fig. 6D).
FIGURE 6.
NRP2 mediates endothelial cell tube formation in hypoxia. A, HUVECs (5 × 105 cells) were infected with either a green fluorescent protein shRNA or two different NRP2 shRNAs (5 × 106 lentiviruses/infection), and NRP1 and NRP2 expression was assayed at 72 h after infection by immunoblotting. B, at 72 h after infection, 104 cells were seeded onto preformed Matrigel (1 mg/ml), and photomicrographs (magnification, ×40) were taken after 24 h in hypoxia (0.5% O2) with an inverted-phase microscope. C, endothelial tube formation was quantified by determining the mean number of junctions formed per field (±S.D.). Significance was assessed by Student's t test by comparison with noninfected parental cells. D, NRP expression was assessed by immunoblotting before (normoxia) and after (hypoxia) the Matrigel assay.
DISCUSSION
The key finding reported in this study is that metabolic stress conditions characteristic of the tumor microenvironment, including hypoxia and nutrient deprivation, stimulate the rapid loss of surface expression and degradation of NRP1 but not of NRP2 in both endothelial and carcinoma cells. More specifically, the data provided suggest that NRP1 is internalized and degraded by autophagy in response to metabolic stress and that NRP2 is resistant to such degradation. We also report that NRP2 can function in the absence of NRP1 to mediate endothelial tube formation in hypoxia. These findings argue for inherent differences in the regulation of NRP1 and NRP2 which could impact their contribution to endothelial and cancer biology.
Our data support a mechanism for cellular response to metabolic stress that involves the rapid internalization of surface NRP1 and its degradation in lysosomes or autophagosomes, even in cells such as HUVECs that express both NRP1 and NRP2. This mechanism is supported by the recent finding that sulfated polysaccharides induce NRP1 internalization and degradation in lysosomes (32). An important issue that derives from these data is the nature of the mechanism that is stimulated by metabolic stress and that targets NRP1 but not NRP2 for degradation. Neither of these NRPs has a potential lysosomal targeting sequence (KFERQ), which is selectively recognized by the molecular chaperone HSC73 (33). Moreover, we were unable to coimmunoprecipitate HSC73 protein with either NRP1 or NRP2 (data not shown). The cytoplasmic domain of NRP1 does contain a PDZ-binding domain (SEA) at its COOH terminus which binds to synectin, also known as GIPC (34). Synectin/GIPC can bind to RGS19, a regulator of G protein signaling which has been implicated in endocytosis and membrane trafficking (35, 36). Although this putative mechanism could account for the differences we observed between NRP1 and NRP2, it is challenged by the fact that the NRP2a isoform also contains the same PDZ-binding sequence at its COOH terminus, and our finding that exogenous expression of NRP1 and NRP2, which contained the SEA sequence, in MDA-MB-453 cells resulted in hypoxia-induced degradation of NRP1 but not NRP2 (Fig. 2). Subsequent studies should evaluate the relative PDZ-binding potential of NRP1 and NRP2 and assess other regions of their cytoplasmic domain sequences for involvement in internalization and degradation.
Our finding that NRP1 is targeted for degradation in response to metabolic stress in vitro raises several important issues that bear on NRP function in vivo. Hypoxia and nutrient deprivation select for cells that can survive under such conditions and, as a consequence, facilitate tumor progression (18, 20). Degradation of NRP1 may occur because its functions are not essential for survival or actually impede survival, e.g. apoptotic Semaphorin signaling (37, 38). Alternatively, it is possible that the internalization and degradation of NRP1 could provide a positive signal that is associated with the regulation of growth factor receptors important for tumor survival and progression. In this context, there is evidence that NRP1 can facilitate not only VEGF signaling (23) but also potentiate signaling mediated by other growth factors such as hepatocyte growth factor (5–7). The possibility that diminished NRP1 expression on tumor cells contributes to progression is tempered by reports that have demonstrated increased NRP1 expression in primary and metastatic tumors (for review, see Ref. 34). We are cautious of such studies, at least for breast cancer, because we were unable to detect significant NRP1 staining in breast tumor specimens using several commercially available Abs (data not shown), a finding that is consistent with the report that NRP1 expression in the breast is confined to myoepithelial cells of the normal gland (39). Although there is evidence that NRP1 facilitates tumor growth and progression (13, 14, 17, 34), there are also data that NRP1 expression in a pancreatic carcinoma cell line impedes tumor growth and spread (40). The use of transgenic mouse tumor models in which either NRP1 or NRP2 have been deleted should provide more definitive insight into the relative contributions of these receptors within the context of the tumor microenvironment.
Our data on HUVECs need to be considered with respect to the response of endothelial cells to metabolic stress. Ischemia, impaired blood flow caused by vasoconstriction and trauma, results in hypoxia and nutrient deprivation (20, 30), conditions that can promote NRP1 degradation in vitro. Such conditions also stimulate VEGF expression and secretion. Interestingly, there is evidence that VEGF and VEGF receptor 2 are internalized in response to endothelial wounding and that internalization is necessary for wound recovery (41). Based on these observations, the hypothesis can be proposed that the induction of VEGF expression that occurs in response to metabolic stress also promotes the internalization and degradation of NRP1. It is also possible that wound recovery involves the reexpression of NRP1, as evidenced by the report that neovascularization in response to ischemia results in increased NRP1 expression (42).
In summary, our data reveal that NRP1 is labile to internalization and lysosomal degradation in response to metabolic stress, a behavior that distinguishes it from NRP2. This property of NRP1 may be intrinsic to its functional regulation and that of other surface receptors with which it associates. At present, these data do not diminish ongoing programs aimed at using NRP1-specific Abs as adjuvant therapy for cancer and tumor angiogenesis (14), but they do suggest that further studies on NRP1 expression and function within the tumor microenvironment are warranted. Nonetheless, the data do support a potentially important role for NRP2 in cancer and angiogenesis, a possibility that is substantiated by recent studies (11, 15).
Acknowledgments
We thank Francoise Bono, Stephen Ethier, Alex Kolodkin, and Jonathan Raper for providing reagents, and Eric Baehrecke and Don Senger for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant CA89209. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NRP, neuropilin; AP, alkaline phosphatase; FBS, fetal bovine serum; HUVEC, human umbilical vein endothelial cell; VEGF, vascular endothelial growth factor.
References
- 1.Kolodkin, A. L., Levengood, D. V., Rowe, E. G., Tai, Y. T., Giger, R. J., and Ginty, D. D. (1997) Cell 90 753–762 [DOI] [PubMed] [Google Scholar]
- 2.He, Z., and Tessier-Lavigne, M. (1997) Cell 90 739–751 [DOI] [PubMed] [Google Scholar]
- 3.Soker, S., Takashima, S., Miao, H. Q., Neufeld, G., and Klagsbrun, M. (1998) Cell 92 735–745 [DOI] [PubMed] [Google Scholar]
- 4.Neufeld, G., Kessler, O., and Herzog, Y. (2002) Adv. Exp. Med. Biol. 515 81–90 [DOI] [PubMed] [Google Scholar]
- 5.Hu, B., Guo, P., Bar-Joseph, I., Imanishi, Y., Jarzynka, M. J., Bogler, O., Mikkelsen, T., Hirose, T., Nishikawa, R., and Cheng, S. Y. (2007) Oncogene 26 5577–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita, A., Gotze, T., and Korc, M. (2007) Cancer Res. 67 10309–10316 [DOI] [PubMed] [Google Scholar]
- 7.Sulpice, E., Plouet, J., Berge, M., Allanic, D., Tobelem, G., and Merkulova-Rainon, T. (2008) Blood 111 2036–2045 [DOI] [PubMed] [Google Scholar]
- 8.Bachelder, R. E., Crago, A., Chung, J., Wendt, M. A., Shaw, L. M., Robinson, G., and Mercurio, A. M. (2001) Cancer Res. 61 5736–5740 [PubMed] [Google Scholar]
- 9.Wang, L., Dutta, S. K., Kojima, T., Xu, X., Khosravi-Far, R., Ekker, S. C., and Mukhopadhyay, D. (2007) PLoS ONE 2 e1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao, H. Q., Lee, P., Lin, H., Soker, S., and Klagsbrun, M. (2000) FASEB J. 14 2532–2539 [DOI] [PubMed] [Google Scholar]
- 11.Gray, M. J., Van Buren, G., Dallas, N. A., Xia, L., Wang, X., Yang, A. D., Somcio, R. J., Lin, Y. G., Lim, S., Fan, F., Mangala, L. S., Arumugam, T., Logsdon, C. D., Lopez-Berestein, G., Sood, A. K., and Ellis, L. M. (2008) J. Natl. Cancer Inst. 100 109–120 [DOI] [PubMed] [Google Scholar]
- 12.Bielenberg, D. R., Pettaway, C. A., Takashima, S., and Klagsbrun, M. (2006) Exp. Cell Res. 312 584–593 [DOI] [PubMed] [Google Scholar]
- 13.Hong, T. M., Chen, Y. L., Wu, Y. Y., Yuan, A., Chao, Y. C., Chung, Y. C., Wu, M. H., Yang, S. C., Pan, S. H., Shih, J. Y., Chan, W. K., and Yang, P. C. (2007) Clin. Cancer Res. 13 4759–4768 [DOI] [PubMed] [Google Scholar]
- 14.Pan, Q., Chanthery, Y., Liang, W. C., Stawicki, S., Mak, J., Rathore, N., Tong, R. K., Kowalski, J., Yee, S. F., Pacheco, G., Ross, S., Cheng, Z., Le Couter, J., Plowman, G., Peale, F., Koch, A. W., Wu, Y., Bagri, A., Tessier-Lavigne, M., and Watts, R. J. (2007) Cancer Cell 11 53–67 [DOI] [PubMed] [Google Scholar]
- 15.Caunt, M., Mak, J., Liang, W. C., Stawicki, S., Pan, Q., Tong, R. K., Kowalski, J., Ho, C., Reslan, H. B., Ross, J., Berry, L., Kasman, I., Zlot, C., Cheng, Z., Le Couter, J., Filvaroff, E. H., Plowman, G., Peale, F., French, D., Carano, R., Koch, A. W., Wu, Y., Watts, R. J., Tessier-Lavigne, M., and Bagri, A. (2008) Cancer Cell 13 331–342 [DOI] [PubMed] [Google Scholar]
- 16.Bachelder, R. E., Wendt, M. A., and Mercurio, A. M. (2002) Cancer Res. 62 7203–7206 [PubMed] [Google Scholar]
- 17.Parikh, A. A., Fan, F., Liu, W. B., Ahmad, S. A., Stoeltzing, O., Reinmuth, N., Bielenberg, D., Bucana, C. D., Klagsbrun, M., and Ellis, L. M. (2004) Am. J. Pathol. 164 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, J. M. (1999) Cancer Res. 59 5863–5870 [PubMed] [Google Scholar]
- 19.Pennacchietti, S., Michieli, P., Galluzzo, M., Mazzone, M., Giordano, S., and Comoglio, P. M. (2003) Cancer Cell 3 347–361 [DOI] [PubMed] [Google Scholar]
- 20.Sutherland, R. M. (1988) Science 240 177–184 [DOI] [PubMed] [Google Scholar]
- 21.Calvani, M., Rapisarda, A., Uranchimeg, B., Shoemaker, R. H., and Melillo, G. (2006) Blood 107 2705–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathupala, S. P., Rempel, A., and Pedersen, P. L. (2001) J. Biol. Chem. 276 43407–43412 [DOI] [PubMed] [Google Scholar]
- 23.Guttmann-Raviv, N., Kessler, O., Shraga-Heled, N., Lange, T., Herzog, Y., and Neufeld, G. (2006) Cancer Lett 231 1–11 [DOI] [PubMed] [Google Scholar]
- 24.Miao, H. Q., Soker, S., Feiner, L., Alonso, J. L., Raper, J. A., and Klagsbrun, M. (1999) J. Cell Biol. 146 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, H., Bagri, A., Zupicich, J. A., Zou, Y., Stoeckli, E., Pleasure, S. J., Lowenstein, D. H., Skarnes, W. C., Chedotal, A., and Tessier-Lavigne, M. (2000) Neuron 25 43–56 [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. H., and Goldberg, A. L. (1998) Trends Cell Biol. 8 397–403 [DOI] [PubMed] [Google Scholar]
- 27.Gerard, K. W., Hipkiss, A. R., and Schneider, D. L. (1988) J. Biol. Chem. 263 18886–18890 [PubMed] [Google Scholar]
- 28.Bowman, E. J., Siebers, A., and Altendorf, K. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 7972–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuchi, T., Aikawa, K., Arai, H., and Inoue, K. (1993) J. Biol. Chem. 268 27345–27348 [PubMed] [Google Scholar]
- 30.Levine, B., and Kroemer, G. (2008) Cell 132 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seglen, P. O., and Gordon, P. B. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narazaki, M., Segarra, M., and Tosato, G. (2008) Blood 111 4126–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuervo, A. M., and Dice, J. F. (1998) J. Mol. Med. 76 6–12 [DOI] [PubMed] [Google Scholar]
- 34.Pellet-Many, C., Frankel, P., Jia, H., and Zachary, I. (2008) Biochem. J. 411 211–226 [DOI] [PubMed] [Google Scholar]
- 35.De Vries, L., Lou, X., Zhao, G., Zheng, B., and Farquhar, M. G. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12340–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramow-Newerly, M., Roy, A. A., Nunn, C., and Chidiac, P. (2006) Cell. Signal. 18 579–591 [DOI] [PubMed] [Google Scholar]
- 37.Castro-Rivera, E., Ran, S., Thorpe, P., and Minna, J. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11432–11437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bielenberg, D. R., and Klagsbrun, M. (2007) Cancer Metastasis Rev. 26 421–431 [DOI] [PubMed] [Google Scholar]
- 39.Stephenson, J. M., Banerjee, S., Saxena, N. K., Cherian, R., and Banerjee, S. K. (2002) Int. J. Cancer 101 409–414 [DOI] [PubMed] [Google Scholar]
- 40.Gray, M. J., Wey, J. S., Belcheva, A., McCarty, M. F., Trevino, J. G., Evans, D. B., Ellis, L. M., and Gallick, G. E. (2005) Cancer Res. 65 3664–3670 [DOI] [PubMed] [Google Scholar]
- 41.Santos, S. C., Miguel, C., Domingues, I., Calado, A., Zhu, Z., Wu, Y., and Dias, S. (2007) Exp. Cell Res. 313 1561–1574 [DOI] [PubMed] [Google Scholar]
- 42.Ishihama, H., Ohbayashi, M., Kurosawa, N., Kitsukawa, T., Matsuura, O., Miyake, Y., and Muramatsu, T. (2001) Investig. Ophthalmol. Vis. Sci. 42 1172–1178 [PubMed] [Google Scholar]